miRNA Expression Signatures Induced by Chicken Astrovirus Infection in Chickens
Abstract
:1. Introduction
2. Results
2.1. Clinical Features of CAstV-Infected Chickens
2.2. Analysis of miRNA Expression
2.3. Gene Ontology and KEGG Pathway Enrichment Analyses of Target Genes
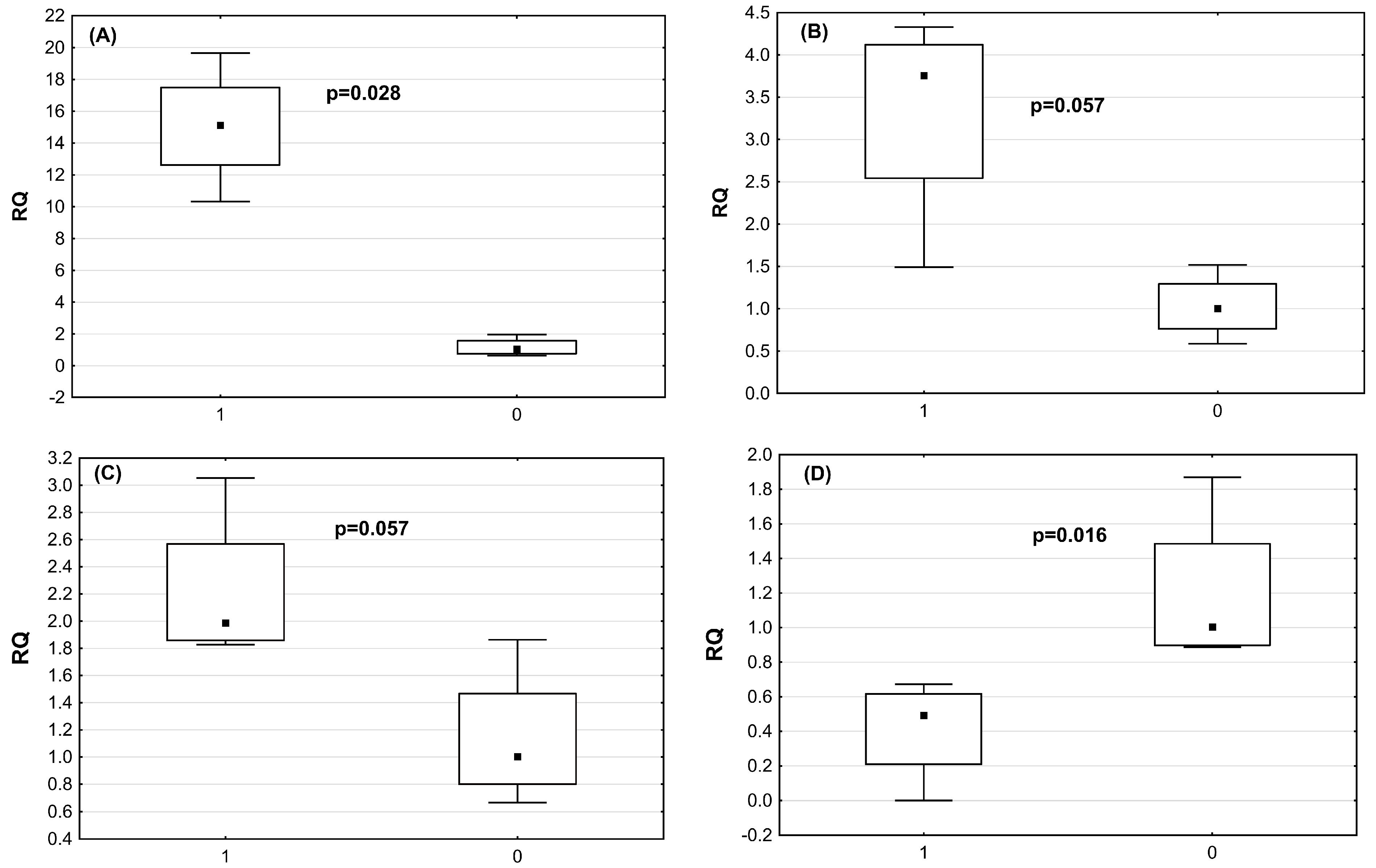
2.4. Verification of DE miRNAs by qRT-PCRmiRNA
3. Discussion
4. Materials and Methods
4.1. Virus and Animals
4.2. Animal Experiments
4.3. Tissue Homogenization and Total RNA Isolation
4.4. RNA Quantification and Quality Control
4.5. Small RNA Sequencing
4.6. Raw Data Analysis
4.7. Accession Number
4.8. Real-Time PCR Validation of RNA Reads
4.9. Statistical Analysis of qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sajewicz-Krukowska, J.; Domanska-Blicharz, K. Nearly full-length genome sequence of a novel astrovirus isolated from chickens with ‘white chicks’ condition. Arch. Virol. 2016, 161, 2581–2587. [Google Scholar] [CrossRef]
- Sajewicz-Krukowska, J.; Pać, K.; Lisowska, A.; Pikuła, A.; Minta, Z.; Króliczewska, B.; Domańska-Blicharz, K. Astrovirus-induced “white chicks” condition—Field observation, virus detection and preliminary characterization. Avian Pathol. 2016, 45, 2–12. [Google Scholar] [CrossRef]
- Kang, K.I.; El-Gazzar, M.; Sellers, H.S.; Dorea, F.; Williams, S.M.; Kim, T.; Collett, S.; Mundt, E. Investigation into the aetiology of runting and stunting syndrome in chickens. Avian Pathol. 2012, 41, 41–50. [Google Scholar] [CrossRef]
- Raji, A.A.; Omar, A.R. Pathogenesis of Chicken Astrovirus Related Illnesses. Front. Vet. Sci. 2022, 9, 899901. [Google Scholar] [CrossRef]
- Baxendale, W.; Mebatsion, T. The isolation and characterisation of astroviruses from chickens. Avian Pathol. 2004, 33, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Palomino-Tapia, V.; Mitevski, D.; Inglis, T.; van der Meer, F.; Martin, E.; Brash, M.; Provost, C.; Gagnon, C.A.; Abdul-Careem, M.F. Chicken Astrovirus (CAstV) Molecular Studies Reveal Evidence of Multiple Past Recombination Events in Sequences Originated from Clinical Samples of White Chick Syndrome (WCS) in Western Canada. Viruses 2020, 12, 1096. [Google Scholar] [CrossRef] [PubMed]
- Herbert, J.A.; Panagiotou, S. Immune Response to Viruses. Encycl. Infect. Immun. 2022, 2022, 429–444. [Google Scholar]
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995. [Google Scholar] [CrossRef]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay Between Viral-Derived miRNAs and Host Immunity During Infection. Front. Immunol. 2020, 10, 3079. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Viral and cellular messenger RNA targets of viral microRNAs. Nature 2009, 457, 421–425. [Google Scholar] [CrossRef]
- Umbach, J.L.; Cullen, B.R. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009, 23, 1151–1164. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Vaddadi, K.; Gandikota, C.; Huang, C.; Liang, Y.; Liu, L. Cellular microRNAs target SARS-CoV-2 spike protein and restrict viral replication. Am. J. Physiol. Cell Physiol. 2023, 325, C420–C428. [Google Scholar] [CrossRef]
- Lindsay, M.A. microRNAs and the immune response. Trends Immunol. 2008, 29, 343–351. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Stik, G.; Dambrine, G.; Pfeffer, S.; Rasschaert, D. The oncogenic microRNA OncomiR-21 overexpressed during Marek’s disease lymphomagenesis is transactivated by the viral oncoprotein. Meq. J. Virol. 2013, 87, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Sandford, E.E.; Orr, M.; Balfanz, E.; Bowerman, N.; Li, X.; Zhou, H.; Johnson, T.J.; Kariyawasam, S.; Liu, P.; Nolan, L.K.; et al. Spleen transcriptome response to infection with avian pathogenic Escherichia coli in broiler chickens. BMC Genom. 2011, 12, 469. [Google Scholar] [CrossRef] [PubMed]
- Olah, I.; Vervelde, L. Structure of the avian lymphoid system. In Avian Immunol; Schat, K.A., Kaspers, B., Kaiser, P., Eds.; Elsevier Academic Press Inc.: Amsterdam, The Netherlands, 2014; pp. 11–44. [Google Scholar]
- Chen, Q.; Tong, C.; Ma, S.; Zhou, L.; Zhao, L.; Zhao, X. Involvement of MicroRNAs in Probiotics-Induced Reduction of the Cecal Inflammation by Salmonella Typhimurium. Front. Immunol. 2017, 8, 704. [Google Scholar] [CrossRef]
- Hong, Y.; Truong, A.D.; Lee, J.; Vu, T.H.; Lee, S.; Song, K.D.; Lillehoj, H.S.; Hong, Y.H. Exosomal miRNA profiling from H5N1 avian influenza virus-infected chickens. Vet. Res. 2021, 52, 36. [Google Scholar] [CrossRef]
- Khanduri, A.; Sahu, A.R.; Wani, S.A.; Khan, R.I.N.; Pandey, A.; Saxena, S.; Malla, W.A.; Mondal, P.; Rajak, K.K.; Muthuchelvan, D.; et al. Dysregulated miRNAome and Proteome of PPRV Infected Goat PBMCs Reveal a Coordinated Immune Response. Front. Immunol. 2018, 9, 2631. [Google Scholar] [CrossRef]
- Yang, J.; Huang, X.; Liu, Y.; Zhao, D.; Han, K.; Zhang, L.; Li, Y.; Liu, Q. Analysis of the microRNA expression profiles of chicken dendritic cells in response to H9N2 avian influenza virus infection. Vet. Res. 2020, 51, 132. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Mu, Z.; Nie, F.; Chang, X.; Duan, H.; Li, H.; Zhang, J.; Zhou, J.; Ji, Y.; Li, M. Thymic transcriptome analysis after Newcastle disease virus inoculation in chickens and the influence of host small RNAs on NDV replication. Sci. Rep. 2021, 11, 10270. [Google Scholar] [CrossRef]
- Hicks, J.A.; Liu, H.-C. Impact of HVT Vaccination on Splenic miRNA Expression in Marek’s Disease Virus Infections. Genes 2019, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Harris-Arnold, A.; Arnold, C.P.; Schaffert, S.; Hatton, O.; Krams, S.M.; Esquivel, C.O.; Martinez, O.M. Epstein-Barr virus modulates host cell microRNA-194 to promote IL-10 production and B lymphoma cell survival. Am. J. Transplant. 2015, 15, 2814–2824. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Lai, C.; Gu, H.; Zhao, L.; Xia, M.; Yang, P.; Wang, X. miR-194 Inhibits Innate Antiviral Immunity by Targeting FGF2 in Influenza H1N1 Virus Infection. Front. Microbiol. 2017, 8, 2187. [Google Scholar] [CrossRef]
- Peng, X.; Gao, Q.S.; Zhou, L.; Chen, Z.H.; Lu, S.; Huang, H.J.; Zhan, C.Y.; Xiang, M. MicroRNAs in avian influenza virus H9N2-infected and non-infected chicken embryo fibroblasts. Genet. Mol. Res. 2015, 14, 9081–9091. [Google Scholar] [CrossRef]
- Tian, H.; He, Z. miR-215 Enhances HCV Replication by Targeting TRIM22 and Inactivating NF-κB Signaling. Yonsei Med. J. 2018, 59, 511–518. [Google Scholar] [CrossRef]
- Wang, Y.; Brahmakshatriya, V.; Lupiani, B.; Reddy, S.M.; Soibam, B.; Benham, A.L.; Gunaratne, P.; Liu, H.C.; Trakooljul, N.; Ing, N.; et al. Integrated analysis of microRNA expression and mRNA transcriptome in lungs of avian influenza virus infected broilers. BMC Genom. 2012, 13, 278. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, J.; Su, J.; Liu, Y.; Guo, J.; Zhang, Y.; Lu, C.; Xing, S.; Guan, Y.; Li, Y.; et al. MicroRNAs in the immune organs of chickens and ducks indicate divergence of immunity against H5N1 avian influenza. FEBS Lett. 2015, 589, 419–425. [Google Scholar] [CrossRef]
- Yang, X.; Gao, W.; Liu, H.; Li, J.; Chen, D.; Yuan, F.; Zhang, Z.; Wang, H. MicroRNA transcriptome analysis in chicken kidneys in response to differing virulent infectious bronchitis virus infections. Arch. Virol. 2017, 162, 3397–3405. [Google Scholar] [CrossRef] [PubMed]
- O’Dowd, K.; Emam, M.; El Khili, M.R.; Emad, A.; Ibeagha-Awemu, E.M.; Gagnon, C.A.; Barjesteh, N. Distinct miRNA Profile of Cellular and Extracellular Vesicles Released from Chicken Tracheal Cells Following Avian Influenza Virus Infection. Vaccines 2020, 8, 438. [Google Scholar] [CrossRef]
- Hicks, J.A.; Trakooljul, N.; Liu, H.C. Alterations in cellular and viral microRNA and cellular gene expression in Marek’s disease virus-transformed T-cell lines treated with sodium butyrate. Poult. Sci. 2019, 98, 642–652. [Google Scholar] [CrossRef]
- Huang, X.; Li, Y.; Wang, X.; Zhang, J.; Wang, L.; Zhou, H.; Jiang, Y.; Cui, W.; Qiao, X.; Li, Y.; et al. Genome-wide identification of chicken bursae of Fabricius miRNAs in response to very virulent infectious bursal disease virus. Arch. Virol. 2022, 167, 1855–1864. [Google Scholar] [CrossRef]
- Correa-Medina, M.; Bravo-Egana, V.; Rosero, S.; Ricordi, C.; Edlund, H.; Diez, J.; Pastori, R.L. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr. Patterns GEP. 2009, 9, 193–199. [Google Scholar] [CrossRef]
- Lopez-Beas, J.; Capilla-Gonzalez, V.; Aguilera, Y.; Mellado, N.; Lachaud, C.C.; Martin, F.; Smani, T.; Soria, B.; Hmadcha, A. miR-7 Modulates hESC Differentiation into Insulin-Producing Beta-like Cells and Contributes to Cell Maturation. Mol. Ther. Nucleic Acids. 2018, 12, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Wang, J.; Wang, J.; Wu, J.; Song, J.; Zhang, C.Y.; Zhang, C.; Wang, C.; Wang, J.J. Increased serum miR-7 is a promising biomarker for type 2 diabetes mellitus and its microvascular complications. Diabetes Res. Clin. Pract. 2017, 130, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, D.; Zhang, S.; Wei, X.; Song, J.; Zhang, Y.; Jin, M.; Shen, Z.; Wang, X.; Feng, Z.; et al. Host-virus interaction: The antiviral defense function of small interfering RNAs can be enhanced by host microRNA-7 in vitro. Sci. Rep. 2015, 5, 9722. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, W.; Xu, Z.; Pan, J.; Zhao, Z.; Ren, Q. Eriocheir sinensis microRNA-7 targets crab Myd88 to enhance white spot syndrome virus replication. Fish. Shellfish Immunol. 2018, 79, 274–283. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, L.; Du, J.; Hu, X.; Xie, Y.; Wu, J.; Lin, X.; Yin, N.; Sun, M.; Li, H. MicroRNA-7 Inhibits Rotavirus Replication by Targeting Viral NSP5 In Vivo and In Vitro. Viruses 2020, 12, 209. [Google Scholar] [CrossRef]
- Lee, H.J.; Palkovits, M.; Young, W.S., 3rd. miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc. Natl. Acad. Sci. USA 2006, 103, 15669–15674. [Google Scholar] [CrossRef] [PubMed]
- Sajewicz-Krukowska, J.; Jastrzębski, J.P.; Grzybek, M.; Domańska-Blicharz, K.; Tarasiuk, K.; Marzec-Kotarska, B. Transcriptome Sequencing of the Spleen Reveals Antiviral Response Genes in Chickens Infected with CAstV. Viruses 2021, 13, 2374. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Jiang, H.; Sun, J.; Diao, Y.; Tang, Y.; Hu, J. Integrated Analysis of miRNA and mRNA Expression Profiles in Spleen of Specific Pathogen-Free Chicken Infected with Avian Reticuloendotheliosis Virus Strain SNV. Int. J. Mol. Sci. 2019, 20, 1041. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Asaf, M.; Kumar, A.; Kulkarni, D.D.; Sood, R.; Bhatia, S.; Bhushan, B.; Raut, A.A. Differential miRNA expression profiling of highly pathogenic avian influenza virus H5N1 infected chicken lungs reveals critical microRNAs, biological pathways and genes involved in the molecular pathogenesis. Virol. Sin. 2022, 37, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, W.-Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.-F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Sig. Transd. 2015, 35, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.C.; Cheung, C.-Y.; Law, A.H.; Mok, C.K.; Peiris, M.; Lau, A.S. p38 mitogen-activated protein kinase-dependent hyperinduction of tumor necrosis factor alpha expression in response to avian influenza virus H5N1. J. Virol. 2005, 79, 10147–10154. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Zhang, Y.; Luo, Z.; Li, P.; Liu, L.; Wang, C.; Wang, H.; Li, H.; Ma, Y. Autophagy mediates avian influenza H5N1 pseudotyped particle-induced lung inflammation through NF-κB and p38 MAPK signaling pathways. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 306, L183–L195. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.; Lee, S.M.; Cheung, C.-Y.; Ng, I.H.; Poon, L.L.; Guan, Y.; Ip, N.Y.; Lau, A.S.; Peiris, J.M. Induction of proinflammatory cytokines in primary human macrophages by influenza A virus (H5N1) is selectively regulated by IFN regulatory factor 3 and p38 MAPK. J. Immunol. 2009, 182, 1088–1098. [Google Scholar] [CrossRef]
- Xing, Z.; Cardona, C.J.; Anunciacion, J.; Adams, S.; Dao, N. Roles of the ERK MAPK in the regulation of proinflammatory and apoptotic responses in chicken macrophages infected with H9N2 avian influenza virus. J. Gen. Virol. 2010, 91, 343–351. [Google Scholar] [CrossRef]
- Chu, Z.; Ma, J.; Wang, C.; Lu, K.; Li, X.; Liu, H.; Wang, X.; Xiao, S.; Yang, Z. Newcastle Disease Virus V Protein Promotes Viral Replication in HeLa Cells through the Activation of MEK/ERK Signaling. Viruses 2018, 10, 489. [Google Scholar] [CrossRef]
- Moser, L.A.; Schultz-Cherry, S. Suppression of astrovirus replication by an ERK1/2 inhibitor. J. Virol. 2008, 82, 7475–7482. [Google Scholar] [CrossRef]
- Rodgers, S.J.; Ferguson, D.T.; Mitchell, C.A.; Ooms, L.M. Regulation of PI3K effector signalling in cancer by the phosphoinositide phosphatases. Biosci. Rep. 2017, 37, BSR20160432. [Google Scholar] [CrossRef]
- Ehrhardt, C.; Wolff, T.; Pleschka, S.; Planz, O.; Beermann, W.; Bode, J.G.; Schmolke, M.; Ludwig, S. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 2007, 81, 3058–3067. [Google Scholar] [CrossRef]
- Hirata, N.; Suizu, F.; Matsuda-Lennikov, M.; Edamura, T.; Bala, J.; Noguchi, M. Inhibition of Akt kinase activity suppresses entry and replication of influenza virus. Biochem. Biophys. Res. Commun. 2014, 450, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Cao, W.; Liao, M. The PI3K/Akt pathway is involved in early infection of some exogenous avian leukosis viruses. J. Gen. Virol. 2011, 92, 1688–1697. [Google Scholar] [CrossRef]
- Kang, Y.; Yuan, R.; Zhao, X.; Xiang, B.; Gao, S.; Gao, P.; Dai, X.; Feng, M.; Li, Y.; Xie, P.; et al. Transient activation of the PI3K/Akt pathway promotes Newcastle disease virus replication and enhances anti-apoptotic signaling responses. Oncotarget 2017, 8, 23551–23563. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, J.; He, M.; Luo, Q.; Liu, F.; Chen, R. Marek’s Disease Virus Activates the PI3K/Akt Pathway Through Interaction of Its Protein Meq With the P85 Subunit of PI3K to Promote Viral Replication. Front. Microbiol. 2018, 9, 2547. [Google Scholar] [CrossRef] [PubMed]
- Tange, S.; Zhou, Y.; Nagakui-Noguchi, Y.; Imai, T.; Nakanishi, A. Initiation of human astrovirus type 1 infection was blocked by inhibitors of phosphoinositide 3-kinase. Virol. J. 2013, 10, 153. [Google Scholar] [CrossRef]
- Mercer, J.; Schelhaas, M.; Helenius, A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010, 79, 803–833. [Google Scholar] [CrossRef]
- Méndez, E.; Muñoz-Yañez, C.; Sánchez-San Martín, C.; Aguirre-Crespo, G.; Baños-Lara Mdel, R.; Gutierrez, M.; Espinosa, R.; Acevedo, Y.; Arias, C.F.; López, S. Characterization of human astrovirus cell entry. J. Virol. 2014, 88, 2452–2460. [Google Scholar] [CrossRef]
- Kim, T.H.; Zhou, H. Overexpression of Chicken IRF7 Increased Viral Replication and Programmed Cell Death to the Avian Influenza Virus Infection Through TGF-Beta/FoxO Signaling Axis in DF-1. Front. Genet. 2018, 9, 415. [Google Scholar] [CrossRef]
- Hargest, V.; Bub, T.; Neale, G.; Schultz-Cherry, S. Astrovirus-induced epithelial-mesenchymal transition via activated TGF-β increases viral replication. PLoS Pathog. 2022, 18, e1009716. [Google Scholar] [CrossRef] [PubMed]
- Moser, L.A.; Carter, M.; Schultz-Cherry, S. Astrovirus increases epithelial barrier permeability independently of viral replication. J. Virol. 2007, 81, 11937–11945. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Iwasaki, A. Autophagy and antiviral immunity. Curr. Opin. Immunol. 2008, 20, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Ge, X.; Guo, X.; Yang, H. Autophagy promotes the replication of encephalomyocarditis virus in host cells. Autophagy 2011, 7, 613–628. [Google Scholar] [CrossRef]
- Kimmey, J.M.; Stallings, C.L. Bacterial pathogens versus autophagy: Implications for therapeutic interventions. Trends Mol. Med. 2016, 22, 1060–1076. [Google Scholar] [CrossRef]
- Liao, Z.; Zhang, X.; Song, C.; Lin, W.; Cheng, Y.; Xie, Z.; Chen, S.; Nie, Y.; Li, A.; Zhang, H.; et al. ALV-J inhibits autophagy through the GADD45β/MEKK4/P38MAPK signaling pathway and mediates apoptosis following autophagy. Cell Death Dis. 2020, 11, 684. [Google Scholar] [CrossRef]
- Xie, T.; Feng, M.; Zhang, X.; Li, X.; Mo, G.; Shi, M.; Zhang, X. Chicken CH25H inhibits ALV-J replication by promoting cellular autophagy. Front. Immunol. 2023, 14, 1093289. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, M.L.; Zhao, J. Crosstalk between autophagy and type I interferon responses in innate antiviral immunity. Viruses 2019, 11, 132. [Google Scholar] [CrossRef]
- Qiao, D.; He, Q.; Cheng, X.; Yao, Y.; Nair, V.; Shao, H.; Qin, A.; Qian, K. Regulation of Avian Leukosis Virus Subgroup J Replication by Wnt/β-Catenin Signaling Pathway. Viruses 2021, 13, 1968. [Google Scholar] [CrossRef]
- Liu, M.; Yang, Q.; Han, J. Transcriptomic analysis reveals that coxsackievirus B3 Woodruff and GD strains use similar key genes to induce FoxO signaling pathway activation in HeLa cells. Arch. Virol. 2022, 167, 131–140. [Google Scholar] [CrossRef]
- Ho, J.; Moyes, D.L.; Tavassoli, M.; Naglik, J.R. The Role of ErbB Receptors in Infection. Trends Microbiol. 2017, 25, 942–952. [Google Scholar] [CrossRef]
- Taylor, M.P.; Koyuncu, O.O.; Enquist, L.W. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 2011, 9, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.B.; Dembo, M.; Hanks, S.K.; Wang, Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc. Natl. Acad. Sci. USA 2001, 98, 11295–11300. [Google Scholar] [CrossRef] [PubMed]
- Karam, B.S.; Morris, R.S.; Bramante, C.T.; Puskarich, M.; Zolfaghari, E.J.; Lotfi-Emran, S.; Ingraham, N.E.; Charles, A.; Odde, D.J.; Tignanelli, C.J. mTOR inhibition in COVID-19: A commentary and review of efficacy in RNA viruses. J. Med. Virol. 2021, 93, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Girardi, E.; López, P.; Pfeffer, S. On the importance of host microRNAs during viral infection. Front. Genet. 2018, 9, 439. [Google Scholar] [CrossRef]
- Smyth, V.J.; Jewhurst, H.L.; Wilkinson, D.S.; Adair, B.M.; Gordon, A.W.; Todd, D. Development and evaluation of real-time TaqMan(R) RT-PCR assays for the detection of avian nephritis virus and chicken astrovirus in chickens. Avian Pathol. 2010, 39, 467–474. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control TOOL for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 2 February 2021).
- Cutadapt 1.16. Available online: https://cutadapt.readthedocs.org/en/stable/ (accessed on 2 October 2019).
- miRBase v21. Available online: http://www.mirbase.org/ (accessed on 2 October 2019).
- RNAcentral 10.0. Available online: https://rnacentral.org/ (accessed on 2 October 2019).
- miRDeep2. Available online: https://www.mdc-berlin.de/content/mirdeep2-documentation (accessed on 2 October 2019).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bioconductor Open Source Software for Bioinformatics. Available online: https://bioconductor.org (accessed on 3 February 2021).
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Huang Da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- The Gene Ontology Home Page. Available online: http://geneontology.org/ (accessed on 1 August 2012).
- KEGG. Available online: http://www.genome.jp/kegg/ (accessed on 10 January 2009).
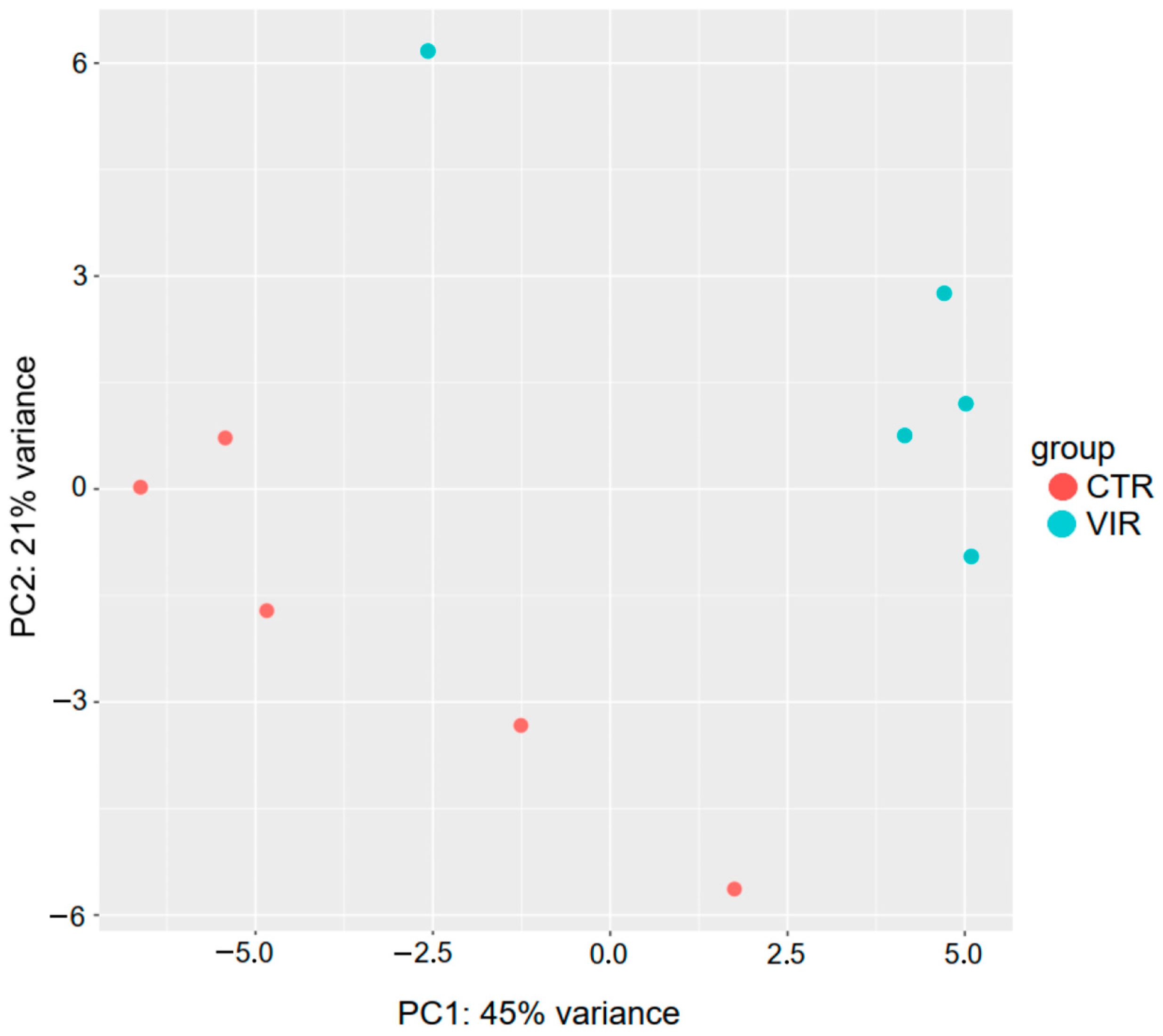


| Base Mean | log 2 Fold Change | p-Value | p adj | reg | |
|---|---|---|---|---|---|
| miR-7b | 164.4544 | −9.9228 | 4.88002383607237 × 10−27 | 8.05203932951941 × 10−25 | down |
| miR-1664-3p | 147.0916 | −1.3340 | 2.37707360550244 × 10−13 | 8.71593655350895 × 10−12 | down |
| miR-122-5p | 127.9187 | −0.9030 | 9.76991651583061 × 10−5 | 0.00140177063053222 | down |
| miR-128-1-5p | 243.0959 | −0.8677 | 3.35882982217558 × 10−10 | 9.23678201098283 × 10−9 | down |
| miR-148a-5p | 1892.0338 | −0.5535 | 4.95174600295029 × 10−9 | 1.256981677672 × 10−7 | down |
| miR-1388a-5p | 322,547.0625 | −0.4547 | 1.22866160658981 × 10−6 | 2.38504900102727 × 10−5 | down |
| let-7b | 11,727.14297 | −0.3930 | 0.00587255446460657 | 0.0307609995765106 | down |
| miR-212-3p | 1008.2254 | −0.3917 | 0.000191195025360761 | 0.00252377433476205 | down |
| miR-181a | 361,275.1404 | −0.3852 | 0.000837453238149873 | 0.00674341931614952 | down |
| miR-212-5p | 919.3084 | −0.3038 | 0.00123092502988673 | 0.00902678355250267 | down |
| miR-7 | 728.0242 | 1.1319 | 4.08074456514255 × 10−16 | 4.48881902165681 | up |
| miR-3536 | 435.0424 | 1.4528 | 0.0072657815031363 | 0.0368878137851535 | up |
| miR-1563 | 404.4242 | 1.7188 | 0.000539888145993531 | 0.00470226549287548 | up |
| miR-215-5p | 883.0175 | 1.7482 | 0.00106070451263859 | 0.00814029044583105 | up |
| miR-3528 | 168.3393 | 1.9790 | 0.00275325105712132 | 0.0185423030377558 | up |
| miR-194 | 46.1090 | 2.0418 | 0.00376268623521676 | 0.023878585723491 | up |
| miR-3535 | 199.8722 | 2.2906 | 0.00033749035146587 | 0.0032756416465805 | up |
| miR-1434 | 31.2215 | 2.4343 | 2.61622382369712 × 10−7 | 5.39596163637531 × 10−6 | up |
| miR-3533 | 14.7673 | 2.8310 | 6.44514082042924 × 10−8 | 1.51921176481546 × 10−6 | up |
| miR-2954 | 13,896.9314 | 3.2233 | 5.95623585116667 × 10−102 | 1.965557830885 × 10−99 | up |
| Description | p-Value | Fold Enrichment | FDR |
|---|---|---|---|
| GO:0006357~regulation of transcription from RNA polymerase II promoter | 8.28583322838162 × 10−8 | 1.3399 | 3.7253106194803763 × 10−4 |
| GO:0035556~intracellular signal transduction | 3.545900675891955 × 10−7 | 1.6623 | 7.971184719405115 × 10−4 |
| GO:0043087~regulation of GTPase activity | 1.8096098121506414 × 10−5 | 2.2200 | 0.020594082232785374 |
| GO:0006468~protein phosphorylation | 1.8322137217780583 × 10−5 | 1.5982 | 0.020594082232785374 |
| GO:0007411~axon guidance | 3.0001284374567666 × 10−5 | 1.8560 | 0.026977154909611247 |
| GO:0000209~protein polyubiquitination | 1.2557755955090277 × 10−4 | 1.9001 | 0.0864692158518967 |
| GO:0018105~peptidyl-serine phosphorylation | 1.3462733784770392 × 10−4 | 1.7754 | 0.0864692158518967 |
| GO:0007399~nervous system development | 2.4972724679447954 × 10−4 | 1.6531 | 0.1403467126984975 |
| GO:0051965~positive regulation of synapse assembly | 4.989835284296587 × 10−4 | 2.4461 | 0.2492699937577495 |
| GO:0071526~semaphorin-plexin signaling pathway | 9.198871146956833 × 10−4 | 2.8591 | 0.4135812467671792 |
| Description | p-Value | Fold Enrichment | FDR |
|---|---|---|---|
| GO:0004674~protein serine/threonine kinase activity | 3.0866587938833846 × 10−7 | 1.6959 | 4.2256358888263535 × 10−4 |
| GO:0000978~RNA polymerase II core promoter proximal region sequence-specific DNA binding | 1.1901488065126979 × 10−6 | 1.3545 | 8.146568580579417 × 10−4 |
| GO:0046872~metal ion binding | 1.0696663302522198 × 10−4 | 1.2135 | 0.03864123656365064 |
| GO:0003700~transcription factor activity, sequence-specific DNA binding | 1.1290353999605739 × 10−4 | 1.4974 | 0.03864123656365064 |
| GO:0042802~identical protein binding | 2.2739749560208233 × 10−4 | 1.2919 | 0.06226143429585014 |
| GO:0001227~transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding | 4.552443546848259 × 10−4 | 1.5698 | 0.10387158692725443 |
| GO:0005085~guanyl-nucleotide exchange factor activity | 6.272465550624852 × 10−4 | 1.6860 | 0.12170592865052825 |
| GO:0004712~protein serine/threonine/tyrosine kinase activity | 7.112106860512972 × 10−4 | 1.4384 | 0.12170592865052825 |
| GO:0005096~GTPase activator activity | 9.016902518531111 × 10−4 | 1.6080 | 0.1325395104834417 |
| GO:0003682~chromatin binding | 0.001034656718005266 | 1.4259 | 0.1325395104834417 |
| Description | p-Value | Fold Enrichment | FDR |
|---|---|---|---|
| GO:0005634~nucleus | 3.8040482560238504 × 10−6 | 1.1461 | 0.0029591683447036292 |
| GO:0098978~glutamatergic synapsę | 7.6071165673615145 × 10−6 | 1.7482 | 0.0029591683447036292 |
| GO:0005829~cytosol | 2.433178408974373 × 10−5 | 1.1812 | 0.006310042673940207 |
| GO:0005794~Golgi apparatus | 4.615516814134699 × 10−5 | 1.3954 | 0.007716152286538233 |
| GO:0005886~plasma membranę | 4.958966765127399 × 10−5 | 1.1752 | 0.007716152286538233 |
| GO:0005654~nucleoplasm | 4.6747909613586004 × 10−4 | 1.1800 | 0.060616456132283184 |
| GO:0005942~phosphatidylinositol 3-kinase complex | 8.362418083447217 × 10−4 | 2.6013 | 0.09294230384174193 |
| GO:0043197~dendritic spine | 0.0010418476883945682 | 1.9510 | 0.10131968769637176 |
| GO:0016020~membrane | 0.0025990445446487298 | 1.2330 | 0.22467296174852353 |
| GO:0005737~cytoplasm | 0.004274525839628792 | 1.0893 | 0.3115246085292393 |
| Description | p-Value | Fold Enrichment | FDR |
|---|---|---|---|
| gga04010:MAPK signaling pathway | 6.9619020229201035 × 10−9 | 1.5804 | 1.1278281277130568 × 10−6 |
| gga04070:Phosphatidylinositol signaling system | 1.4035027273706892 × 10−6 | 1.8709 | 1.1368372091702583 × 10−4 |
| gga04144:Endocytosis | 3.5017161152942025 × 10−6 | 1.4888 | 1.8909267022588694 × 10−4 |
| gga04520:Adherens junction | 1.0685138134129477 × 10−5 | 1.7929 | 4.327480944322438 × 10−4 |
| gga04140:Autophagy-animal | 4.6602982670722855 × 10−5 | 1.5888 | 0.0015099366385314204 |
| gga04310:Wnt signaling pathway | 2.5808668477564584 × 10−4 | 1.4834 | 0.006968340488942438 |
| gga04068:FoxO signaling pathway | 0.001287555905712632 | 1.4785 | 0.026319355164882662 |
| gga04012:ErbB signaling pathway | 0.0012997212427102549 | 1.6218 | 0.026319355164882662 |
| gga00562:Inositol phosphate metabolism | 0.0018404595627396826 | 1.6353 | 0.03312827212931429 |
| gga04510:Focal adhesion | 0.004997193963046481 | 1.3189 | 0.07461497775631122 |
| gga04910:Insulin signaling pathway | 0.005066449106910021 | 1.4165 | 0.07461497775631122 |
| gga04150:mTOR signaling pathway | 0.008692914782026601 | 1.3490 | 0.11735434955735911 |
| gga04810:Regulation of actin cytoskeleton | 0.011161569231401824 | 1.2753 | 0.12952353934521796 |
| gga04912:GnRH signaling pathway | 0.01119339228909291 | 1.4628 | 0.12952353934521796 |
| gga04114:Oocyte meiosis | 0.01204386517678131 | 1.4082 | 0.13007374390923815 |
| gga03250:Viral life cycle—HIV-1 | 0.01976756804864089 | 1.5644 | 0.20014662649248902 |
| gga04261:Adrenergic signaling in cardiomyocytes | 0.02273549527770102 | 1.3176 | 0.21665589617573913 |
| gga04625:C-type lectin receptor signaling pathway | 0.0242064397986349 | 1.3955 | 0.2178579581877141 |
| gga04216:Ferroptosis | 0.030580226017975658 | 1.6819 | 0.26073666394273987 |
| gga00564:Glycerophospholipid metabolism | 0.03379259593242035 | 1.3510 | 0.27372002705260484 |
| miRNA | Astrovirus-Infected Chickens RQ Median (Mean, SD) | Non-Infected Chickens RQ Median (Mean, SD) | p-Value |
|---|---|---|---|
| miR-2954 | 15.1 (15.0; +/−3.8) | 1.04 (1.17; +/−0.58) | p = 0.028 |
| miR-3533 | 3.75 (3.33; +/−0.3) | 1.0 (1.0; +/−38) | p = 0.57 |
| miR-7 | 1.99 (2.21; =/−0.57) | 1.0 (1.14; +/−0.5) | p = 0.57 |
| miR-1664 | 0.5 (0.41; +/−0.29) | 1.1 (1.24; +/−1.86) | p = 0.016 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sajewicz-Krukowska, J.; Mirosław, P.; Jastrzębski, J.P.; Domańska-Blicharz, K.; Tarasiuk, K.; Marzec-Kotarska, B. miRNA Expression Signatures Induced by Chicken Astrovirus Infection in Chickens. Int. J. Mol. Sci. 2023, 24, 15128. https://doi.org/10.3390/ijms242015128
Sajewicz-Krukowska J, Mirosław P, Jastrzębski JP, Domańska-Blicharz K, Tarasiuk K, Marzec-Kotarska B. miRNA Expression Signatures Induced by Chicken Astrovirus Infection in Chickens. International Journal of Molecular Sciences. 2023; 24(20):15128. https://doi.org/10.3390/ijms242015128
Chicago/Turabian StyleSajewicz-Krukowska, Joanna, Paweł Mirosław, Jan P. Jastrzębski, Katarzyna Domańska-Blicharz, Karolina Tarasiuk, and Barbara Marzec-Kotarska. 2023. "miRNA Expression Signatures Induced by Chicken Astrovirus Infection in Chickens" International Journal of Molecular Sciences 24, no. 20: 15128. https://doi.org/10.3390/ijms242015128
APA StyleSajewicz-Krukowska, J., Mirosław, P., Jastrzębski, J. P., Domańska-Blicharz, K., Tarasiuk, K., & Marzec-Kotarska, B. (2023). miRNA Expression Signatures Induced by Chicken Astrovirus Infection in Chickens. International Journal of Molecular Sciences, 24(20), 15128. https://doi.org/10.3390/ijms242015128







