Epigenetic Activation of TUSC3 Sensitizes Glioblastoma to Temozolomide Independent of MGMT Promoter Methylation Status
Abstract
1. Introduction
2. Results
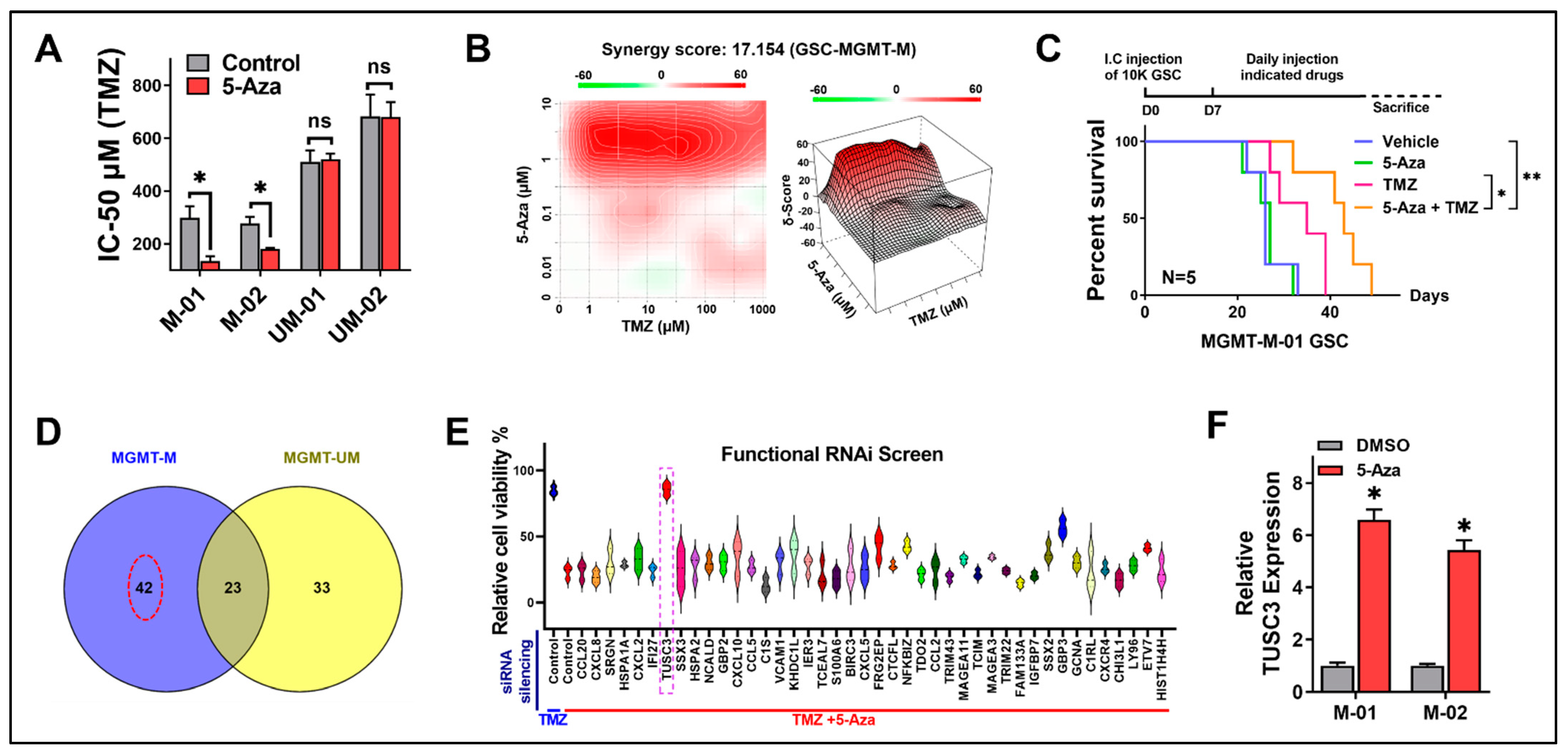
2.1. GSCs Are Sensitized by 5-Aza to TMZ Treatment through Activation of TUSC3
2.2. TUSC3 Expression Is Epigenetically Regulated and Positively Correlated with GBM Overall Survival
2.3. Activation of TUSC3 Enhances TMZ Sensitivity in Both MGMT-M and MGMT-UM GSCs
2.4. Synergy of 5-Aza with MGMT Silencing to Influence TMZ Sensitivity in MGMT-UM GSCs
2.5. Pharmacological Epigenetic Reactivation of TUSC3 Synergizes with TMZ, Leading to Positive Therapeutic Outcomes
3. Discussion
4. Methods and Materials
4.1. Human Samples and Processing
4.2. Cell Culture and Reagents
4.3. Gene Overexpression in GSCs
4.4. Gene Silencing by CRISPR/Cas9 System
4.5. Western Blot Analysis
4.6. Real-Time PCR
4.7. Cell Viability Assay, Proliferation Curve and IC-50 Evaluation
4.8. DNMT1 Activity Assay
4.9. Trans-Well Migration and Invasion Assays
4.10. Drug-Response Synergy Index Calculation
4.11. Genomic DNA Extraction and Bisulfite DNA Sequencing Analysis
4.12. Microarray
4.13. siRNA Knockdown
4.14. Rescue Experiments
4.15. Mice and GBM Xenograft Model
4.16. Illumina Methylation 850 EPIC Arrays
4.17. Bioinformatics Analysis
4.18. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ballman, K.V.; Buckner, J.C.; Brown, P.D.; Giannini, C.; Flynn, P.J.; LaPlant, B.R.; Jaeckle, K.A. The relationship between six-month progression-free survival and 12-month overall survival end points for phase II trials in patients with glioblastoma multiforme. Neuro Oncol. 2007, 9, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.E.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. Dose-dense temozolomide for newly diagnosed glioblastoma: A randomized phase III clinical trial. J. Clin. Oncol. 2013, 31, 4085–4091. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Godard, S.; Dietrich, P.Y.; Regli, L.; Ostermann, S.; Otten, P.; Van Melle, G.; de Tribolet, N.; Stupp, R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004, 10, 1871–1874. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Binabaj, M.M.; Bahrami, A.; ShahidSales, S.; Joodi, M.; Joudi Mashhad, M.; Hassanian, S.M.; Anvari, K.; Avan, A. The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J. Cell Physiol. 2018, 233, 378–386. [Google Scholar] [CrossRef]
- Rao, A.M.; Quddusi, A.; Shamim, M.S. The significance of MGMT methylation in Glioblastoma Multiforme prognosis. J. Pak. Med. Assoc. 2018, 68, 1137–1139. [Google Scholar]
- Dong, Z.; Cui, H. Epigenetic modulation of metabolism in glioblastoma. Semin. Cancer Biol. 2019, 57, 45–51. [Google Scholar] [CrossRef]
- Uddin, M.S.; Mamun, A.A.; Alghamdi, B.S.; Tewari, D.; Jeandet, P.; Sarwar, M.S.; Ashraf, G.M. Epigenetics of glioblastoma multiforme: From molecular mechanisms to therapeutic approaches. Semin. Cancer Biol. 2022, 83, 100–120. [Google Scholar] [CrossRef]
- Gusyatiner, O.; Hegi, M.E. Glioma epigenetics: From subclassification to novel treatment options. Semin. Cancer Biol. 2018, 51, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Ryu, H.W.; Won, H.R.; Kwon, S.H. Advances in epigenetic glioblastoma therapy. Oncotarget 2017, 8, 18577–18589. [Google Scholar] [CrossRef]
- Wu, Q.; Berglund, A.E.; Etame, A.B. The Impact of Epigenetic Modifications on Adaptive Resistance Evolution in Glioblastoma. Int. J. Mol. Sci. 2021, 22, 8324. [Google Scholar] [CrossRef]
- Clavel, M.; Monfardini, S.; Fosså, S.; Smyth, J.; Renard, J.; Kaye, S.B. 5-Aza-2’-deoxycytidine (NSC 127716) in non-seminomatous testicular cancer. Phase II from the EORTC Early Clinical Trials Cooperative Group and Genito-Urinary Group. Ann. Oncol. 1992, 3, 399–400. [Google Scholar] [CrossRef]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef]
- Kadia, T.M.; Reville, P.K.; Wang, X.; Rausch, C.R.; Borthakur, G.; Pemmaraju, N.; Daver, N.G.; DiNardo, C.D.; Sasaki, K.; Issa, G.C.; et al. Phase II Study of Venetoclax Added to Cladribine Plus Low-Dose Cytarabine Alternating With 5-Azacitidine in Older Patients with Newly Diagnosed Acute Myeloid Leukemia. J. Clin. Oncol. 2022, 40, 3848–3857. [Google Scholar] [CrossRef] [PubMed]
- Oran, B.; de Lima, M.; Garcia-Manero, G.; Thall, P.F.; Lin, R.; Popat, U.; Alousi, A.M.; Hosing, C.; Giralt, S.; Rondon, G.; et al. A phase 3 randomized study of 5-azacitidine maintenance vs observation after transplant in high-risk AML and MDS patients. Blood Adv. 2020, 4, 5580–5588. [Google Scholar] [CrossRef] [PubMed]
- Sessa, C.; ten Bokkel Huinink, W.; Stoter, G.; Renard, J.; Cavalli, F. Phase II study of 5-aza-2’-deoxycytidine in advanced ovarian carcinoma. The EORTC Early Clinical Trials Group. Eur. J. Cancer 1990, 26, 137–138. [Google Scholar] [CrossRef]
- Phan, N.L.; Trinh, N.V.; Pham, P.V. Low concentrations of 5-aza-2’-deoxycytidine induce breast cancer stem cell differentiation by triggering tumor suppressor gene expression. OncoTargets Ther. 2016, 9, 49–59. [Google Scholar] [CrossRef]
- Tsai, H.C.; Li, H.; Van Neste, L.; Cai, Y.; Robert, C.; Rassool, F.V.; Shin, J.J.; Harbom, K.M.; Beaty, R.; Pappou, E.; et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012, 21, 430–446. [Google Scholar] [CrossRef] [PubMed]
- Borodovsky, A.; Salmasi, V.; Turcan, S.; Fabius, A.W.; Baia, G.S.; Eberhart, C.G.; Weingart, J.D.; Gallia, G.L.; Baylin, S.B.; Chan, T.A.; et al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget 2013, 4, 1737–1747. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.S.; da Costa Rosa, M.; Borodovsky, A.; Festuccia, W.T.; Chan, T.; Riggins, G.J. Demethylation and epigenetic modification with 5-azacytidine reduces IDH1 mutant glioma growth in combination with temozolomide. Neuro-Oncol. 2019, 21, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Garshasbi, M.; Hadavi, V.; Habibi, H.; Kahrizi, K.; Kariminejad, R.; Behjati, F.; Tzschach, A.; Najmabadi, H.; Ropers, H.H.; Kuss, A.W. A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am. J. Hum. Genet. 2008, 82, 1158–1164. [Google Scholar] [CrossRef] [PubMed]
- Horak, P.; Tomasich, E.; Vaňhara, P.; Kratochvílová, K.; Anees, M.; Marhold, M.; Lemberger, C.E.; Gerschpacher, M.; Horvat, R.; Sibilia, M.; et al. TUSC3 loss alters the ER stress response and accelerates prostate cancer growth in vivo. Sci. Rep. 2014, 4, 3739. [Google Scholar] [CrossRef]
- Kratochvílová, K.; Horak, P.; Ešner, M.; Souček, K.; Pils, D.; Anees, M.; Tomasich, E.; Dráfi, F.; Jurtíková, V.; Hampl, A.; et al. Tumor suppressor candidate 3 (TUSC3) prevents the epithelial-to-mesenchymal transition and inhibits tumor growth by modulating the endoplasmic reticulum stress response in ovarian cancer cells. Int. J. Cancer 2015, 137, 1330–1340. [Google Scholar] [CrossRef]
- Mohorko, E.; Glockshuber, R.; Aebi, M. Oligosaccharyltransferase: The central enzyme of N-linked protein glycosylation. J. Inherit. Metab. Dis. 2011, 34, 869–878. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, X.; Shen, J.; Zhao, H.; Yu, X.; Chen, Y.; Zhuang, Z.; Deng, X.; Feng, H.; Wang, Y.; et al. Decreased TUSC3 Promotes Pancreatic Cancer Proliferation, Invasion and Metastasis. PLoS ONE 2016, 11, e0149028. [Google Scholar] [CrossRef]
- Jiang, Z.; Guo, M.; Zhang, X.; Yao, L.; Shen, J.; Ma, G.; Liu, L.; Zhao, L.; Xie, C.; Liang, H.; et al. TUSC3 suppresses glioblastoma development by inhibiting Akt signaling. Tumour Biol. 2016, 37, 12039–12047. [Google Scholar] [CrossRef]
- Yuan, J.; Yu, X.; Wang, A.; Li, Y.; Liu, F.; Wang, Y.; Sun, S.; Bing, X.; Liu, Y.; Du, J. Tumor suppressor candidate 3: A novel grading tool and predictor of clinical malignancy in human gliomas. Oncol. Lett. 2018, 15, 5655–5661. [Google Scholar] [CrossRef]
- Cheng, Z.X.; Yin, W.B.; Wang, Z.Y. MicroRNA-132 induces temozolomide resistance and promotes the formation of cancer stem cell phenotypes by targeting tumor suppressor candidate 3 in glioblastoma. Int. J. Mol. Med. 2017, 40, 1307–1314. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e411. [Google Scholar] [CrossRef]
- McElhinney, R.S.; Donnelly, D.J.; McCormick, J.E.; Kelly, J.; Watson, A.J.; Rafferty, J.A.; Elder, R.H.; Middleton, M.R.; Willington, M.A.; McMurry, T.B.; et al. Inactivation of O6-alkylguanine-DNA alkyltransferase. 1. Novel O6-(hetarylmethyl)guanines having basic rings in the side chain. J. Med. Chem. 1998, 41, 5265–5271. [Google Scholar] [CrossRef] [PubMed]
- Baer, M.R.; Kogan, A.A.; Bentzen, S.M.; Mi, T.; Lapidus, R.G.; Duong, V.H.; Emadi, A.; Niyongere, S.; O’Connell, C.L.; Youngblood, B.A.; et al. Phase I Clinical Trial of DNA Methyltransferase Inhibitor Decitabine and PARP Inhibitor Talazoparib Combination Therapy in Relapsed/Refractory Acute Myeloid Leukemia. Clin. Cancer Res. 2022, 28, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Khan, O.A.; Ranson, M.; Michael, M.; Olver, I.; Levitt, N.C.; Mortimer, P.; Watson, A.J.; Margison, G.P.; Midgley, R.; Middleton, M.R. A phase II trial of lomeguatrib and temozolomide in metastatic colorectal cancer. Br. J. Cancer 2008, 98, 1614–1618. [Google Scholar] [CrossRef] [PubMed]
- Momparler, R.L.; Rivard, G.E.; Gyger, M. Clinical trial on 5-aza-2’-deoxycytidine in patients with acute leukemia. Pharmacol. Ther. 1985, 30, 277–286. [Google Scholar] [CrossRef]
- Sabharwal, A.; Corrie, P.G.; Midgley, R.S.; Palmer, C.; Brady, J.; Mortimer, P.; Watson, A.J.; Margison, G.P.; Middleton, M.R. A phase I trial of lomeguatrib and irinotecan in metastatic colorectal cancer. Cancer Chemother. Pharmacol. 2010, 66, 829–835. [Google Scholar] [CrossRef]
- Wang, E.S.; Montesinos, P.; Minden, M.D.; Lee, J.H.; Heuser, M.; Naoe, T.; Chou, W.C.; Laribi, K.; Esteve, J.; Altman, J.K.; et al. Phase 3 trial of gilteritinib plus azacitidine vs azacitidine for newly diagnosed FLT3mut+ AML ineligible for intensive chemotherapy. Blood 2022, 140, 1845–1857. [Google Scholar] [CrossRef]
- Bearzatto, A.; Szadkowski, M.; Macpherson, P.; Jiricny, J.; Karran, P. Epigenetic regulation of the MGMT and hMSH6 DNA repair genes in cells resistant to methylating agents. Cancer Res. 2000, 60, 3262–3270. [Google Scholar]
- Chen, G.D.; Qian, D.Y.; Li, Z.G.; Fan, G.Y.; You, K.L.; Wu, Y.L. Down-regulation of p16 and MGMT promotes the anti-proliferative and pro-apoptotic effects of 5-Aza-dC and radiation on cervical cancer cells. Cell Biochem. Funct. 2017, 35, 488–496. [Google Scholar] [CrossRef]
- do Amaral, G.; Planello, A.C.; Borgato, G.; de Lima, D.G.; Guimarães, G.N.; Marques, M.R.; de Souza, A.P. 5-Aza-CdR promotes partial MGMT demethylation and modifies expression of different genes in oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 127, 425–432. [Google Scholar] [CrossRef]
- Lima, D.G.; do Amaral, G.; Planello, A.C.; Borgato, G.B.; Guimarães, G.N.; de Souza, A.P. Combined therapy with cisplatin and 5-AZA-2CdR modifies methylation and expression of DNA repair genes in oral squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2022, 15, 131–144. [Google Scholar] [PubMed]
- Yang, J.; Zhu, X.B.; He, L.X.; Gu, Z.W.; Jin, M.Z.; Ji, W.Y. Clinical significance of epigenetic silencing and re-expression of O6-methylguanine-DNA methyltransferase using epigenetic agents in laryngeal carcinoma. Oncol. Lett. 2015, 9, 35–42. [Google Scholar] [CrossRef][Green Version]
- Cioccoloni, G.; Bonmassar, L.; Pagani, E.; Caporali, S.; Fuggetta, M.P.; Bonmassar, E.; D’Atri, S.; Aquino, A. Influence of fatty acid synthase inhibitor orlistat on the DNA repair enzyme O6-methylguanine-DNA methyltransferase in human normal or malignant cells in vitro. Int. J. Oncol. 2015, 47, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Berglund, A.E.; MacAulay, R.J.; Etame, A.B. A Novel Role of BIRC3 in Stemness Reprogramming of Glioblastoma. Int. J. Mol. Sci. 2021, 23, 297. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Berglund, A.E.; Wang, D.; MacAulay, R.J.; Mulé, J.J.; Etame, A.B. Paradoxical epigenetic regulation of XAF1 mediates plasticity towards adaptive resistance evolution in MGMT-methylated glioblastoma. Sci. Rep. 2019, 9, 14072. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Fortin, J.P.; Labbe, A.; Lemire, M.; Zanke, B.W.; Hudson, T.J.; Fertig, E.J.; Greenwood, C.M.; Hansen, K.D. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014, 15, 503. [Google Scholar] [CrossRef] [PubMed]
- Triche, T.J., Jr.; Weisenberger, D.J.; Van Den Berg, D.; Laird, P.W.; Siegmund, K.D. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013, 41, e90. [Google Scholar] [CrossRef]
- Welsh, E.A.; Eschrich, S.A.; Berglund, A.E.; Fenstermacher, D.A. Iterative rank-order normalization of gene expression microarray data. BMC Bioinform. 2013, 14, 153. [Google Scholar] [CrossRef]
- Creed, J.; Gerke, T.; Berglund, A. MatSurv: Survival analysis and visualization in MATLAB. J. Open Source Softw. 2020, 5, 1830. [Google Scholar] [CrossRef]




| ID | Growth | SOX2 | CD133 | MGMT | Nestin |
|---|---|---|---|---|---|
| MGMT-M-01 | Spheres | + | + | hypermethylated | + |
| MGMT-M-02 | Spheres | + | + | hypermethylated | + |
| MGMT-UM-01 | Spheres | + | + | hypomethylated | + |
| MGMT-UM-02 | Spheres | + | + | hypomethylated | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Berglund, A.E.; Macaulay, R.J.; Etame, A.B. Epigenetic Activation of TUSC3 Sensitizes Glioblastoma to Temozolomide Independent of MGMT Promoter Methylation Status. Int. J. Mol. Sci. 2023, 24, 15179. https://doi.org/10.3390/ijms242015179
Wu Q, Berglund AE, Macaulay RJ, Etame AB. Epigenetic Activation of TUSC3 Sensitizes Glioblastoma to Temozolomide Independent of MGMT Promoter Methylation Status. International Journal of Molecular Sciences. 2023; 24(20):15179. https://doi.org/10.3390/ijms242015179
Chicago/Turabian StyleWu, Qiong, Anders E. Berglund, Robert J. Macaulay, and Arnold B. Etame. 2023. "Epigenetic Activation of TUSC3 Sensitizes Glioblastoma to Temozolomide Independent of MGMT Promoter Methylation Status" International Journal of Molecular Sciences 24, no. 20: 15179. https://doi.org/10.3390/ijms242015179
APA StyleWu, Q., Berglund, A. E., Macaulay, R. J., & Etame, A. B. (2023). Epigenetic Activation of TUSC3 Sensitizes Glioblastoma to Temozolomide Independent of MGMT Promoter Methylation Status. International Journal of Molecular Sciences, 24(20), 15179. https://doi.org/10.3390/ijms242015179






