Is the Neuropeptide PEN a Ligand of GPR83?
Abstract
1. Introduction
2. Results
2.1. Overexpression of GPR83 and the Deletion Variant GPR83del in Human Cell Lines
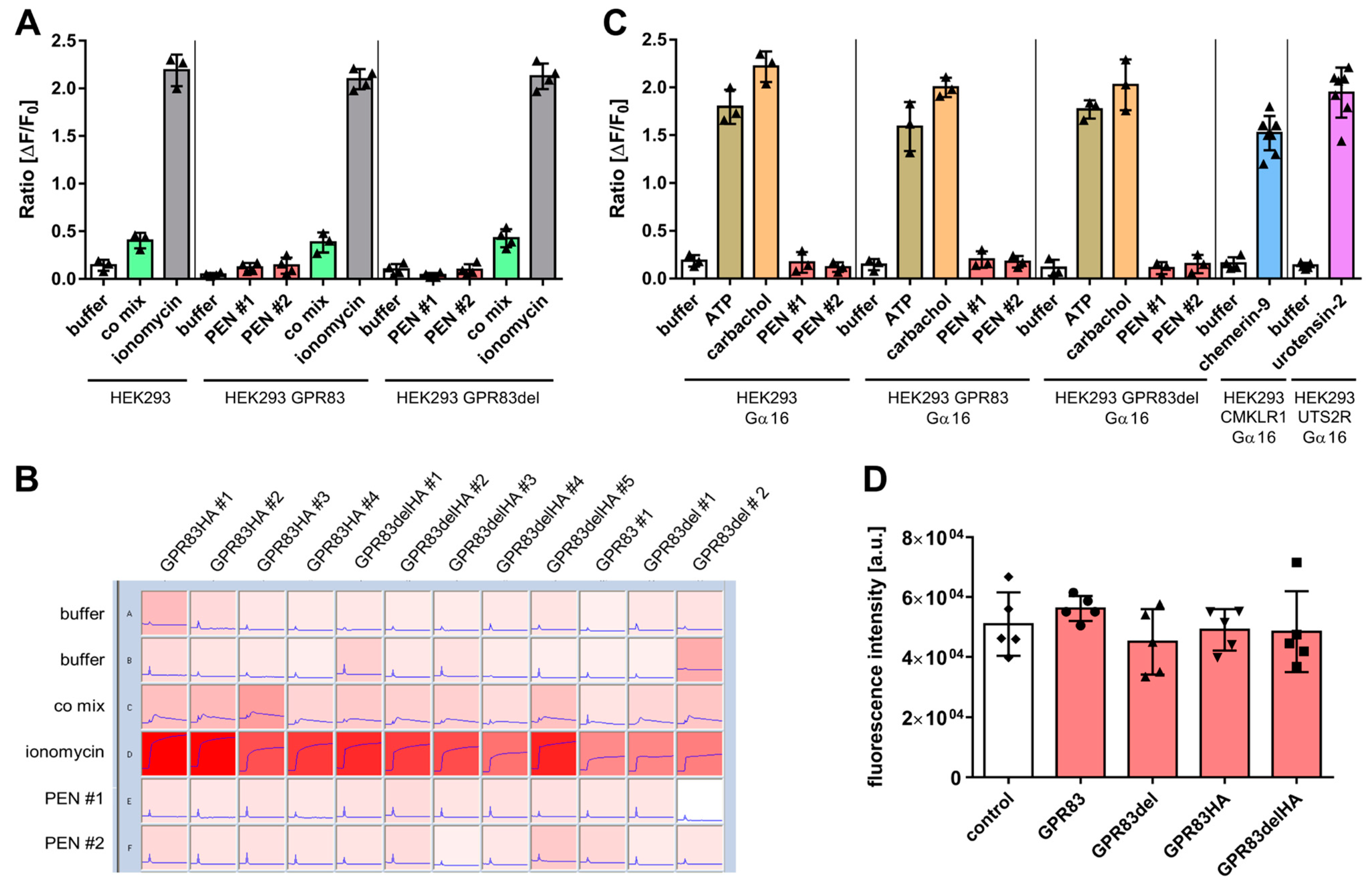
2.2. Intracellular Calcium Mobilization
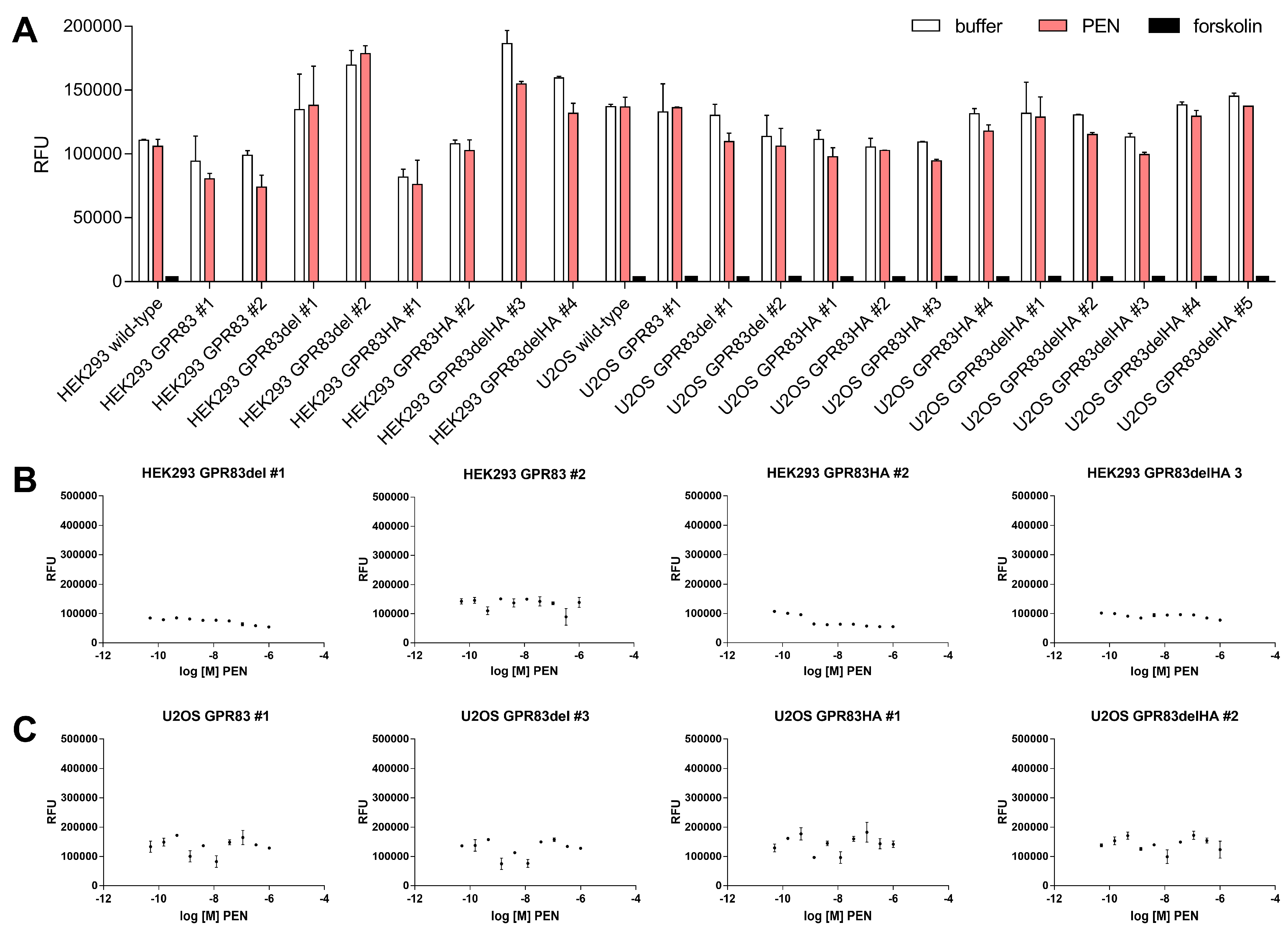
2.3. Ligand-Dependent cAMP Formation
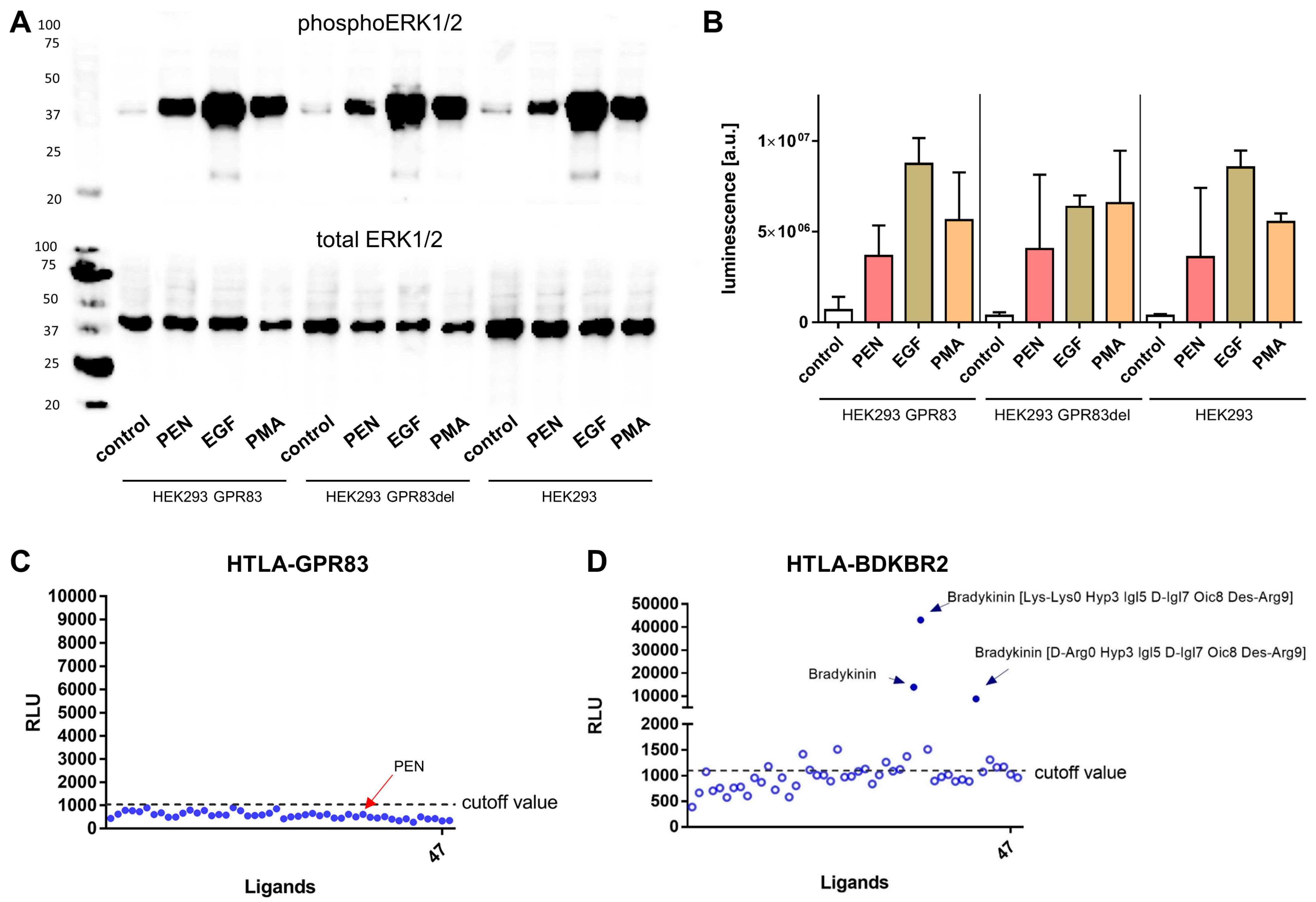
2.4. ERK1/2-Phosphorylation and β-Arrestin Activation
2.5. Radioligand Binding
3. Discussion
4. Materials and Methods
4.1. Reagents and Cell Culture
4.2. Peptide Synthesis
4.3. RNA Isolation and Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
- hGPR83-Fwd: 5′-TCAAGAACCAGCGAATGCAC-3′
- hGPR83-Rev: 5′-TGCTGTTCACAAAGCGAACC-3′
- hGAPDH-Fwd: 5′-TGCACCACCAACTGCTTAGC-3′
- hGAPDH-Rev: 5′-GGCATGGACTGTGGTCATGAG-3′
- hUBC-Fwd: 5′-ATTTGGGTCGCAGTTCTTG-3′
- hUBC-Rev: 5′-TGCCTTGACATTCTCGATGGT-3′
4.4. Immunofluorescence
4.5. Calcium Mobilization Assay
4.6. cAMP Assay
4.7. ERK1/2 Phosphorylation Assay
4.8. TANGO Assay
4.9. Radioiodination
4.10. Radioligand Binding Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dubins, J.S.; Sanchez-Alavez, M.; Zhukov, V.; Sanchez-Gonzalez, A.; Moroncini, G.; Carvajal-Gonzalez, S.; Hadcock, J.R.; Bartfai, T.; Conti, B. Downregulation of GPR83 in the hypothalamic preoptic area reduces core body temperature and elevates circulating levels of adiponectin. Metabolism 2012, 61, 1486–1493. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.D.; Muller, A.; Yi, C.X.; Habegger, K.M.; Meyer, C.W.; Gaylinn, B.D.; Finan, B.; Heppner, K.; Trivedi, C.; Bielohuby, M.; et al. The orphan receptor Gpr83 regulates systemic energy metabolism via ghrelin-dependent and ghrelin-independent mechanisms. Nat. Commun. 2013, 4, 1968. [Google Scholar] [CrossRef] [PubMed]
- Fakira, A.K.; Lueptow, L.M.; Trimbake, N.A.; Devi, L.A. PEN Receptor GPR83 in Anxiety-like Behaviors: Differential Regulation in Global vs Amygdalar Knockdown. Front. Neurosci. 2021, 15, 675769. [Google Scholar] [CrossRef]
- Lueptow, L.M.; Devi, L.A.; Fakira, A.K. Targeting the Recently Deorphanized Receptor GPR83 for the Treatment of Immunological, Neuroendocrine and Neuropsychiatric Disorders. Prog. Mol. Biol. Transl. Sci. 2018, 159, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Mack, S.M.; Gomes, I.; Devi, L.A. Neuropeptide PEN and Its Receptor GPR83: Distribution, Signaling, and Regulation. ACS Chem. Neurosci. 2019, 10, 1884–1891. [Google Scholar] [CrossRef]
- Hansen, W.; Loser, K.; Westendorf, A.M.; Bruder, D.; Pfoertner, S.; Siewert, C.; Huehn, J.; Beissert, S.; Buer, J. G protein-coupled receptor 83 overexpression in naive CD4+CD25− T cells leads to the induction of Foxp3+ regulatory T cells in vivo. J. Immunol. 2006, 177, 209–215. [Google Scholar] [CrossRef]
- Lu, L.F.; Gavin, M.A.; Rasmussen, J.P.; Rudensky, A.Y. G protein-coupled receptor 83 is dispensable for the development and function of regulatory T cells. Mol. Cell Biol. 2007, 27, 8065–8072. [Google Scholar] [CrossRef] [PubMed]
- Toms, C.; Jessup, H.; Thompson, C.; Baban, D.; Davies, K.; Powrie, F. Gpr83 expression is not required for the maintenance of intestinal immune homeostasis and regulation of T-cell-dependent colitis. Immunology 2008, 125, 302–312. [Google Scholar] [CrossRef]
- Choi, S.; Hachisuka, J.; Brett, M.A.; Magee, A.R.; Omori, Y.; Iqbal, N.U.; Zhang, D.; DeLisle, M.M.; Wolfson, R.L.; Bai, L.; et al. Parallel ascending spinal pathways for affective touch and pain. Nature 2020, 587, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.; Bobeck, E.N.; Margolis, E.B.; Gupta, A.; Sierra, S.; Fakira, A.K.; Fujita, W.; Muller, T.D.; Muller, A.; Tschop, M.H.; et al. Identification of GPR83 as the receptor for the neuroendocrine peptide PEN. Sci. Signal. 2016, 9, ra43. [Google Scholar] [CrossRef]
- Wardman, J.H.; Berezniuk, I.; Di, S.; Tasker, J.G.; Fricker, L.D. ProSAAS-derived peptides are colocalized with neuropeptide Y and function as neuropeptides in the regulation of food intake. PLoS ONE 2011, 6, e28152. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.R.; Hauser, A.S.; Vedel, L.; Strachan, R.T.; Huang, X.P.; Gavin, A.C.; Shah, S.D.; Nayak, A.P.; Haugaard-Kedstrom, L.M.; Penn, R.B.; et al. Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors. Cell 2019, 179, 895–908.e821. [Google Scholar] [CrossRef]
- Parobchak, N.; Rao, S.; Negron, A.; Schaefer, J.; Bhattacharya, M.; Radovick, S.; Babwah, A.V. Uterine Gpr83 mRNA is highly expressed during early pregnancy and GPR83 mediates the actions of PEN in endometrial and non-endometrial cells. FS Sci. 2020, 1, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Sallee, N.A.; Lee, E.; Leffert, A.; Ramirez, S.; Brace, A.D.; Halenbeck, R.; Kavanaugh, W.M.; Sullivan, K.M.C. A Pilot Screen of a Novel Peptide Hormone Library Identified Candidate GPR83 Ligands. SLAS Discov. 2020, 25, 1047–1063. [Google Scholar] [CrossRef]
- Mack, S.M.; Gomes, I.; Fakira, A.K.; Duarte, M.L.; Gupta, A.; Fricker, L.; Devi, L.A. GPR83 engages endogenous peptides from two distinct precursors to elicit differential signaling. Mol. Pharmacol. 2022, 102, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Z.; Wang, Y.F.; Shao, X.X.; Liu, Y.L.; Xu, Z.G.; Wang, S.L.; Guo, Z.Y. FAM237A, rather than peptide PEN and proCCK56-63, binds to and activates the orphan receptor GPR83. FEBS J. 2023, 290, 3461–3479. [Google Scholar] [CrossRef]
- Muller, A.; Berkmann, J.C.; Scheerer, P.; Biebermann, H.; Kleinau, G. Insights into Basal Signaling Regulation, Oligomerization, and Structural Organization of the Human G-Protein Coupled Receptor 83. PLoS ONE 2016, 11, e0168260. [Google Scholar] [CrossRef]
- Kroeze, W.K.; Sassano, M.F.; Huang, X.P.; Lansu, K.; McCorvy, J.D.; Giguere, P.M.; Sciaky, N.; Roth, B.L. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 2015, 22, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.; Kleinau, G.; Piechowski, C.L.; Muller, T.D.; Finan, B.; Pratzka, J.; Gruters, A.; Krude, H.; Tschop, M.; Biebermann, H. G-protein coupled receptor 83 (GPR83) signaling determined by constitutive and zinc(II)-induced activity. PLoS ONE 2013, 8, e53347. [Google Scholar] [CrossRef] [PubMed]
- Soletto, L.; Hernandez-Balfago, S.; Rocha, A.; Scheerer, P.; Kleinau, G.; Cerda-Reverter, J.M. Melanocortin Receptor Accessory Protein 2-Induced Adrenocorticotropic Hormone Response of Human Melanocortin 4 Receptor. J. Endocr. Soc. 2019, 3, 314–323. [Google Scholar] [CrossRef]
- Wang, M.; Wang, X.; Jiang, B.; Zhai, Y.; Zheng, J.; Yang, L.; Tai, X.; Li, Y.; Fu, S.; Xu, J.; et al. Identification of MRAP protein family as broad-spectrum GPCR modulators. Clin. Transl. Med. 2022, 12, e1091. [Google Scholar] [CrossRef] [PubMed]
- Luttrell, L.M.; Lefkowitz, R.J. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell Sci. 2002, 115, 455–465. [Google Scholar] [CrossRef]
- Gurevich, V.V.; Gurevich, E.V. Arrestin-mediated signaling: Is there a controversy? World J. Biol. Chem. 2018, 9, 25–35. [Google Scholar] [CrossRef]
- Rozenfeld, R.; Devi, L.A. Receptor heteromerization and drug discovery. Trends Pharmacol. Sci. 2010, 31, 124–130. [Google Scholar] [CrossRef]
- Rozenfeld, R.; Devi, L.A. Exploring a role for heteromerization in GPCR signalling specificity. Biochem. J. 2011, 433, 11–18. [Google Scholar] [CrossRef]
- Wilson, S.J.; McGinley, K.; Huang, A.J.; Smyth, E.M. Heterodimerization of the alpha and beta isoforms of the human thromboxane receptor enhances isoprostane signaling. Biochem. Biophys. Res. Commun. 2007, 352, 397–403. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ellis, J.; Pediani, J.D.; Canals, M.; Milasta, S.; Milligan, G. Orexin-1 receptor-cannabinoid CB1 receptor heterodimerization results in both ligand-dependent and -independent coordinated alterations of receptor localization and function. J. Biol. Chem. 2006, 281, 38812–38824. [Google Scholar] [CrossRef]
- Evers, B.M.; Townsend, C.M., Jr.; Upp, J.R.; Allen, E.; Hurlbut, S.C.; Kim, S.W.; Rajaraman, S.; Singh, P.; Reubi, J.C.; Thompson, J.C. Establishment and characterization of a human carcinoid in nude mice and effect of various agents on tumor growth. Gastroenterology 1991, 101, 303–311. [Google Scholar] [CrossRef]
- Barnea, G.; Strapps, W.; Herrada, G.; Berman, Y.; Ong, J.; Kloss, B.; Axel, R.; Lee, K.J. The genetic design of signaling cascades to record receptor activation. Proc. Natl. Acad. Sci. USA 2008, 105, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Bandholtz, S.; Erdmann, S.; von Hacht, J.L.; Exner, S.; Krause, G.; Kleinau, G.; Grotzinger, C. Urolinin: The First Linear Peptidic Urotensin-II Receptor Agonist. J. Med. Chem. 2016, 59, 10100–10112. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giesecke, Y.; Asimi, V.; Stulberg, V.; Kleinau, G.; Scheerer, P.; Koksch, B.; Grötzinger, C. Is the Neuropeptide PEN a Ligand of GPR83? Int. J. Mol. Sci. 2023, 24, 15117. https://doi.org/10.3390/ijms242015117
Giesecke Y, Asimi V, Stulberg V, Kleinau G, Scheerer P, Koksch B, Grötzinger C. Is the Neuropeptide PEN a Ligand of GPR83? International Journal of Molecular Sciences. 2023; 24(20):15117. https://doi.org/10.3390/ijms242015117
Chicago/Turabian StyleGiesecke, Yvonne, Vahid Asimi, Valentina Stulberg, Gunnar Kleinau, Patrick Scheerer, Beate Koksch, and Carsten Grötzinger. 2023. "Is the Neuropeptide PEN a Ligand of GPR83?" International Journal of Molecular Sciences 24, no. 20: 15117. https://doi.org/10.3390/ijms242015117
APA StyleGiesecke, Y., Asimi, V., Stulberg, V., Kleinau, G., Scheerer, P., Koksch, B., & Grötzinger, C. (2023). Is the Neuropeptide PEN a Ligand of GPR83? International Journal of Molecular Sciences, 24(20), 15117. https://doi.org/10.3390/ijms242015117







