QTLs and Candidate Loci Associated with Drought Tolerance Traits of Kaybonnet x ZHE733 Recombinant Inbred Lines Rice Population
Abstract
:1. Introduction
2. Results
2.1. Analysis of Parents K/Z RIL Population–Drought Resistant and Drought Sensitive
2.2. Variation in Morphological Traits of K/Z RILs under Drought Stress Conditions
2.3. Genetic Variation for Grain Yield Components under Reproductive-Stage Drought Stress
2.4. Variation in Root Architectural Traits under ABA Conditions
2.5. Correlation of Morphological Traits and Grain Yield Components under WW and DS Conditions with Root Architectural Traits under ABA Conditions
2.6. Relationships of Root Anatomy and Drought Resistance in Tolerant and Sensitive Lines
2.7. High-Density Genetic Linkage Map with GBS Markers
2.8. QTL Mapping of Morphological and Yield Traits under Reproductive-Stage Drought Stress Conditions and Root Architectural Traits under ABA Conditions
2.9. Candidate Genes Underlying QTL Regions
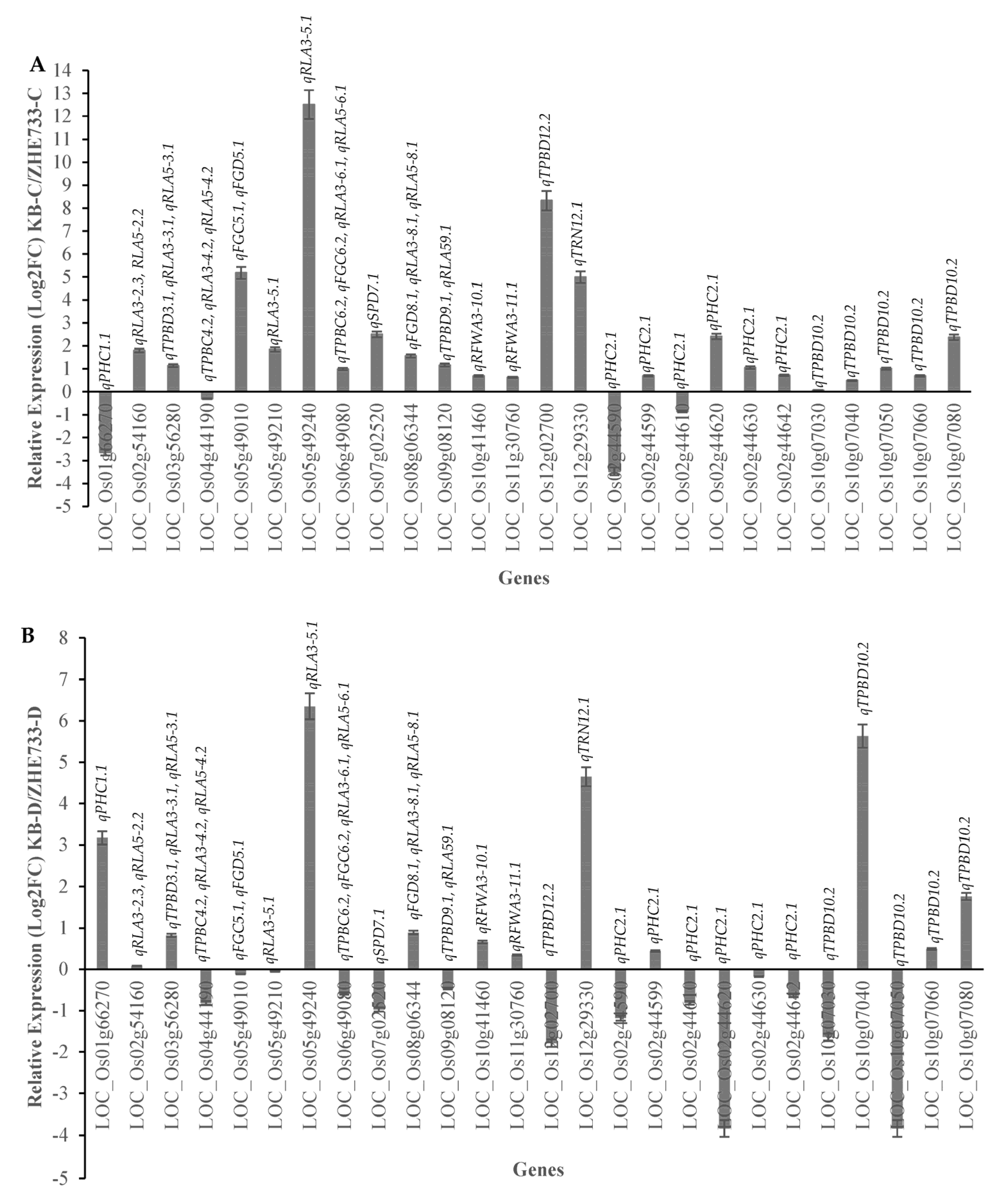
2.10. RT-qPCR Validation of the Key Functional Genes Identified within the QTL Regions Regulating Drought-Related Traits and ABA Sensitivity
2.11. Genetic Diversity in 26 Loci across K/Z RIL Population
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Drought Stress Treatment at the Reproductive Stage
4.3. Screening for ABA Sensitivity
4.4. Measurements of Root Anatomical Phenes
4.5. Genotyping
4.6. SNP Identification
4.7. Linkage Map Construction and QTL Mapping
4.8. Identification of Candidate Genes within the QTL Regions
4.9. RT-qPCR Validation of the Key Functional Genes Identified within the QTL Regions Regulating Drought-Related Traits and ABA Sensitivity
4.10. Analysis of Genetic Diversity in the K/Z RIL Population
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edwards, D.; Batley, J. Plant genome sequencing: Applications for crop improvement. Plant Biotech. J. 2010, 8, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the UN (FAO). FAOSTAT Database. 2018. Available online: http://www.fao.org/faostat/en/#data (accessed on 2 February 2023).
- Quick Stats, USDA-NASS. State Agriculture Overview: Arkansas. 2016. Available online: https://www.nass.usda.gov/Quick_Stats/Ag_Overview/stateOverview.php?state=ARKANSAS (accessed on 7 February 2023).
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef]
- Tuong, T.P.; Bouman, B.A.M. Rice production in water-scarce environments. In Water Productivity in Agriculture: Limits and Opportunities for Improvement; Kijne, J.W., Barker, R., Molden, D., Eds.; CABI Publishing: Wallingford, UK, 2003; pp. 53–67. [Google Scholar]
- Farooq, M.; Basra, S.M.A.; Wahid, A.; Ahmad, N.; Saleem, B.A. Improving the drought tolerance in rice (Oryza sativa L.) by exogenous application of salicylic acid. J. Agron. Crop Sci. 2009, 195, 237–246. [Google Scholar] [CrossRef]
- Rahimi, J.; Ebrahimpour, M.; Khalili, A. Spatial changes of extended De Martonne climatic zones affected by climate change in Iran. Theor. Appl. Climatol. 2013, 112, 409–418. [Google Scholar] [CrossRef]
- Zhao, T.; Dai, A. Uncertainties in historical changes and future projections of drought. Part II: Model-simulated historical and future drought changes. Clim. Chang. 2017, 144, 535–548. [Google Scholar] [CrossRef]
- Ito, O.; O’Toole, J.C.; Hardy, B. Genetic Improvement of Rice for Water-Limited Environments; International Rice Research Institute: Los Baños, Philippines, 1999; 353p. [Google Scholar]
- Bunnag, S.; Pongthai, P. Selection of rice (Oryza sativa L.) cultivars tolerant to drought stress at the vegetative stage under field conditions. Am. J. Plant Sci. 2013, 4, 1701–1708. [Google Scholar] [CrossRef]
- Hsiao, T.C. The soil-plant-atmosphere continuum in relation to drought and crop production. In Drought Resistance in Crops, with Emphasis on Rice; International Rice Research Institute: Los Baños, Philippines, 1982; pp. 39–52. [Google Scholar]
- O’Toole, J.C. Adaptation of rice to drought-prone environments. In Drought Resistance in Crops with Emphasis on Rice; International Rice Research Institute: Los Baños, Philippines, 1982; pp. 195–213. [Google Scholar]
- Sadeghi, S.M.; Danesh, R.K. Effects of water deficit role at different stages of reproductive growth on yield components of rice. World Appl. Sci. J. 2011, 13, 2021–2026. [Google Scholar]
- Bouman, B.A.M.; Hengsdijk, H.; Hardy, B.; Bindraban, P.S.; Tuong, T.P.; Ladha, J.K. Water-wise rice production. In Proceedings of the International Workshop on Water-wise Rice Production, Los Baños, Philippines, 8–11 April 2002; International Rice Research Institute: Los Baños, Philippines, 2002; 356p. [Google Scholar]
- Dixit, S.; Singh, A.; Cruz, M.; Maturan, P.T.; Amante, M.; Kumar, A. Multiple major QTL lead to stable yield performance of rice cultivars across varying drought intensities. BMC Genet. 2014, 15, 16. [Google Scholar] [CrossRef]
- Venuprasad, R.; Dalid, C.O.; Del Valle, M.; Zhao, D.; Espiritu, M.; Sta Cruz, M.T. Identification and characterization of large-effect quantitative trait loci for grain yield under lowland drought stress in rice using bulk-segregant analysis. Theor. Appl. Genet. 2009, 120, 177–190. [Google Scholar] [CrossRef]
- Kumar, A.; Thomas, J.; Yingling, S.; Dwiningsih, Y.; Ramegowda, V.; Gaspar, J.; Henry, C.; Counce, P.; Siebenmorgen, T.J.; Moldenhauer, K.A.K.; et al. Screening of diverse rice cultivars for heat tolerance and grain quality under high nighttime temperature. BR Wells Rice Res. Stud.-Ark. Agric. Exp. Stn. Univ. Ark. System. 2017, 643, 61–67. [Google Scholar]
- Zhu, X.; Shen, W.; Huang, J.; Zhang, T.; Zhang, X.; Cui, Y.; Sang, X.; Ling, Y.; Li, Y.; Wang, N.; et al. Mutation of the OsSAC1 gene, which encodes an endoplasmic reticulum protein with an unknown function, causes sugar accumulation in rice leaves. Plant Cell Physiol. 2018, 59, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Dwiningsih, Y.; Kumar, A.; Thomas, J.; Ruiz, C.; Alkahtani, J.; Baisakh, N.; Pereira, A. Quantitative trait loci and candidate gene identification for chlorophyll content in RIL rice population under drought conditions. Indones. J. Nat. Pigment. 2021, 3, 54–64. [Google Scholar] [CrossRef]
- McCouch, S.R.; Zhao, K.; Wright, M.; Tung, C.W.; Ebana, K.; Thomson, M. Development of genome-wide SNP assays for rice. Breed. Sci. 2010, 60, 524–535. [Google Scholar] [CrossRef]
- Dwiningsih, Y. Molecular Genetic Analysis of Drought Resistance and Productivity Traits of Rice Genotypes; University of Arkansas: Fayetteville, AR, USA, 2020. [Google Scholar]
- Thomson, M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotech. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Mardani, Z.; Rabiei, B.; Sabouri, H.; Sabouri, A. Mapping of QTLs for germination characteristics under non-stress and drought stress in rice. Rice Sci. 2013, 20, 391–399. [Google Scholar] [CrossRef]
- Barnabás, B.; Jager, K.; Feher, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef]
- Lilley, J.M.; Ludlow, M.M.; McCouch, S.R.; O’Toole, J.C. Locating QTLs for osmotic adjustment and dehydration tolerance in rice. J. Exp. Bot. 1996, 47, 1427–1436. [Google Scholar] [CrossRef]
- Courtois, B.; McLaren, G.; Sinha, P.K.; Prasad, K.; Yadav, R.; Shen, L. Mapping QTL associated with drought avoidance in upland rice. Mol. Breed. 2000, 6, 55–66. [Google Scholar] [CrossRef]
- Price, A.H.; Young, E.M.; Tomos, A.D. Quantitative trait loci associated with stomatal conductance, leaf rolling and heading date mapped in upland rice (Oryza sativa L.). New Phytol. 1997, 137, 83–91. [Google Scholar] [CrossRef]
- Price, A.; Steele, K.; Moore, B.; Jones, R. Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes: II. Mapping quantitative trait loci for root morphology and distribution. Field Crop Res. 2002, 76, 25–43. [Google Scholar] [CrossRef]
- Kanbar, A.; Shashidhar, H.E.; Hittalmani, S. Mapping of QTL associated with root and related traits in DH population of rice (Oryza sativa L). Indian J. Genet. 2002, 62, 287–290. [Google Scholar]
- Xu, Y.; This, D.; Pausch, R.; Vonhof, W.; Coburn, J.; Comstock, J. Leaf-level water use efficiency determined by carbon isotope discrimination in rice seedlings: Genetic variation associated with population structure and QTL mapping. Theor. Appl. Genet. 2009, 118, 1065–1081. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.M.; Boopathi, N.M.; Kumar, S.; Ramasubramanian, T.; Chengsong, Z.; Jeyaprakash, P.; Senthil, A.; Babu, R.C. Molecular mapping and location of QTLs for drought-resistance traits in indica rice (Oryza sativa L.) lines adapted to target environments. Acta Physiol. Plant 2010, 32, 355–364. [Google Scholar] [CrossRef]
- Rabello, A.R.; Guimarães, C.M.; Rangel, P.H.N.; da Silva, F.R.; Seixas, D.; de Souza, E. Identification of drought-responsive genes in roots of upland rice (Oryza sativa L.). BMC Genom. 2008, 9, 485. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.H.; Fu, B.Y.; Xu, H.X.; Li, Y.S. Proteomic analysis of PEG-simulated drought stress responsive proteins of rice leaves using a pyramiding rice line at the seedling stage. Bot. Stud. 2010, 51, 137–145. [Google Scholar]
- Sahoo, K.K.; Tripathi, A.K.; Pareek, A.; Singla-Pareek, S.L. Taming drought stress in rice through genetic engineering of transcription factors and protein kinases. Plant Stress. 2013, 7, 60–72. [Google Scholar]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef]
- Schroeder, J.I.; Kwak, J.M.; Allen, G.J. Guard cell abscisic acid signaling and engineering drought hardiness in plants. Nature 2001, 410, 327–330. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, 15–45. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, 165–183. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y. The role of ABF family bZIP class transcription factors in stress response. Physiol. Plant 2006, 126, 519–527. [Google Scholar] [CrossRef]
- Berri, S.; Abbruscato, P.; Faivre-Rampant, O.; Brasileiro, A.C.; Fumasoni, I.; Satoh, K. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Brown, K.M.; Lynch, J.P. Root cortical aerenchyma improves the drought tolerance of maize (Zea mays L.). Plant Cell Environ. 2010, 33, 740–749. [Google Scholar] [CrossRef]
- Kumar, A.; Yingling, S.; Thomas, J.; Ruiz, C.; Dwiningsih, Y.; Gupta, C.; Couce, P.; Siebenmorgen, T.J.; Moldenhauer, K.A.K.; Pereira, A. Screening of Diverse Japonica Rice Genotypes for Grain Yield and Quality under High Nightime Temperature; B.R. Wells Rice Research Studies; Arkansas Agricultural Experiment Station, University of Arkansas System: Fayetteville, AR, USA, 2018; pp. 50–56. [Google Scholar]
- Islam, A.S.F.; Ali, M.R.; Gregorio, G.B.; Islam, M.R. Genetic diversity analysis of stress tolerant rice (Oryza sativa L.). Afr. J. Biotech. 2012, 11, 15123–15129. [Google Scholar]
- Fang, N.; Wei, X.; Shen, L.; Yu, Y.; Li, M.; Yin, C. Fine mapping of a panicle blast resistance gene Pb-bd1 in Japonica landrace Bodao and its application in rice breeding. Rice 2019, 12, 18. [Google Scholar] [CrossRef]
- Gibert, A.; Gray, E.F.; Westoby, M.; Wright, I.; Falster, D.S. Data from: On the Link between Functional Traits and Growth Rate: Meta-Analysis Shows Effects Change with Plant Size, as Predicted; Dryad Digital Repository: Durham, NC, USA, 2016. [Google Scholar] [CrossRef]
- De Souza, L.M.; Le Guen, V.; Cerqueira-Silva, C.B.M.; Silva, C.C.; Mantello, C.C.; Conson, A.R.O. Genetic diversity strategy for the management and use of rubber genetic resources: More than 1,000 wild and cultivated accessions in a 100-genotype core collection. PLoS ONE 2015, 10, e0134607. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Bhattarai, U.; Subudhi, P.K. Identification of drought responsive QTLs during vegetative growth stage of rice using a saturated GBS-based SNP linkage map. Euphytica 2018, 214, 38. [Google Scholar] [CrossRef]
- Sabar, M.; Shabir, G.; Shah, S.M.; Aslam, K.; Naveed, S.A.; Arif, M. Identification and mapping of QTLs associated with drought tolerance traits in rice by a cross between Super Basmati and IR55419-04. Breed. Sci. 2019, 69, 169–178. [Google Scholar] [CrossRef]
- Barik, S.R.; Pandit, E.; Pradhan, S.K.; Mohanty, S.P.; Mohapatra, T. Genetic mapping of morpho-physiological traits involved during reproductive stage drought tolerance in rice. PLoS ONE 2019, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Melandri, G.; Prashar, A.; McCouch, S.R.; Linden, G.; Jones, H.G.; Kadam, N. Association mapping and genetic dissection of drought-induced canopy temperature differences in rice. J. Exp. Bot. 2020, 71, 1614–1627. [Google Scholar] [CrossRef] [PubMed]
- Barik, S.R.; Pandit, E.; Mohanty, S.P.; Nayak, D.K.; Pradhan, S.K. Genetic mapping of physiological traits associated with terminal stage drought tolerance in rice. BMC Genet. 2020, 21, 76. [Google Scholar] [CrossRef]
- Dwiningsih, Y.; Kumar, A.; Thomas, J.; Ruiz, C.; Alkahtani, J.; Al-hashimi, A.; Pereira, A. Identification of genomic regions controlling chalkiness and grain characteristics in a recombinant inbred line rice population based on high-throughput SNP markers. Genes 2021, 12, 1690. [Google Scholar] [CrossRef]
- Studer, A.J.; Doebley, J.F. Do large effect QTL fractionate? A case study at the maize domestication QTL teosinte branched1. Genetics 2011, 188, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. Regulation of Apetala2/ethylene response factors in plants. Front. Plant Sci. 2017, 8, 150. [Google Scholar] [CrossRef]
- Abiri, R.; Shaharuddin, N.A.; Maziah, M.; Norhana, Z.; Yusof, B.; Atabaki, N. Role of ethylene and the APETALA 2/ethylene response factor superfamily in rice under various abiotic and biotic stress conditions. Environ. Exp. Bot. 2017, 134, 33–34. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Chen, H.; Chen, S.Y.; Zhang, J.S. Role of ethylene in plant growth and responses to stresses. In Phytohormones: A Window to Metabolism Signaling and Biotechnological Applications; Springer Science + Business Media: New York, NY, USA, 2014; pp. 81–118. [Google Scholar]
- Nan, N.; Wang, J.; Shi, Y.; Qian, Y.; Jiang, L.; Huang, S. Rice plastidial NAD-dependent malate dehydrogenase 1 negatively regulates salt stress response by reducing the vitamin B6 content. Plant Biotechnol. J. 2020, 18, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Sasi, S.; Venkatesh, J.; Daneshi, R.F.; Gururani, M.A. Photosystem II extrinsic proteins and their putative role in abiotic stress tolerance in higher plants. Plants 2018, 7, 100. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Li, C.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Huang, X.; Chao, D.; Gao, J.; Zhu, M.; Shi, M.; Lin, H. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes. Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef]
- Ciftci-Yilmaz, S.; Morsy, M.R.; Song, L.; Coutu, A.; Krizek, B.A.; Lewis, M.W. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar]
- Mukhopadhyay, A.; Vij, S.; Tyagi, A.K. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. USA 2004, 101, 6309–6314. [Google Scholar] [CrossRef]
- Sakamoto, H.; Maruyama, K.; Sakuma, Y.; Meshi, T.; Iwabuchi, M.; Shinozaki, K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004, 136, 2734–2746. [Google Scholar] [CrossRef]
- Nawaz, G.; Kang, H. Rice OsRH58, a chloroplast DEAD-box RNA helicase, improves salt or drought stress tolerance in Arabidopsis by affecting chloroplast translation. BMC Plant Biol. 2019, 19, 17. [Google Scholar] [CrossRef]
- Vashisht, A.A.; Tuteja, N. Stress responsive DEAD-box helicases: A new pathway to engineer plant stress tolerance. J. Photochem. Photobiol. 2006, 84, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Macovi, A.; Vaid, N.; Tula, S.; Tuteja, N. A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal Behav. 2012, 7, 1138–1143. [Google Scholar] [CrossRef]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 2007, 115, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Cai, W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 2012, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 19, 6. [Google Scholar] [CrossRef]
- Liang, Y.; Kang, K.; Gan, L.; Ning, S.; Xiong, J.; Song, S. Drought-responsive genes, late embryogenesis abundant group3 (LEA3) and vicinal oxygen chelate, function in lipid accumulation in Brassica napus and Arabidopsis mainly via enhancing photosynthetic efficiency and reducing ROS. Plant Biotechnol. J. 2019, 17, 2123–2142. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C. The role of the Late Embryogenesis-Abundant (LEA) protein family in development and the abiotic stress response: A comprehensive expression analysis of Potato (Solanum Tuberosum). Genes 2019, 10, 148. [Google Scholar] [CrossRef]
- Kamarudin, Z.S.; Yusop, M.R.; Ismail, M.R.; Mohamed, M.; Harun, A.R.; Yusuff, O. LEA gene expression assessment in advanced mutant rice genotypes under drought stress. Int. J. Genom. 2019, 2019, 8406036. [Google Scholar] [CrossRef]
- Hu, L.; He, H.; Zhu, C.; Peng, X.; Fu, J.; He, X. Genome-wide identification and phylogenetic analysis of the chalcone synthase gene family in rice. J. Plant Res. 2017, 130, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2013, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Yao, H.; Peng, X.; Wang, R.; Li, F.; Wang, Z. Overexpression of Chalcone synthase improves flavonoid accumulation and drought tolerance in Tobacco. Preprints 2019, 2019060103. [Google Scholar] [CrossRef]
- Jiang, S.; Ramachandran, S. Identification and molecular characterization of myosin gene family in Oryza sativa genome. Plant Cell Physiol. 2004, 5, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Paszkowski, J. Regulation of rice root development by a retrotransposon acting as a microRNA sponge. eLife 2017, 6, 30038. [Google Scholar] [CrossRef]
- Greub, C.P. A Morpho-Physiological Analysis of Diverse Drought Resistant and Sensitive Rice Genotypes to Identify Distinguishing Anatomical Root Phenes. Master’s Thesis, University of Arkansas, Fayetteville, AR, USA, 2015; p. 1281. [Google Scholar]
- Huang, L.; Zhang, F.; Zhang, F.; Wang, W.; Zhou, Y.; Fu, B. Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genom. 2014, 15, 1026. [Google Scholar] [CrossRef]
- Hsiao, T.C. Rapid changes in levels of polyribosomes in Zea mays in response to water stress. PI Physiol. 1970, 46, 281–285. [Google Scholar] [CrossRef]
- Specht, J.E.; Chase, K.; Macrander, M.; Graef, G.L.; Chung, J.; Markwell, J.P. Soybean response to water: A QTL analysis of drought tolerance. Crop Sci. 2001, 41, 493–509. [Google Scholar] [CrossRef]
- Farooq, M.; Siddique, K.H.M.; Rehman, H.; Aziz, T.; Lee, D.J. Rice direct seeding: Experiences, challenges and opportunities. Soil. Till Res. 2011, 111, 87–98. [Google Scholar] [CrossRef]
- Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 2015, 18, 684. [Google Scholar] [CrossRef]
- Kumar, A.; Yingling, S.; Ruiz, C.; Dwiningsih, Y.; Gupta, C.; Thomas, J.; Counce, P.; Moldenhauer, K.A.K.; Siebenmorgen, T.J.; Pereira, A. Screening of Indica and Japonica Rice Subspecies for Grain Yield and Quality under High Nightime Temperature; Research Series—Arkansas Agricultural Experiment Station; Arkansas Agricultural Experiment Station, University of Arkansas System: Fayetteville, AR, USA, 2019; pp. 61–66. [Google Scholar]
- Mantovani, A.; Iglesias, R.R. Factors limiting seed germination of terrestrial bromeliads in the sandy coastal plains (Restinga) of Maricá, Rio de Janeiro, Brazil. Rodriguésia 2008, 59, 135–150. [Google Scholar] [CrossRef]
- Islam, M.T.; Salam, M.A. Growth and yield performance of some rice genotypes under different soil moisture regimes. Bangladesh J. Trg. Dev. 1994, 7, 57–62. [Google Scholar]
- Manickavelu, A.; Nadarajan, N.; Ganesh, S.K.; Gnanamalar, R.P.; Chandra Babu, R. Drought tolerance in rice: Morphological and molecular genetic consideration. Plant Growth Regul. 2006, 50, 121–138. [Google Scholar] [CrossRef]
- Sarvestani, Z.T.; Pirdashti, H.; Sanavy, S.A.; Balouchi, H. Study of water stress effects in different growth stages on yield and yield components of different rice (Oryza sativa L.) cultivars. Pak. J. Biol. Sci. 2008, 11, 1303–1309. [Google Scholar] [PubMed]
- Mostajeran, A.; Rahimi-Eichi, V. Effects of drought stress on growth and yield of rice (Oryza sativa L.) cultivars and accumulation of proline and soluble sugars in sheath and blades of their different ages leaves. Am. Eur. J. Agric. Environ. Sci. 2009, 5, 264–272. [Google Scholar]
- Ji, K.; Wang, Y.; Sun, W.; Lou, Q.; Mei, H.; Shen, S.; Chen, H. Drought-responsive mechanisms in rice genotypes with contrasting drought tolerance during reproductive stage. J. Plant Physiol. 2012, 169, 336–344. [Google Scholar] [CrossRef]
- Ashfaq, M.; Haider, M.S.; Khan, A.S.; Allah, S.U. Breeding potential of the basmati rice germplasm under water stress condition. Afr. J. Biotechnol. 2012, 11, 6647–6657. [Google Scholar]
- Sokoto, M.B.; Muhammad, A. Response of rice varieties to water stress in Sokoto, Sudan Savannah, Nigeria. J. Biosci. Med. 2014, 2, 68–74. [Google Scholar] [CrossRef]
- Kamoshita, A.; Babu, R.C.; Boopathi, N.M.; Fukai, S. Phenotypic and genotypic analysis of drought-resistance traits for development of rice cultivars adapted to rainfed environments. Field Crop Res. 2008, 109, 1–23. [Google Scholar] [CrossRef]
- Palanog, A.D.; Swamy, B.P.M.; Shamsudin, N.A.A. Grain yield QTLs with consistent-effect under reproductive stage drought stress in rice. Field Crops Res. 2014, 161, 46–54. [Google Scholar] [CrossRef]
- Dwiningsih, Y.; Rahmaningsih, M.; Alkahtani, J. Development of single nucleotide polymorphism (SNP) markers in tropical crops. Adv. Sustain. Sci. Eng. Technol. 2020, 2, 2. [Google Scholar] [CrossRef]
- Venuprasad, R.; Impa, S.M.; Veeresh Gowda, R.P.; Atlin, G.N.; Serraj, R. Rice near-isogenic-lines (NILs) contrasting for grain yield under lowland drought stress. Field Crops Res. 2011, 123, 38–46. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef]
- Duan, J.; Li, J.; Guo, S.; Kang, Y. Exogenous spermidine affects polyamine metabolism in salinity-stressed Cucumis sativus roots and enhances short-term salinity tolerance. J. Plant Physiol. 2008, 165, 1620–1635. [Google Scholar] [CrossRef]
- Todorov, D.; Alexieva, V.V.; Karanov, E. Effect of putrescine, 4-PU-30, and abscisic acid on maize plants grown under normal, drought, and re-watering conditions. J. Plant Growth Regul. 1998, 17, 197–203. [Google Scholar] [CrossRef]
- Kanbar, A.; Toorchi, M.; Shashidhar, H.E. Relationship between root and yield morphological characters in rainfed low land rice (Oryza sativa L.). Cereal Res. Commun. 2009, 37, 261–268. [Google Scholar] [CrossRef]
- Chimungu, J.G.; Brown, K.M.; Lynch, J.P. Large root cortical cell size improves drought tolerance in maize. Plant Physiol. 2014, 166, 2166–2178. [Google Scholar] [CrossRef]
- Chimungu, J.G.; Brown, K.M.; Lynch, J.P. Reduced root cortical cell file number improves drought tolerance in maize. Plant Physiol. 2014, 166, 1943–1955. [Google Scholar] [CrossRef]
- Chimungu, J.G.; Loades, K.W.; Lynch, J.P. Root anatomical phenes predict root penetration ability and biomechanical properties in maize (Zea mays). J. Exp. Bot. 2015, 66, 3151–3162. [Google Scholar] [CrossRef]
- Chimungu, J.G.; Maliro, M.; Nalivata, P.; Kanyama-Phiri, G.; Brown, K.; Lynch, J. Utility of root cortical aerenchyma under water limited conditions in tropical maize. Field Crops Res. 2015, 171, 86–98. [Google Scholar] [CrossRef]
- Kadam, N.N.; Yin, X.; Bindraban, P.S.; Struik, P.C.; Jagadish, K.S. Does morphological and anatomical plasticity during the vegetative stage make wheat more tolerant of water deficit stress than rice? Plant Physiol. 2015, 167, 1389–1401. [Google Scholar] [CrossRef]
- Prince, S.J.; Murphy, M.; Mutava, R.N.; Durnell, L.A.; Valliyodan, B.; Shannon, J.G.; Nguyen, H.T. Root xylem plasticity to improve water use and yield in water-stressed soybean. J. Exp. Bot. 2017, 68, 2027–2036. [Google Scholar] [CrossRef]
- Peña-Valdivia, C.B.; Sánchez-Urdaneta, A.B.; Rangel, J.M.; Muñoz, J.J.; García-Nava, R.; Velázquez, R.C. Anatomical root variations in response to water deficit: Wild and domesticated common bean. Biol. Res. 2010, 43, 417–427. [Google Scholar] [CrossRef]
- Purushothaman, R.; Zaman-Allah, M.; Mallikarjuna, N.; Pannirselvam, R.; Krishnamurthy, L.; Gowda, C.L.L. Root anatomical traits and their possible contribution to drought tolerance in grain legumes. Plant Prod. Sci. 2013, 16, 1–8. [Google Scholar] [CrossRef]
- Lynch, J.P.; Ho, M.D. Rhizoeconomics: Carbon costs of phosphorus acquisition. Plant Soil. 2005, 269, 45–56. [Google Scholar] [CrossRef]
- Palta, J.A.; Gregory, P.J. Drought affects the fluxes of carbon to roots and soil in 13C pulse-labelled plants of wheat. Soil Biol. Biochem. 1997, 29, 1395–1403. [Google Scholar] [CrossRef]
- Sharp, R.E.; Davies, W.J. Solute regulation and growth by roots and shoots of water-stressed maize plants. Planta 1979, 147, 43–49. [Google Scholar] [CrossRef]
- Jaramillo, R.E.; Nord, E.A.; Chimungu, J.G.; Brown, K.M.; Lynch, J.P. Root cortical burden influences drought tolerance in maize. Ann. Bot. 2013, 112, 429–437. [Google Scholar] [CrossRef]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Steudle, E.; Peterson, C.A. How does water get through roots? J. Exp. Bot. 1998, 49, 775–778. [Google Scholar] [CrossRef]
- Richards, R.A.; Passioura, J.B. A breeding program to reduce the diameter of the major xylem vessel in the seminal roots of wheat and its effect on grain yield in rain-fed environments. Aust. J. Agric. Res. 1989, 40, 943–950. [Google Scholar] [CrossRef]
- Evans, D.E. Aerenchyma formation. New Phytol. 2004, 161, 35–49. [Google Scholar] [CrossRef]
- Rieger, M.; Litvin, P. Root system hydraulic conductivity in species with contrasting root anatomy. J. Exp. Bot. 1999, 50, 201–209. [Google Scholar] [CrossRef]
- Baisakh, N.; Yabes, J.; Gutierrez, A.; Mangu, V.; Ma, P.; Famoso, A. Genetic mapping identifies consistent quantitative trait loci for yield traits of rice under greenhouse drought conditions. Genes 2020, 11, 62. [Google Scholar] [CrossRef]
- Mu, P.; Li, Z.; Li, C. QTL mapping of the root traits and their correlation analysis with drought resistance using DH lines from paddy and upland rice cross. Chin. Sci. Bull. 2003, 48, 2718–2724. [Google Scholar] [CrossRef]
- Courtois, B.; Shen, L.; Petalcorin, W.; Carandang, S.; Mauleon, R.; Li, Z. Locating QTLs controlling constitutive root traits in the rice population IAC 165 × Co39. Euphytica 2003, 3, 335–345. [Google Scholar] [CrossRef]
- Lanceras, J.C.; Pantuwan, G.; Jongdee, B.; Toojinda, T. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol. 2004, 135, 384–399. [Google Scholar] [CrossRef]
- Bernier, J.; Kumar, A.; Ramaiah, V.; Spaner, D.; Atlin, G. A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci. 2007, 2, 507–518. [Google Scholar] [CrossRef]
- Bernier, J.; Serraj, R.; Kumar, A. The large-effect drought resistance QTL qtl12.1 increases water uptake in upland rice. Field Crops Res. 2015, 2, 139–146. [Google Scholar] [CrossRef]
- Mishra, K.K.; Vikram, P.; Yadaw, R.B. QDTY12.1: A locus with a consistent effect on grain yield under drought in rice. BMC Genet. 2013, 14, 12. [Google Scholar] [CrossRef]
- Yadaw, R.B.; Dixit, S.; Raman, A.; Mishra, K.K.; Vikram, P.; Swamy, B.P.M. A QTL for high grain yield under lowland drought in the background of popular rice variety Sabitri from Nepal. Field Crops Res. 2013, 144, 281–287. [Google Scholar] [CrossRef]
- Zhou, L.; Lie, S.; Wu, W.; Chen, D.; Zhan, X.; Zhu, A.; Zhang, Y.; Cheng, S.; Cao, L.; Lou, X.; et al. Dissection of genetic architecture of rice plant height and heading date by multiple-strategy-based association studies. Sci. Rep. 2016, 6, 29718. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Jian, S.U.N.; Li, C.X.; Liu, H.L.; Wang, J.G.; Zhao, H.W.; Zou, D.T. Genetic dissection of the developmental behavior of plant height in rice under different water supply conditions. J. Integr. Agric. 2016, 12, 2688–2702. [Google Scholar] [CrossRef]
- Yadav, S.; Sandhu, N.; Singh, V.K.; Catolos, M.; Kumar, A. Genotyping-by-sequencing based QTL mapping for rice grain yield under reproductive stage drought stress tolerance. Sci. Rep. 2019, 9, 14326. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Chen, Y.; Ji, Z.; Liang, Y.; Zheng, A.; Wen, Z. Control of plant height by 24 alleles at 12 quantitative trait loci in rice. Crop Breed. Appl. Biotechnol. 2019, 19, 200–207. [Google Scholar] [CrossRef]
- Xu, P.; Yang, J.; Ma, Z.; Yu, D.; Zhou, J.; Tao, D. Identification and validation of aerobic adaptation QTLs in upland rice. Life 2020, 10, 65. [Google Scholar] [CrossRef]
- Yadav, S.; Sandhu, N.; Majumder, R.R.; Dixit, S.; Kumar, S.; Singh, S.P. Epistatic interactions of major effect drought QTLs with genetic background loci determine grain yield of rice under drought stress. Sci. Rep. 2019, 9, 2616. [Google Scholar] [CrossRef]
- Takahashi, Y.; Shomura, A.; Sasaki, T.; Yano, M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 2001, 98, 7922–7927. [Google Scholar] [CrossRef]
- Xu, J.; Li, J.Z.; Zheng, X.W.; Zou, L.X. QTL mapping of the root traits in rice. Acta Genet. Sin. 2001, 28, 433–438. [Google Scholar]
- Lou, Q.; Chen, L.; Mei, H.; Wei, H.; Feng, F.; Wang, P. Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice. J. Exp. Bot. 2015, 66, 4749–4757. [Google Scholar] [CrossRef]
- Kitomi, Y.; Kanno, N.; Kawai, S.; Mizubayashi, T.; Fukuoka, S.; Uga, Y. QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1. Rice 2015, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Gimhani, D.R.; Abhayawickrama, B.P.; Saliha, M.S.F.; Kottearachchi, N.S. Mapping and validation of quantitative trait loci for root architectural traits in rice (Oryza sativa) under non-stress and salinity stress conditions. Trop. Agric. Res. 2018, 3, 241–254. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bansal, K.C.; Aggarwal, P.K.; Datta, S.K.; Craufurd, P.Q. Agricultural biotechnology for crop improvement in a variable climate: Hope or hype? Trends Plant Sci. 2011, 16, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Vikram, P.; Swamy, B.P.M.; Dixit, S.; Singh, R.; Singh, B.P.; Miro, B. Drought susceptibility of modern rice varieties: An effect of linkage of drought tolerance with undesirable traits. Sci. Rep. 2015, 5, 14799. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Courtois, B.; Huang, N.; McLaren, G. Mapping genes controlling root morphology and root distribution in a double-haploid population of rice. Theor. Appl. Genet. 1997, 94, 619–632. [Google Scholar] [CrossRef]
- Pan, Y.; Seymour, G.B.; Lu, C. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012, 31, 349–360. [Google Scholar] [CrossRef]
- Agrawal, L.; Gupta, S.; Mishra, S.K.; Pandey, G.; Kumar, S.; Chauhan, P.S. Elucidation of complex nature of PEG induced drought-stress response in rice root using comparative proteomics approach. Front. Plant Sci. 2016, 7, 1466. [Google Scholar] [CrossRef]
- Guo, R.; Shi, L.X.; Jiao, Y.; Li, M.X.; Zhong, X.L.; Gu, F.X. Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB Plants 2018, 10, ply016. [Google Scholar] [CrossRef]
- Lu, Y. Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front. Plant Sci. 2016, 7, 168. [Google Scholar] [CrossRef]
- Shen, H.; Liu, C.; Zhang, Y.; Meng, X. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 2012, 80, 241–253. [Google Scholar] [CrossRef]
- Yan, Y.; Jia, H.; Wang, F.; Wang, C.; Shuchang, L.; Xingqi, G. Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to Rhizoctonia solani infection in transgenic Nicotiana benthamiana. Front. Physiol. 2015, 6, 265. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, 92913. [Google Scholar] [CrossRef]
- Kikuchi, S.; Satoh, K.; Nagata, T.; Kawagashira, N.; Doi, K.; Kishimoto, N. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 2003, 301, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K. The abiotic stress responsive NAC-type transcription factor OsNAC5 regulates stress inducible genes and stress tolerance in rice. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef]
- Shim, J.S.; Oh, N.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 2018, 9, 310. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Mandal, S.N.; Parida, S.K.; Prasad, M. Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS ONE 2013, 8, 64594. [Google Scholar] [CrossRef]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M. Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 2009, 229, 1065–1075. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Bioph Res. Commun. 2009, 379, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.; Choi, Y.D. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Hayano-Kanashiro, C.; Calderon-Vazquez Ibarra-Laclette, E.; Herrera-Estrella, L.; Simpson, J. Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS ONE 2009, 4, 10. [Google Scholar] [CrossRef]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Dai, X.; Xu, Y.; Guo, J.; Liu, Y.; Chen, N. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009, 150, 244–256. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Quan, R.; Hu, S.; Zhang, Z.; Zhang, H.; Zhang, Z.; Huang, R. Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 2010, 8, 476–488. [Google Scholar] [CrossRef]
- IRRI. SES (Standard Evaluation System for Rice). International Network for Genetic Evaluation of Rice; Genetic Resource Center: Los Baños, Philippines, 2013. [Google Scholar]
- Burton, A.L.; Williams, M.; Lynch, J.P.; Brown, K.M. RootScan: Software for high-throughput analysis of root anatomical traits. Plant Soil. 2012, 357, 189–203. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in bi-parental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943, 2, 172–175. [Google Scholar] [CrossRef]
- Solis, J.; Gutierrez, A.; Mangu, V.; Sanchez, E.; Bedre, R.; Linscombe, S. Genetic mapping of quantitative trait loci for grain yield under drought in rice under controlled greenhouse conditions. Front. Chem. 2018, 5, 129. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R. Gene nomenclature system for rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef]
- Ramegowda, V.; Basu, S.; Krishnan, A.; Pereira, A. Rice growth under drought kinase is required for drought tolerance and grain yield under normal and drought stress conditions. Plant Physiol. 2014, 166, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, C.B.; Basu, S.; Pereira, A.; Tseng, T.M.; Zimmer, P.D.; Burgos, N.R. Analysis of stress-responsive gene expression in cultivated and weedy rice differing in cold stress tolerance. PLoS ONE 2015, 7, e0132100. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, G.P.M.; Basu, S.; Ramegowda, V.; Braga, E.B.; Pereira, A. Comparative analysis of gene expression in response to cold stress in diverse rice genotypes. Biochem. Biophys. Res. Commun. 2016, 471, 253–259. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly Media: Sebastopol, CA, USA, 2020. [Google Scholar]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 8 February 2023).


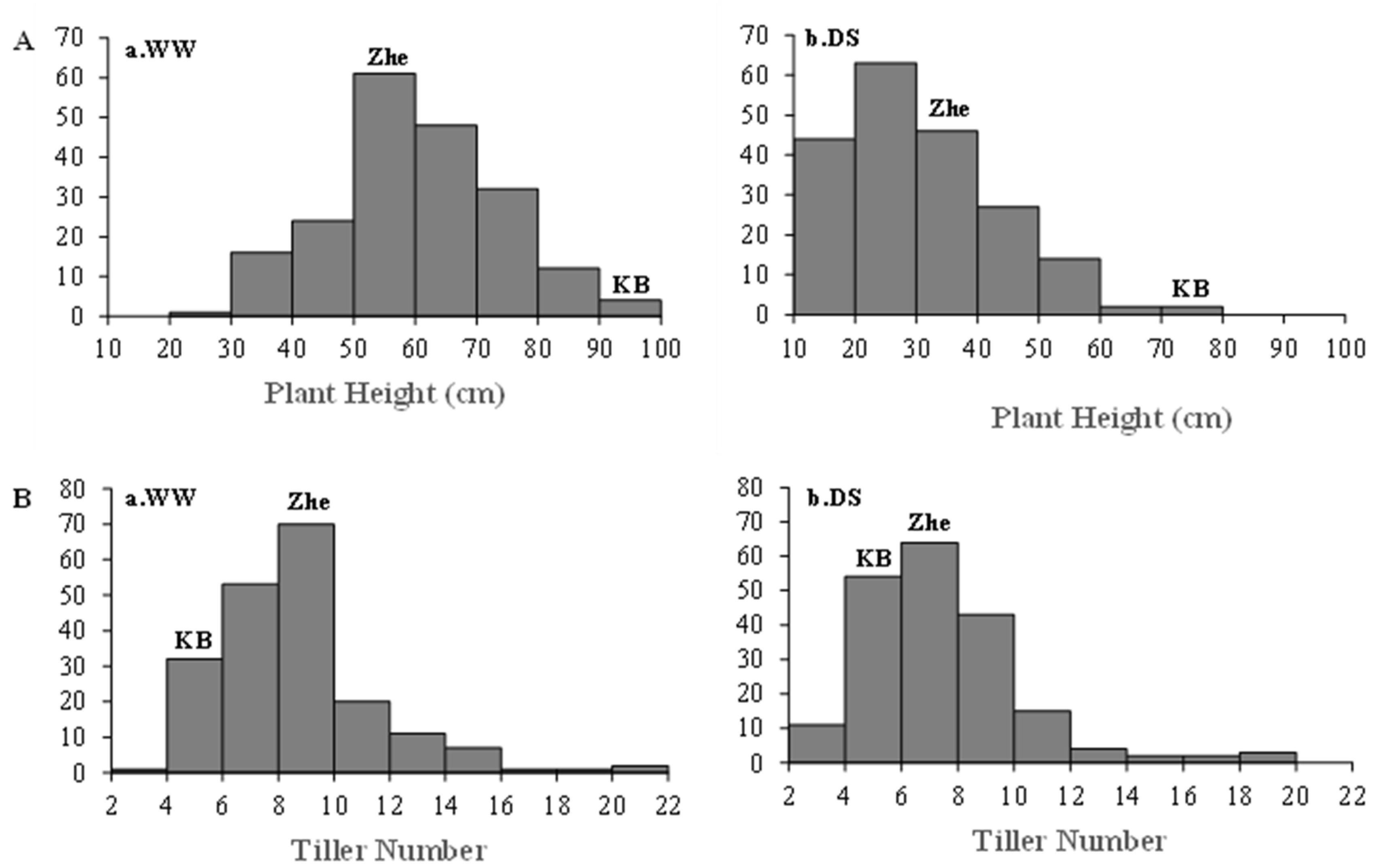
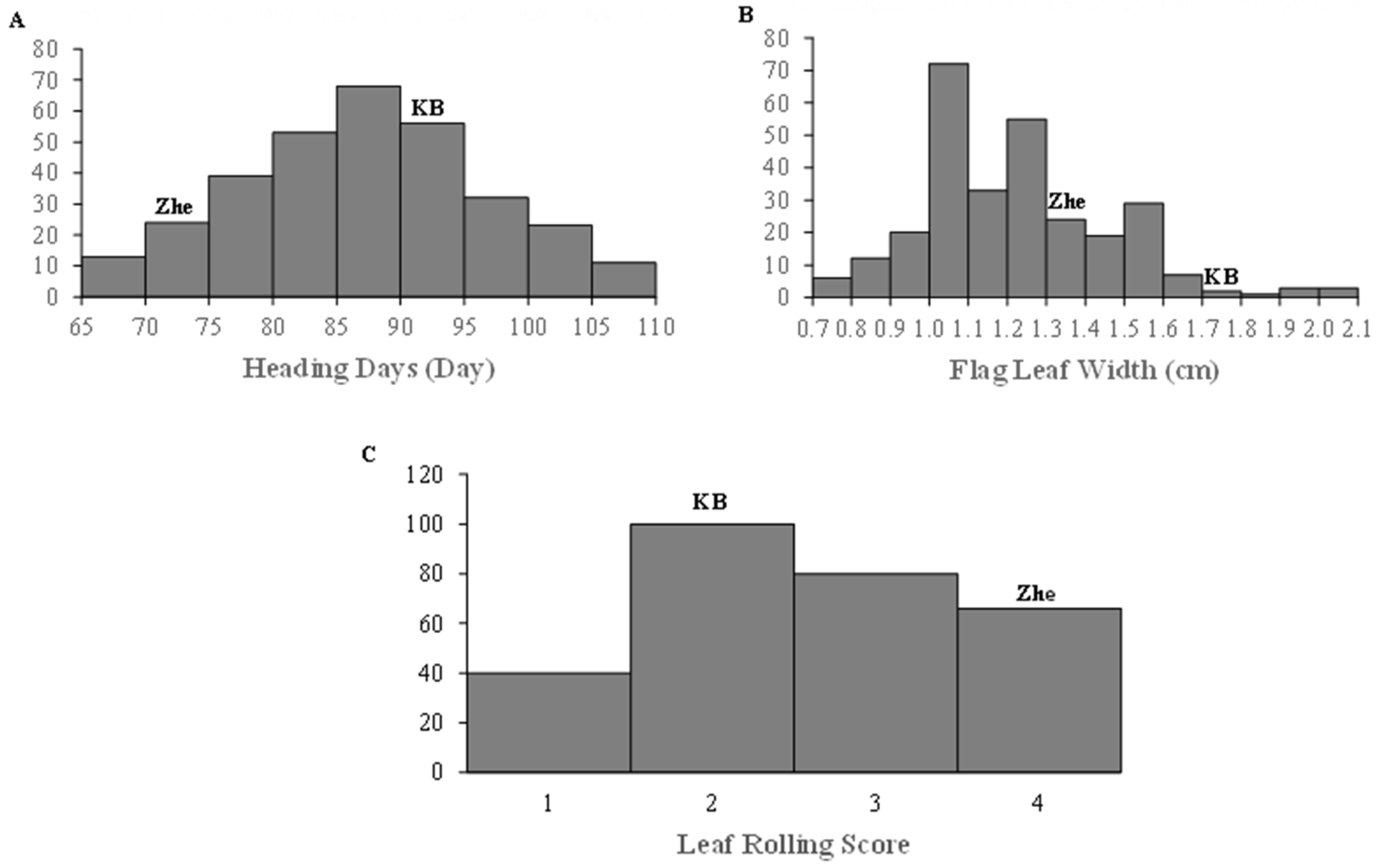
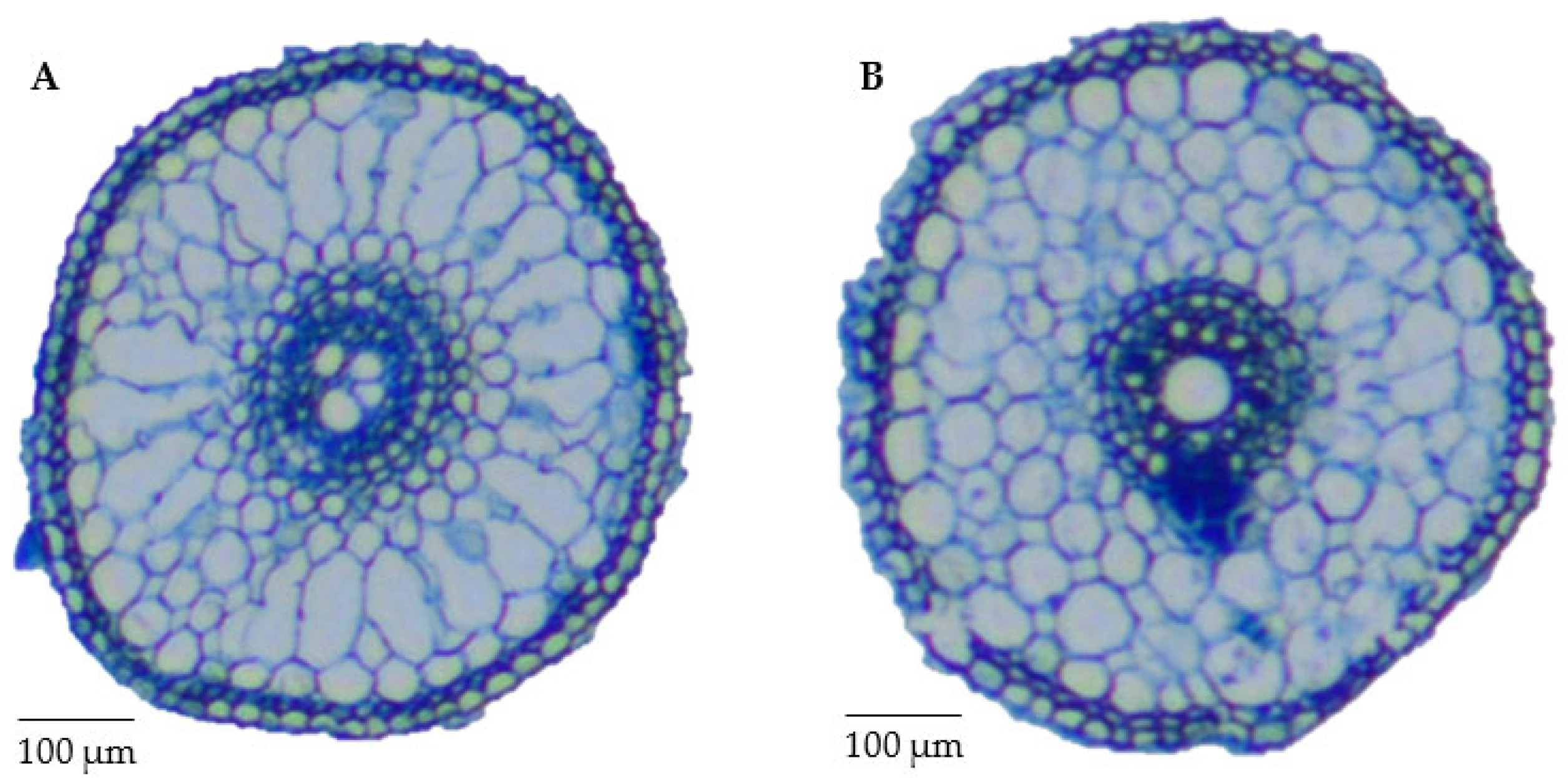
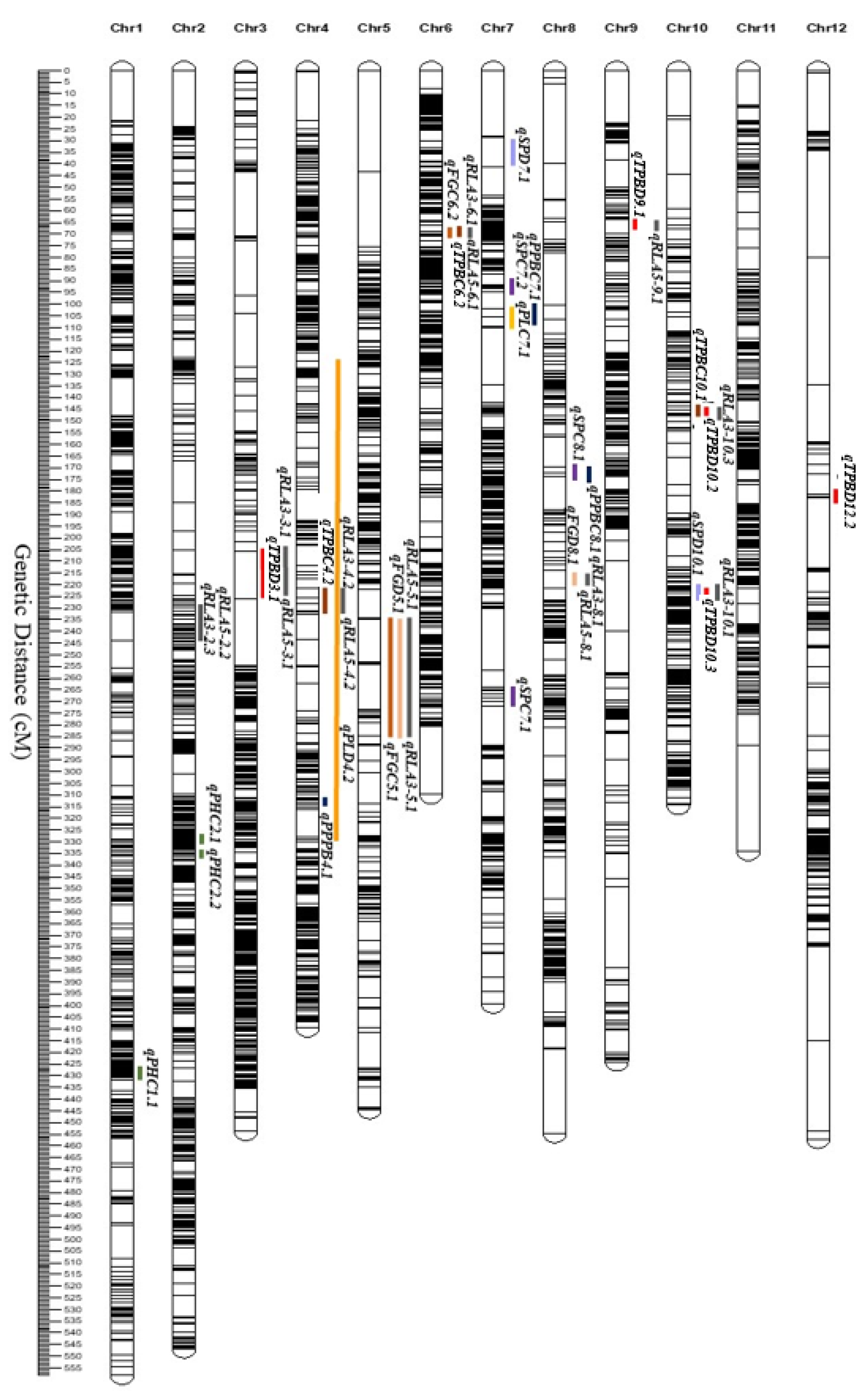
| Traits | Treatments | Parent | K/Z RIL Population | |||||
|---|---|---|---|---|---|---|---|---|
| Kaybonnet | Reduction (%) Kaybonnet under DS | ZHE733 | Reduction (%) ZHE733 under DS | Average | Reduction (%) | Range | ||
| Total plant biomass | WW | 14.80 ± 0.11 | 4.73 | 13.40 ± 0.19 | 51.49 | 20.00 ± 0.24 | 12.50 | 4.00–49.00 |
| DS | 14.10 ± 0.17 | 6.50 ± 0.24 | 17.50 ± 0.24 | 2.00–47.00 | ||||
| Spikelet per panicle number | WW | 104.00 ± 0.22 | 28.85 | 90.00 ± 0.27 | 38.89 | 133.35 ± 0.31 | 38.75 | 45.00–328.00 |
| DS | 74.00 ± 0.15 | 55.00 ± 0.16 | 81.68 ± 0.27 | 26.80–188.40 | ||||
| Filled grain per panicle number | WW | 66.00 ± 0.23 | 24.24 | 43.20 ± 0.18 | 62.96 | 43.07 ± 0.21 | 47.62 | 2.00–176.00 |
| DS | 50.00 ± 0.28 | 16.00 ± 0.12 | 22.56 ± 0.24 | 0.00–97.00 | ||||
| Panicle length (cm) | WW | 21.70 ± 0.16 | 11.06 | 18.60 ± 0.15 | 16.13 | 21.24 ± 0.16 | 15.82 | 11.90–33.30 |
| DS | 19.30 ± 0.12 | 15.60 ± 0.23 | 17.88 ± 0.19 | 11.64–26.62 | ||||
| Primary panicle branch number | WW | 19.00 ± 0.11 | 31.58 | 8.00 ± 0.14 | 12.50 | 11.17 ± 0.13 | 16.92 | 3.00–22.00 |
| DS | 13.00 ± 0.17 | 4.00 ± 0.12 | 9.28 ± 0.12 | 5.40–15.60 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dwiningsih, Y.; Thomas, J.; Kumar, A.; Gupta, C.; Gill, N.; Ruiz, C.; Alkahtani, J.; Baisakh, N.; Pereira, A. QTLs and Candidate Loci Associated with Drought Tolerance Traits of Kaybonnet x ZHE733 Recombinant Inbred Lines Rice Population. Int. J. Mol. Sci. 2023, 24, 15167. https://doi.org/10.3390/ijms242015167
Dwiningsih Y, Thomas J, Kumar A, Gupta C, Gill N, Ruiz C, Alkahtani J, Baisakh N, Pereira A. QTLs and Candidate Loci Associated with Drought Tolerance Traits of Kaybonnet x ZHE733 Recombinant Inbred Lines Rice Population. International Journal of Molecular Sciences. 2023; 24(20):15167. https://doi.org/10.3390/ijms242015167
Chicago/Turabian StyleDwiningsih, Yheni, Julie Thomas, Anuj Kumar, Chirag Gupta, Navdeep Gill, Charles Ruiz, Jawaher Alkahtani, Niranjan Baisakh, and Andy Pereira. 2023. "QTLs and Candidate Loci Associated with Drought Tolerance Traits of Kaybonnet x ZHE733 Recombinant Inbred Lines Rice Population" International Journal of Molecular Sciences 24, no. 20: 15167. https://doi.org/10.3390/ijms242015167
APA StyleDwiningsih, Y., Thomas, J., Kumar, A., Gupta, C., Gill, N., Ruiz, C., Alkahtani, J., Baisakh, N., & Pereira, A. (2023). QTLs and Candidate Loci Associated with Drought Tolerance Traits of Kaybonnet x ZHE733 Recombinant Inbred Lines Rice Population. International Journal of Molecular Sciences, 24(20), 15167. https://doi.org/10.3390/ijms242015167








