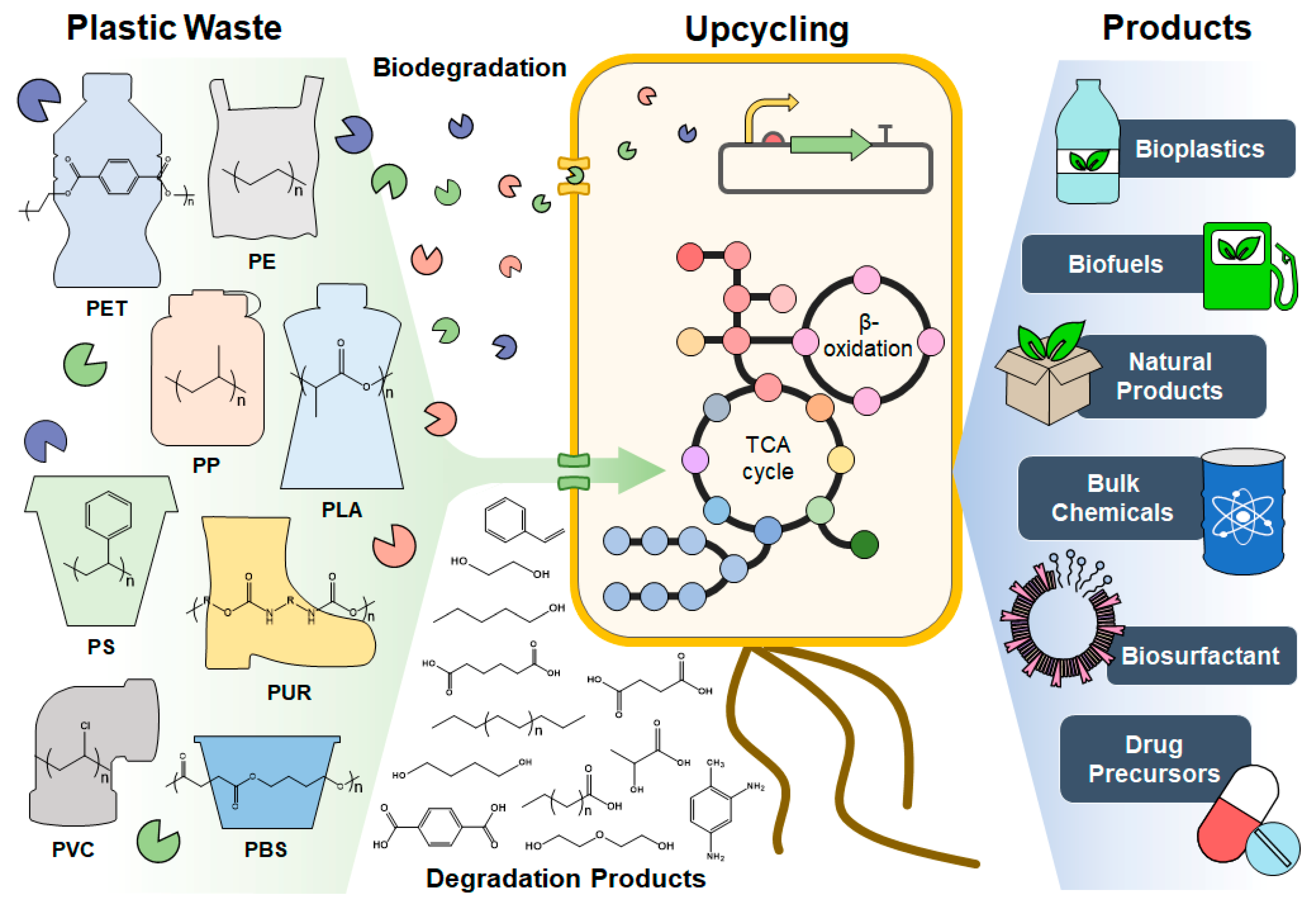
Biotechnological Plastic Degradation and Valorization Using Systems Metabolic Engineering
Abstract
:1. Introduction
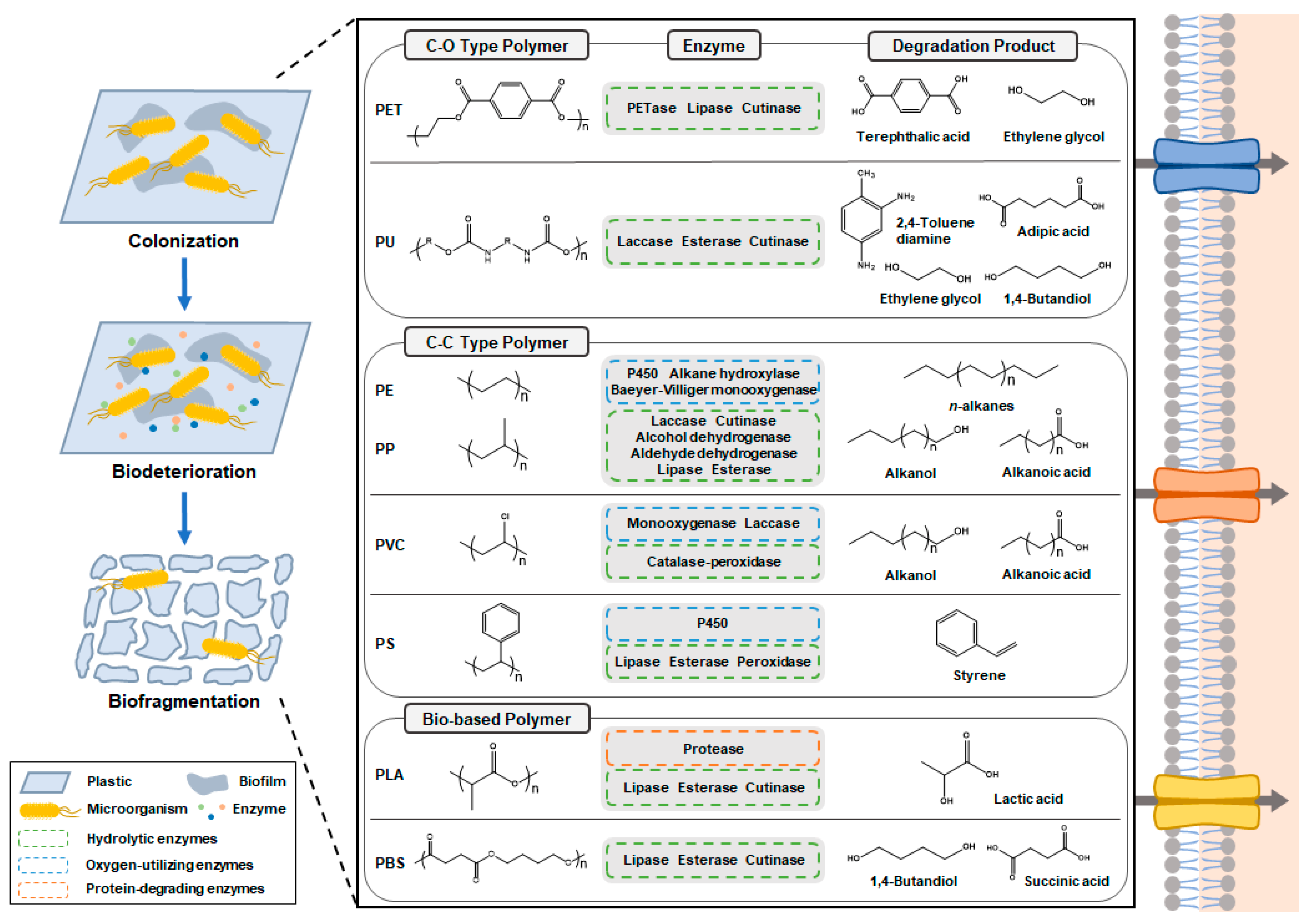
2. Microbial and Enzymatic Degradation of Plastics
2.1. Fossil-Based Plastics
2.1.1. Polyethylene and Polypropylene
2.1.2. Polyvinylchloride
2.1.3. Polyurethane
2.1.4. Polyethylene Terephthalate
2.1.5. Polystyrene
2.2. Bio-Based Plastics
2.2.1. Polylactic Acid
2.2.2. Polybutylene Succinate
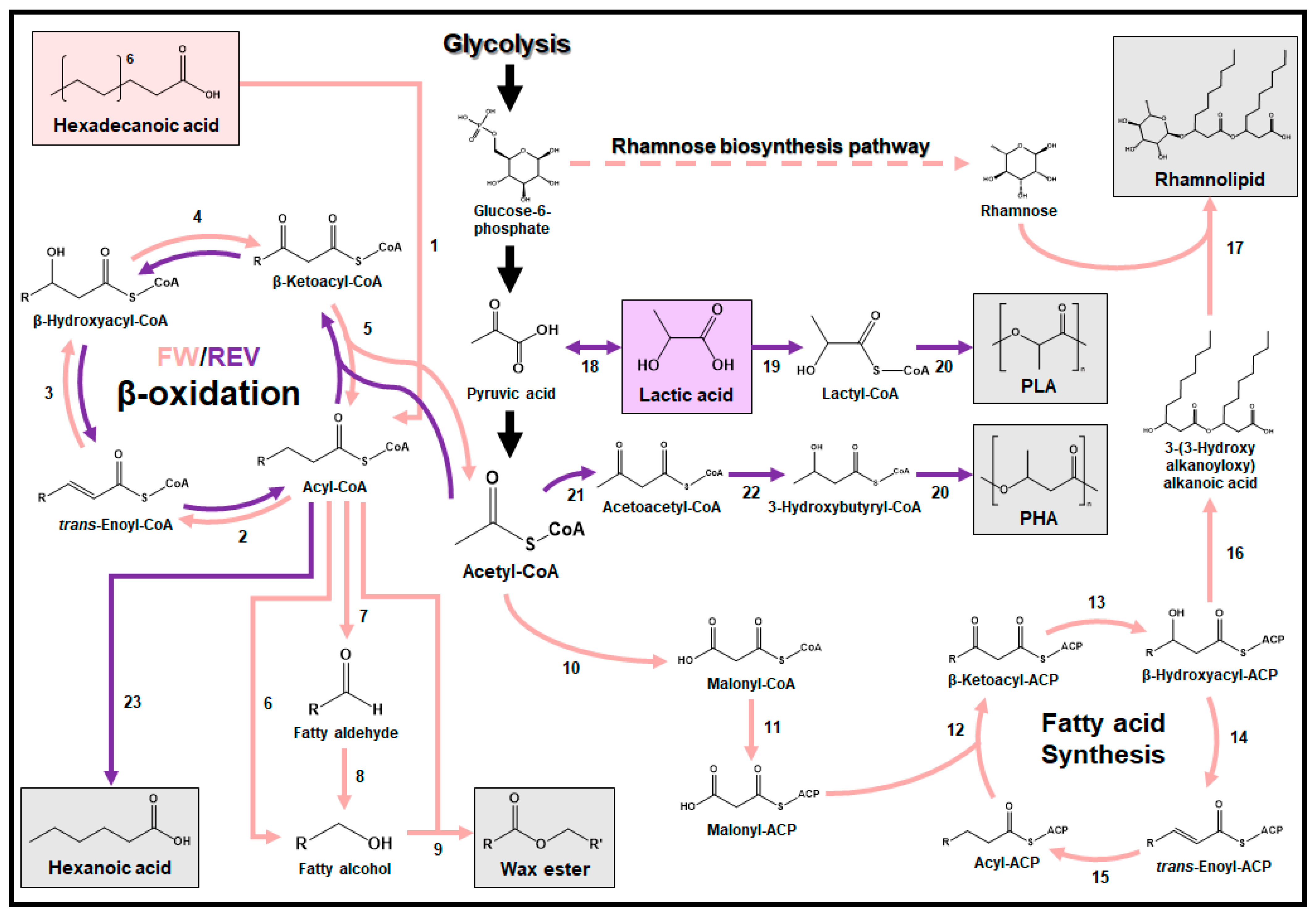
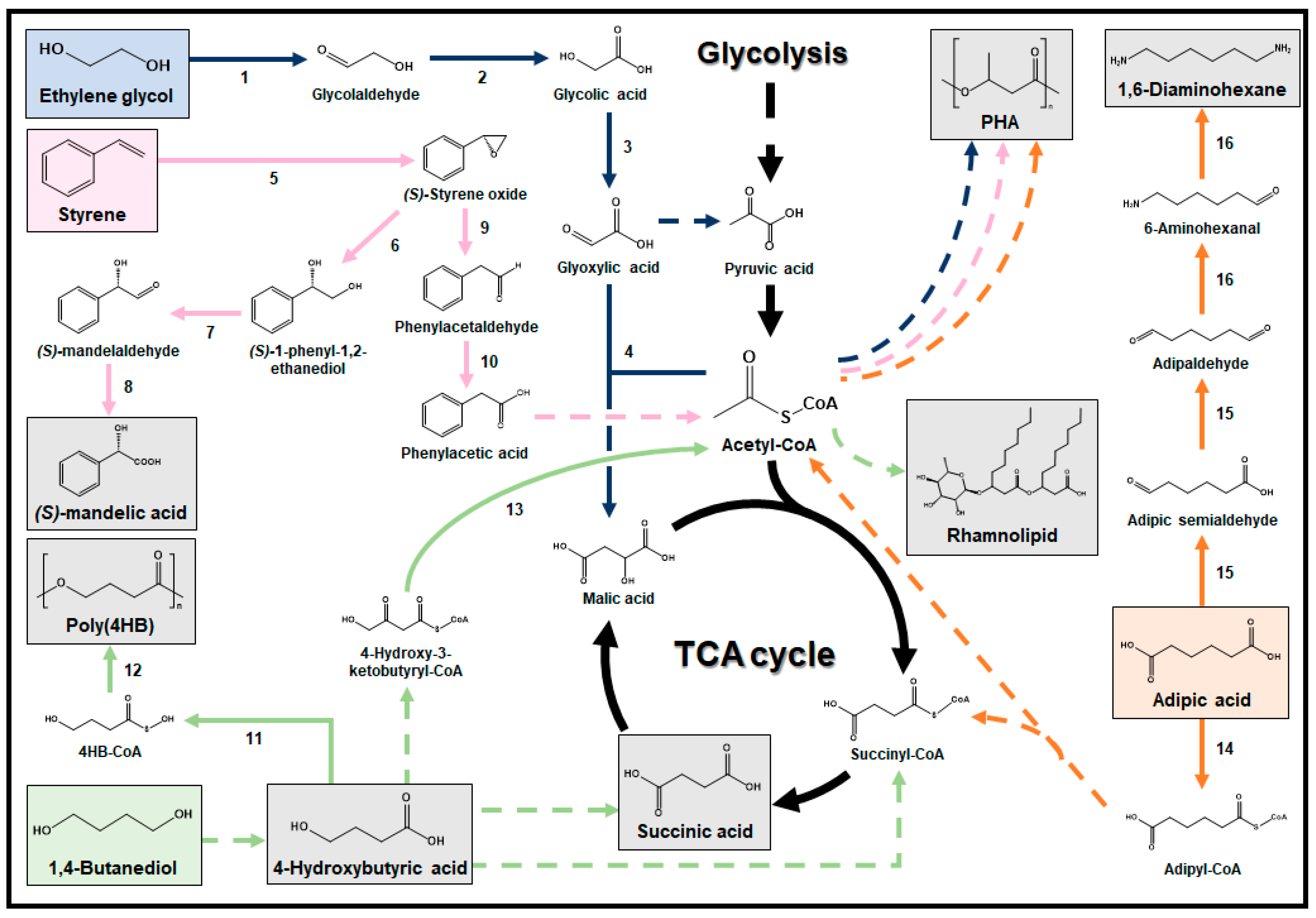
3. Valorization of Plastic Degradation Products into Useful Chemicals and Materials
3.1. Hexadecan(oat)e
3.2. Lactic Acid
3.3. 1,4-Butanediol
3.4. Ethylene Glycol
3.5. Styrene
3.6. Adipic Acid
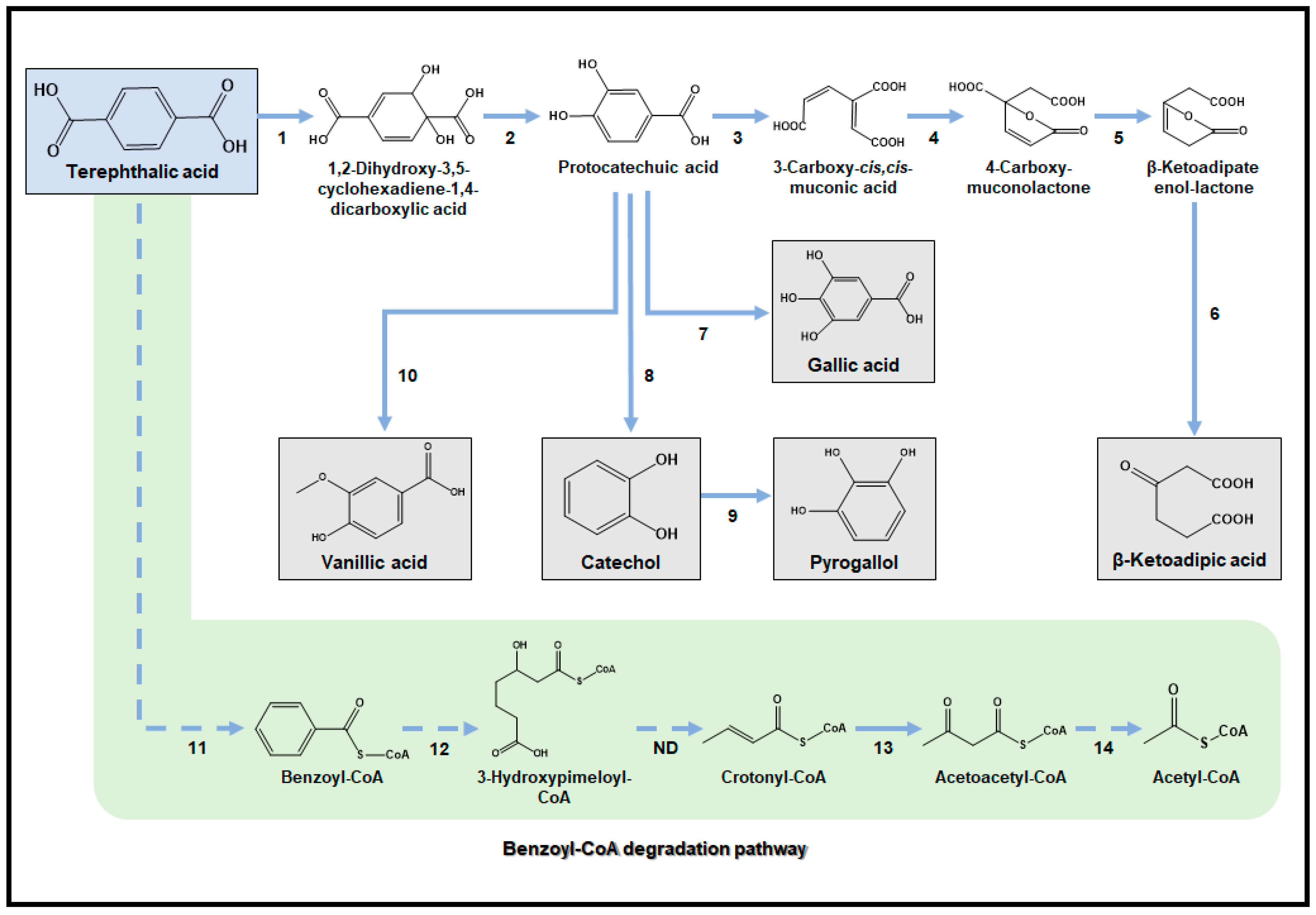
3.7. Terephthalic Acid
4. Future Perspectives on the Development of Plastic Waste Bio-Upcycling Technology
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lomwongsopon, P.; Varrone, C. Critical review on the progress of plastic bioupcycling technology as a potential solution for sustainable plastic waste management. Polymers 2022, 14, 4996. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Plastic biodegradation: Frontline microbes and their enzymes. Sci. Total Environ. 2021, 759, 143536. [Google Scholar] [CrossRef] [PubMed]
- Tiso, T.; Winter, B.; Wei, R.; Hee, J.; de Witt, J.; Wierckx, N.; Quicker, P.; Bornscheuer, U.T.; Bardow, A.; Nogales, J. The metabolic potential of plastics as biotechnological carbon sources–review and targets for the future. Metab. Eng. 2022, 71, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Ragaert, K.; Delva, L.; Van Geem, K. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017, 69, 24–58. [Google Scholar] [CrossRef] [PubMed]
- Abu-Thabit, N.Y.; Pérez-Rivero, C.; Uwaezuoke, O.J.; Ngwuluka, N.C. From waste to wealth: Upcycling of plastic and lignocellulosic wastes to PHAs. J. Chem. Technol. Biotechnol. 2022, 97, 3217–3240. [Google Scholar] [CrossRef]
- Rahimi, A.; García, J.M. Chemical recycling of waste plastics for new materials production. Nat. Rev. Chem. 2017, 1, 0046. [Google Scholar] [CrossRef]
- Ahn, J.H.; Jung, K.H.; Lim, E.S.; Kim, S.M.; Han, S.O.; Um, Y. Recent advances in microbial production of medium chain fatty acid from renewable carbon resources: A comprehensive review. Bioresour. Technol. 2023, 381, 129147. [Google Scholar]
- Zhang, Y.; Pedersen, J.N.; Eser, B.E.; Guo, Z. Biodegradation of polyethylene and polystyrene: From microbial deterioration to enzyme discovery. Biotechnol. Adv. 2022, 60, 107991. [Google Scholar] [CrossRef]
- Yang, J.; Yang, Y.; Wu, W.M.; Zhao, J.; Jiang, L. Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms. Environ. Sci. Technol. 2014, 48, 13776–13784. [Google Scholar] [CrossRef]
- Yun, S.D.; Lee, C.O.; Kim, H.W.; An, S.J.; Kim, S.; Seo, M.J.; Park, C.; Yun, C.H.; Chi, W.S.; Yeom, S.J. Exploring a new biocatalyst from Bacillus thuringiensis JNU01 for polyethylene biodegradation. Environ. Sci. Technol. Lett. 2023, 10, 485–492. [Google Scholar] [CrossRef]
- Nowak, B.; Pająk, J.; Drozd-Bratkowicz, M.; Rymarz, G. Microorganisms participating in the biodegradation of modified polyethylene films in different soils under laboratory conditions. Int. Biodeterior. Biodegrad. 2011, 65, 757–767. [Google Scholar] [CrossRef]
- Lee, J.A.; Ahn, J.H.; Kim, I.; Li, S.; Lee, S.Y. Synthesis, characterization, and application of fully biobased and biodegradable nylon-4, 4 and-5, 4. ACS Sustain. Chem. Eng. 2020, 8, 5604–5614. [Google Scholar] [CrossRef]
- Kundu, D.; Hazra, C.; Chatterjee, A.; Chaudhari, A.; Mishra, S. Biopolymer and biosurfactant-graft-calcium sulfate/polystyrene nanocomposites: Thermophysical, mechanical and biodegradation studies. Polym. Degrad. Stab. 2014, 107, 37–52. [Google Scholar] [CrossRef]
- Kong, D.; Wang, L.; Chen, X.; Xia, W.; Su, L.; Zuo, F.; Yan, Z.; Chen, S.; Wu, J. Chemical-biological degradation of polyethylene combining Baeyer–Villiger oxidation and hydrolysis reaction of cutinase. Green Chem. 2022, 24, 2203–2211. [Google Scholar] [CrossRef]
- Khandare, S.D.; Chaudhary, D.R.; Jha, B. Marine bacterial biodegradation of low-density polyethylene (LDPE) plastic. Biodegradation 2021, 32, 127–143. [Google Scholar] [CrossRef]
- Gao, R.; Sun, C. A marine bacterial community capable of degrading poly (ethylene terephthalate) and polyethylene. J. Hazard. Mater. 2021, 416, 125928. [Google Scholar] [CrossRef]
- Jeon, H.J.; Kim, M.N. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 2015, 103, 141–146. [Google Scholar] [CrossRef]
- Yao, C.; Xia, W.; Dou, M.; Du, Y.; Wu, J. Oxidative degradation of UV-irradiated polyethylene by laccase-mediator system. J. Hazard. Mater. 2022, 440, 129709. [Google Scholar] [CrossRef]
- Magnin, A.; Entzmann, L.; Pollet, E.; Avérous, L. Breakthrough in polyurethane bio-recycling: An efficient laccase-mediated system for the degradation of different types of polyurethanes. Waste Manag. 2021, 132, 23–30. [Google Scholar] [CrossRef]
- Schmidt, J.; Wei, R.; Oeser, T.; Dedavid e Silva, L.A.; Breite, D.; Schulze, A.; Zimmermann, W. Degradation of polyester polyurethane by bacterial polyester hydrolases. Polymers 2017, 9, 65. [Google Scholar] [CrossRef]
- Akutsu, Y.; Nakajima-Kambe, T.; Nomura, N.; Nakahara, T. Purification and properties of a polyester polyurethane-degrading enzyme from Comamonas acidovorans TB-35. Appl. Environ. Microbiol. 1998, 64, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Son, H.F.; Joo, S.; Seo, H.; Sagong, H.Y.; Lee, S.H.; Hong, H.; Kim, K.J. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzym. Microb. Technol. 2020, 141, 109656. [Google Scholar] [CrossRef] [PubMed]
- Tournier, V.; Topham, C.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, A.; Thies, S.; Knieps-Grünhagen, E.; Gertzen, C.; Kobus, S.; Höppner, A.; Ferrer, M.; Gohlke, H.; Smits, S.H.; Jaeger, K.E. A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri–structural and functional insights. Front. Microbiol. 2020, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Billig, S.; Oeser, T.; Birkemeyer, C.; Zimmermann, W. Hydrolysis of cyclic poly (ethylene terephthalate) trimers by a carboxylesterase from Thermobifida fusca KW3. Appl. Microbiol. Biotechnol. 2010, 87, 1753–1764. [Google Scholar] [CrossRef]
- Li, A.; Sheng, Y.; Cui, H.; Wang, M.; Wu, L.; Song, Y.; Yang, R.; Li, X.; Huang, H. Discovery and mechanism-guided engineering of BHET hydrolases for improved PET recycling and upcycling. Nat. Commun. 2023, 14, 4169. [Google Scholar] [CrossRef]
- Sagong, H.Y.; Seo, H.; Kim, T.; Son, H.F.; Joo, S.; Lee, S.H.; Kim, S.; Woo, J.S.; Hwang, S.Y.; Kim, K.J. Decomposition of the PET film by MHETase using exo-PETase function. ACS Catal. 2020, 10, 4805–4812. [Google Scholar] [CrossRef]
- Mohan, A.J.; Sekhar, V.C.; Bhaskar, T.; Nampoothiri, K.M. Microbial assisted high impact polystyrene (HIPS) degradation. Bioresour. Technol. 2016, 213, 204–207. [Google Scholar] [CrossRef]
- Nakamiya, K.; Sakasita, G.; Ooi, T.; Kinoshita, S. Enzymatic degradation of polystyrene by hydroquinone peroxidase of Azotobacter beijerinckii HM121. J. Biosci. Bioeng. 1997, 84, 480–482. [Google Scholar] [CrossRef]
- Akutsu-Shigeno, Y.; Teeraphatpornchai, T.; Teamtisong, K.; Nomura, N.; Uchiyama, H.; Nakahara, T.; Nakajima-Kambe, T. Cloning and sequencing of a poly (DL-lactic acid) depolymerase gene from Paenibacillus amylolyticus strain TB-13 and its functional expression in Escherichia coli. Appl. Environ. Microbiol. 2003, 69, 2498–2504. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Guo, Z.; Li, F.; Chen, S. Purification and characterization of poly (L-lactic acid) depolymerase from Pseudomonas sp. strain DS04-T. Polym. Eng Sci. 2011, 51, 454–459. [Google Scholar] [CrossRef]
- Shi, K.; Su, T.; Wang, Z. Comparison of poly (butylene succinate) biodegradation by Fusarium solani cutinase and Candida antarctica lipase. Polym. Degrad. Stab. 2019, 164, 55–60. [Google Scholar] [CrossRef]
- Hoshino, A.; Isono, Y. Degradation of aliphatic polyester films by commercially available lipases with special reference to rapid and complete degradation of poly (L-lactide) film by lipase PL derived from Alcaligenes sp. Biodegradation 2002, 13, 141–147. [Google Scholar] [CrossRef]
- Lee, C.W.; Kimura, Y.; Chung, J.-D. Mechanism of enzymatic degradation of poly (butylene succinate). Macromol. Res. 2008, 16, 651–658. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, J.; Ruan, Y.; Cai, X.; Li, J.; Zhang, L.; Huang, H. Degradation of polypropylene by the Pseudomonas aeruginosa strains LICME WZH-4 and WGH-6. J. Polym. Environ. 2022, 30, 3949–3958. [Google Scholar] [CrossRef]
- Rajandas, H.; Parimannan, S.; Sathasivam, K.; Ravichandran, M.; Yin, L.S. A novel FTIR-ATR spectroscopy based technique for the estimation of low-density polyethylene biodegradation. Polym. Test. 2012, 31, 1094–1099. [Google Scholar] [CrossRef]
- Gao, R.; Liu, R.; Sun, C. A marine fungus Alternaria alternata FB1 efficiently degrades polyethylene. J. Hazard. Mater. 2022, 431, 128617. [Google Scholar] [CrossRef]
- Skariyachan, S.; Patil, A.A.; Shankar, A.; Manjunath, M.; Bachappanavar, N.; Kiran, S. Enhanced polymer degradation of polyethylene and polypropylene by novel thermophilic consortia of Brevibacillus sps. and Aneurinibacillus sp. screened from waste management landfills and sewage treatment plants. Polym. Degrad. Stab. 2018, 149, 52–68. [Google Scholar] [CrossRef]
- Cacciari, I.; Quatrini, P.; Zirletta, G.; Mincione, E.; Vinciguerra, V.; Lupattelli, P.; Giovannozzi Sermanni, G. Isotactic polypropylene biodegradation by a microbial community: Physicochemical characterization of metabolites produced. Appl. Environ. Microbiol. 1993, 59, 3695–3700. [Google Scholar] [CrossRef]
- Zhang, Z.; Peng, H.; Yang, D.; Zhang, G.; Zhang, J.; Ju, F. Polyvinyl chloride degradation by a bacterium isolated from the gut of insect larvae. Nat. Commun. 2022, 13, 5360. [Google Scholar] [CrossRef]
- Nakajima-Kambe, T.; Onuma, F.; Kimpara, N.; Nakahara, T. Isolation and characterization of a bacterium which utilizes polyester polyurethane as a sole carbon and nitrogen source. FEMS Microbiol. Lett. 1995, 129, 39–42. [Google Scholar] [CrossRef]
- Akutsu-Shigeno, Y.; Adachi, Y.; Yamada, C.; Toyoshima, K.; Nomura, N.; Uchiyama, H.; Nakajima-Kambe, T. Isolation of a bacterium that degrades urethane compounds and characterization of its urethane hydrolase. Appl. Microbiol. Biotechnol. 2006, 70, 422–429. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Kim, H.R.; Lee, H.M.; Yu, H.C.; Jeon, E.; Lee, S.; Li, J.; Kim, D.H. Biodegradation of polystyrene by Pseudomonas sp. isolated from the gut of superworms (larvae of Zophobas atratus). Environ. Sci. Technol. 2020, 54, 6987–6996. [Google Scholar] [CrossRef]
- Kim, H.W.; Jo, J.H.; Kim, Y.B.; Le, T.K.; Cho, C.W.; Yun, C.H.; Chi, W.S.; Yeom, S.J. Biodegradation of polystyrene by bacteria from the soil in common environments. J. Hazard. Mater. 2021, 416, 126239. [Google Scholar] [CrossRef]
- Kumar, A.G.; Hinduja, M.; Sujitha, K.; Rajan, N.N.; Dharani, G. Biodegradation of polystyrene by deep-sea Bacillus paralicheniformis G1 and genome analysis. Sci. Total Environ. 2021, 774, 145002. [Google Scholar] [CrossRef]
- Jarerat, A.; Tokiwa, Y. Poly (L-lactide) degradation by Saccharothrix waywayandensis. Biotechnol. Lett. 2003, 25, 401–404. [Google Scholar] [CrossRef]
- Pranamuda, H.; Tokiwa, Y.; Tanaka, H. Polylactide degradation by an Amycolatopsis sp. Appl. Environ. Microbiol. 1997, 63, 1637–1640. [Google Scholar] [CrossRef]
- Jarerat, A.; Tokiwa, Y. Degradation of poly (L-lactide) by a fungus. Macromol. Biosci. 2001, 1, 136–140. [Google Scholar] [CrossRef]
- Jarerat, A.; Tokiwa, Y.; Tanaka, H. Poly (L-lactide) degradation by Kibdelosporangium aridum. Biotechnol. Lett. 2003, 25, 2035–2038. [Google Scholar] [CrossRef]
- Yeom, S.J.; Le, T.K.; Yun, C.H. P450-driven plastic-degrading synthetic bacteria. Trends Biotechnol. 2022, 40, 166–179. [Google Scholar] [CrossRef]
- Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B.; Throne-Holst, M. Bacterial metabolism of long-chain n-alkanes. Appl. Microbiol. Biotechnol. 2007, 76, 1209–1221. [Google Scholar] [CrossRef]
- Atashgahi, S.; Liebensteiner, M.G.; Janssen, D.B.; Smidt, H.; Stams, A.J.; Sipkema, D. Microbial synthesis and transformation of inorganic and organic chlorine compounds. Front. Microbiol. 2018, 9, 3079. [Google Scholar] [CrossRef]
- Temporiti, M.E.E.; Nicola, L.; Nielsen, E.; Tosi, S. Fungal enzymes involved in plastics biodegradation. Microorganisms 2022, 10, 1180. [Google Scholar] [CrossRef]
- Das, G.; Bordoloi, N.K.; Rai, S.K.; Mukherjee, A.K.; Karak, N. Biodegradable and biocompatible epoxidized vegetable oil modified thermostable poly (vinyl chloride): Thermal and performance characteristics post biodegradation with Pseudomonas aeruginosa and Achromobacter sp. J. Hazard. Mater. 2012, 209, 434–442. [Google Scholar] [CrossRef]
- Ali, M.I.; Ahmed, S.; Robson, G.; Javed, I.; Ali, N.; Atiq, N.; Hameed, A. Isolation and molecular characterization of polyvinyl chloride (PVC) plastic degrading fungal isolates. J. Basic Microbiol. 2014, 54, 18–27. [Google Scholar] [CrossRef]
- Park, W.J.; Hwangbo, M.; Chu, K.H. Plastisphere and microorganisms involved in polyurethane biodegradation. Sci. Total Environ. 2023, 886, 163932. [Google Scholar] [CrossRef]
- Magnin, A.; Pollet, E.; Phalip, V.; Avérous, L. Evaluation of biological degradation of polyurethanes. Biotechnol. Adv. 2020, 39, 107457. [Google Scholar] [CrossRef]
- Liu, J.; He, J.; Xue, R.; Xu, B.; Qian, X.; Xin, F.; Blank, L.M.; Zhou, J.; Wei, R.; Dong, W. Biodegradation and up-cycling of polyurethanes: Progress, challenges, and prospects. Biotechnol. Adv. 2021, 48, 107730. [Google Scholar] [CrossRef]
- Howard, G.T.; Crother, B.; Vicknair, J. Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas chlororaphis. Int. Biodeterior. Biodegrad. 2001, 47, 141–149. [Google Scholar] [CrossRef]
- Nomura, N.; Shigeno-Akutsu, Y.; Nakajima-Kambe, T.; Nakahara, T. Cloning and sequence analysis of a polyurethane esterase of Comamonas acidovorans TB-35. J. Ferment. Bioeng. 1998, 86, 339–345. [Google Scholar] [CrossRef]
- do Canto, V.P.; Thompson, C.E.; Netz, P.A. Polyurethanases: Three-dimensional structures and molecular dynamics simulations of enzymes that degrade polyurethane. J. Mol. Graph. Model. 2019, 89, 82–95. [Google Scholar] [CrossRef]
- Chen, L.; Rong, M.; Yang, L.; Yu, J.; Qu, H.; Meng, Q.; Ni, S.; Xu, Z.; Zhu, X.; Wang, L. Construction of super-hydrophobic hypercrosslinked porous polymers for selectively removing aromatic diamines from the polyurethane bio-hydrolysate. Chem. Eng. J. 2022, 428, 132509. [Google Scholar] [CrossRef]
- Jansen, B.; Schumacher-Perdreau, F.; Peters, G.; Pulverer, G. Evidence for degradation of synthetic polyurethanes by Staphylococcus epidermidis. Zentralbl. Bakteriol. 1991, 276, 36–45. [Google Scholar] [CrossRef]
- Kushwaha, A.; Goswami, L.; Singhvi, M.; Kim, B.S. Biodegradation of poly (ethylene terephthalate): Mechanistic insights, advances, and future innovative strategies. Chem. Eng. J. 2023, 457, 141230. [Google Scholar]
- Pirillo, V.; Pollegioni, L.; Molla, G. Analytical methods for the investigation of enzyme-catalyzed degradation of polyethylene terephthalate. FEBS J. 2021, 288, 4730–4745. [Google Scholar] [CrossRef]
- Meereboer, K.W.; Misra, M.; Mohanty, A.K. Review of recent advances in the biodegradability of polyhydroxyalkanoate (PHA) bioplastics and their composites. Green Chem. 2020, 22, 5519–5558. [Google Scholar] [CrossRef]
- Palm, G.J.; Reisky, L.; Böttcher, D.; Müller, H.; Michels, E.A.; Walczak, M.C.; Berndt, L.; Weiss, M.S.; Bornscheuer, U.T.; Weber, G. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat. Commun. 2019, 10, 1717. [Google Scholar] [CrossRef]
- Franden, M.A.; Jayakody, L.N.; Li, W.J.; Wagner, N.J.; Cleveland, N.S.; Michener, W.E.; Hauer, B.; Blank, L.M.; Wierckx, N.; Klebensberger, J. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018, 48, 197–207. [Google Scholar] [CrossRef]
- Sasoh, M.; Masai, E.; Ishibashi, S.; Hara, H.; Kamimura, N.; Miyauchi, K.; Fukuda, M. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 2006, 72, 1825–1832. [Google Scholar] [CrossRef]
- Hou, L.; Majumder, E.L.W. Potential for and distribution of enzymatic biodegradation of polystyrene by environmental microorganisms. Materials 2021, 14, 503. [Google Scholar] [CrossRef]
- Chow, J.; Perez-Garcia, P.; Dierkes, R.; Streit, W.R. Microbial enzymes will offer limited solutions to the global plastic pollution crisis. Microb. Biotechnol. 2023, 16, 195–217. [Google Scholar] [CrossRef]
- Tomita, K.; Nakajima, T.; Kikuchi, Y.; Miwa, N. Degradation of poly (L-lactic acid) by a newly isolated thermophile. Polym. Degrad. Stab. 2004, 84, 433–438. [Google Scholar] [CrossRef]
- Tokiwa, Y.; Calabia, B.P. Biodegradability and biodegradation of polyesters. J. Polym. Environ. 2007, 15, 259–267. [Google Scholar] [CrossRef]
- Jarerat, A.; Tokiwa, Y.; Tanaka, H. Microbial poly (L-lactide)-degrading enzyme induced by amino acids, peptides, and poly (L-amino acids). J. Polym. Environ. 2004, 12, 139–146. [Google Scholar] [CrossRef]
- Urbanek, A.K.; Mirończuk, A.M.; García-Martín, A.; Saborido, A.; de la Mata, I.; Arroyo, M. Biochemical properties and biotechnological applications of microbial enzymes involved in the degradation of polyester-type plastics. Biochim. Biophys. Acta-Proteins Proteom. 2020, 1868, 140315. [Google Scholar] [CrossRef]
- Shah, A.A.; Eguchi, T.; Mayumi, D.; Kato, S.; Shintani, N.; Kamini, N.R.; Nakajima-Kambe, T. Degradation of aliphatic and aliphatic–aromatic co-polyesters by depolymerases from Roseateles depolymerans strain TB-87 and analysis of degradation products by LC-MS. Polym. Degrad. Stab. 2013, 98, 2722–2729. [Google Scholar] [CrossRef]
- Zhao, J.H.; Wang, X.Q.; Zeng, J.; Yang, G.; Shi, F.H.; Yan, Q. Biodegradation of poly (butylene succinate) in compost. J. Appl. Polym. Sci. 2005, 97, 2273–2278. [Google Scholar] [CrossRef]
- Kim, S.H.; Cho, J.Y.; Hwang, J.H.; Kim, H.J.; Oh, S.J.; Kim, H.J.; Bhatia, S.K.; Yun, J.; Lee, S.H.; Yang, Y.H. Revealing the key gene involved in bioplastic degradation from superior bioplastic degrader Bacillus sp. JY35. Int. J. Biol. Macromol. 2023, 244, 125298. [Google Scholar] [CrossRef]
- Sangale, M.K.; Shahnawaz, M.; Ade, A. A review on biodegradation of polythene: The microbial approach. J. Bioremed. Biodeg. 2012, 3, 1–9. [Google Scholar] [CrossRef]
- Er CT, X.; Sen, L.Z.; Srinophakun, P.; Wei, O.C. Recent advances and challenges in sustainable management of plastic waste using biodegradation approach. Bioresour. Technol. 2023, 374, 128772. [Google Scholar]
- Lee, S.; Lee, Y.R.; Kim, S.J.; Lee, J.S.; Min, K. Recent advances and challenges in the biotechnological upcycling of plastic wastes for constructing a circular bioeconomy. Chem. Eng. J. 2023, 454, 140470. [Google Scholar] [CrossRef]
- Ezeji, T.; Milne, C.; Price, N.D.; Blaschek, H.P. Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol-producing microorganisms. Appl. Microbiol. Biotechnol. 2010, 85, 1697–1712. [Google Scholar] [CrossRef]
- Lu, C.; Leitner, N.; Wijffels, R.H.; Dos Santos VA, M.; Weusthuis, R.A. Microbial production of medium-chain-length α, ω-diols via two-stage process under mild conditions. Bioresour. Technol. 2022, 352, 127111. [Google Scholar] [CrossRef]
- Gregory, G.J.; Wang, C.; Sadula, S.; Koval, S.; Lobo, R.F.; Vlachos, D.G.; Papoutsakis, E.T. Polyethylene valorization by combined chemical catalysis with bioconversion by plastic-enriched microbial consortia. ACS Sustain. Chem. Eng. 2023, 11, 3494–3505. [Google Scholar] [CrossRef]
- Black, P.N.; DiRusso, C.C. Transmembrane movement of exogenous long-chain fatty acids: Proteins, enzymes, and vectorial esterification. Microbiol. Mol. Biol. Rev. 2003, 67, 454–472. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, R.; Zhang, M.; Gao, H. Native efflux pumps of Escherichia coli responsible for short and medium chain alcohol. Biochem. Eng. J. 2018, 133, 149–156. [Google Scholar] [CrossRef]
- Patra, M.; Salonen, E.; Terama, E.; Vattulainen, I.; Faller, R.; Lee, B.W.; Holopainen, J.; Karttunen, M. Under the influence of alcohol: The effect of ethanol and methanol on lipid bilayers. Biophys. J. 2006, 90, 1121–1135. [Google Scholar] [CrossRef]
- Round, J.; Roccor, R.; Li, S.N.; Eltis, L.D. A fatty acyl coenzyme A reductase promotes wax ester accumulation in Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2017, 83, e00902–e00917. [Google Scholar] [CrossRef]
- Wilbanks, B.; Trinh, C.T. Comprehensive characterization of toxicity of fermentative metabolites on microbial growth. Biotechnol. Biofuels 2017, 10, 262. [Google Scholar] [CrossRef]
- Togashi, N.; Shiraishi, A.; Nishizaka, M.; Matsuoka, K.; Endo, K.; Hamashima, H.; Inoue, Y. Antibacterial activity of long-chain fatty alcohols against Staphylococcus aureus. Molecules 2007, 12, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Casillas-Vargas, G.; Ocasio-Malavé, C.; Medina, S.; Morales-Guzmán, C.; Del Valle, R.G.; Carballeira, N.M.; Sanabria-Ríos, D.J. Antibacterial fatty acids: An update of possible mechanisms of action and implications in the development of the next-generation of antibacterial agents. Prog. Lipid Res. 2021, 82, 101093. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cai, B.; Shao, Z. Oil degradation and biosurfactant production by the deep sea bacterium Dietzia maris As-13-3. Front. Microbiol. 2014, 5, 711. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, W.; Gao, J.; Ren, C.; Xu, Y. Revealing the characteristics of glucose-and lactate-based chain elongation for caproate production by Caproicibacterium lactatifermentans through transcriptomic, bioenergetic, and regulatory analyses. Msystems 2022, 7, e00534-22. [Google Scholar] [CrossRef]
- Kang, S.; Kim, H.; Jeon, B.S.; Choi, O.; Sang, B.I. Chain elongation process for caproate production using lactate as electron donor in Megasphaera hexanoica. Bioresour. Technol. 2022, 346, 126660. [Google Scholar] [CrossRef]
- Warnecke, T.; Gill, R.T. Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Microb. Cell Factories 2005, 4, 25. [Google Scholar] [CrossRef]
- Nduko, J.M.; Matsumoto, K.I.; Ooi, T.; Taguchi, S. Effectiveness of xylose utilization for high yield production of lactate-enriched P (lactate-co-3-hydroxybutyrate) using a lactate-overproducing strain of Escherichia coli and an evolved lactate-polymerizing enzyme. Metab. Eng. 2013, 15, 159–166. [Google Scholar] [CrossRef]
- Yamada, M.; Matsumoto, K.I.; Uramoto, S.; Motohashi, R.; Abe, H.; Taguchi, S. Lactate fraction dependent mechanical properties of semitransparent poly(lactate-co-3-hydroxybutyrate)s produced by control of lactyl-CoA monomer fluxes in recombinant Escherichia coli. J. Biotechnol. 2011, 154, 255–260. [Google Scholar] [CrossRef]
- Yang, T.H.; Kim, T.W.; Kang, H.O.; Lee, S.H.; Lee, E.J.; Lim, S.C.; Oh, S.O.; Song, A.J.; Park, S.J.; Lee, S.Y. Biosynthesis of polylactic acid and its copolymers using evolved propionate CoA transferase and PHA synthase. Biotechnol. Bioeng. 2010, 105, 150–160. [Google Scholar] [CrossRef]
- Jung, Y.K.; Kim, T.Y.; Park, S.J.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol. Bioeng. 2010, 105, 161–171. [Google Scholar] [CrossRef]
- Zu, T.N.; Athamneh, A.I.; Senger, R.S. Characterizing the phenotypic responses of Escherichia coli to multiple 4-carbon alcohols with Raman spectroscopy. Fermentation 2016, 2, 3. [Google Scholar] [CrossRef]
- Ahn, J.H.; Seo, H.; Park, W.; Seok, J.; Lee, J.A.; Kim, W.J.; Kim, G.B.; Kim, K.J.; Lee, S.Y. Enhanced succinic acid production by Mannheimia employing optimal malate dehydrogenase. Nat. Commun. 2020, 11, 1970. [Google Scholar] [CrossRef] [PubMed]
- Li, W.J.; Narancic, T.; Kenny, S.T.; Niehoff, P.J.; O’Connor, K.; Blank, L.M.; Wierckx, N. Unraveling 1, 4-butanediol metabolism in Pseudomonas putida KT2440. Front. Microbiol. 2020, 11, 382. [Google Scholar] [CrossRef]
- Qian, X.; Xin, K.; Zhang, L.; Zhou, J.; Xu, A.; Dong, W.; Jiang, M. Integration of ARTP mutation and adaptive laboratory evolution to reveal 1,4-butanediol degradation in Pseudomonas putida KT2440. Microbiol. Spectr. 2023, 11, e04988-22. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Jang, Y.S.; Lee, S.Y. Production of succinic acid by metabolically engineered microorganisms. Curr. Opin. Biotechnol. 2016, 42, 54–66. [Google Scholar] [CrossRef]
- Ito, Y.; Hirasawa, T.; Shimizu, H. Metabolic engineering of Saccharomyces cerevisiae to improve succinic acid production based on metabolic profiling. Biosci. Biotechnol. Biochem. 2014, 78, 151–159. [Google Scholar] [CrossRef]
- Guo, H.; Liu, H.; Jin, Y.; Zhang, R.; Yu, Y.; Deng, L.; Wang, F. Advances in research on the bio-production of 1, 4-butanediol by the engineered microbes. Biochem. Eng. J. 2022, 185, 108478. [Google Scholar] [CrossRef]
- Kämpf, M.M.; Thöny-Meyer, L.; Ren, Q. Biosynthesis of poly(4-hydroxybutyrate) in recombinant Escherichia coli grown on glycerol is stimulated by propionic acid. Int. J. Biol. Macromol. 2014, 71, 8–13. [Google Scholar] [CrossRef]
- Kudo, H.; Ono, S.; Abe, K.; Matsuda, M.; Hasunuma, T.; Nishizawa, T.; Asayama, M.; Nishihara, H.; Chohnan, S. Enhanced supply of acetyl-CoA by exogenous pantothenate kinase promotes synthesis of poly(3-hydroxybutyrate). Microb. Cell Fact. 2023, 22, 75. [Google Scholar] [CrossRef]
- Utomo RN, C.; Li, W.J.; Tiso, T.; Eberlein, C.; Doeker, M.; Heipieper, H.J.; Jupke, A.; Wierckx, N.; Blank, L.M. Defined microbial mixed culture for utilization of polyurethane monomers. ACS Sustain. Chem. Eng. 2020, 8, 17466–17474. [Google Scholar] [CrossRef]
- Dissanayake, L.; Jayakody, L.N. Engineering microbes to bio-upcycle polyethylene terephthalate. Front. Bioeng. Biotechnol. 2021, 9, 656465. [Google Scholar] [CrossRef]
- Moghayedi, M.; Ahmadzadeh, H.; Ghazvini, K.; Goharshadi, E. Neglected antibacterial activity of ethylene glycol as a common solvent. Microb. Pathog. 2017, 107, 457–461. [Google Scholar] [CrossRef]
- Pandit, A.V.; Harrison, E.; Mahadevan, R. Engineering Escherichia coli for the utilization of ethylene glycol. Microb. Cell Factories 2021, 20, 22. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhou, Y.; Wang, T.; Too, H.P.; Wang, D.I.; Li, Z. Highly regio-and enantioselective multiple oxy-and amino-functionalizations of alkenes by modular cascade biocatalysis. Nat. Commun. 2016, 7, 11917. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.P.; Niu, F.X.; Yan, Z.B.; Fong, L.S.; Huang, Y.B.; Liu, J.Z. Recent advances in metabolically engineered microorganisms for the production of aromatic chemicals derived from aromatic amino acids. Front. Bioeng. Biotechnol. 2020, 8, 407. [Google Scholar] [CrossRef]
- Lukito, B.R.; Sekar, B.S.; Wu, S.; Li, Z. Whole cell-based cascade biotransformation for the production of (S)-mandelic acid from styrene, L-phenylalanine, glucose, or glycerol. Adv. Synth. Catal. 2019, 361, 3560–3568. [Google Scholar] [CrossRef]
- Domínguez-Cuevas, P.; González-Pastor, J.E.; Marqués, S.; Ramos, J.L.; de Lorenzo, V. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J. Biol. Chem. 2006, 281, 11981–11991. [Google Scholar] [CrossRef]
- Arshad, A.; Ashraf, B.; Ali, I.; Jamil, N. Biosynthesis of polyhydroxyalkanoates from styrene by Enterobacter spp. isolated from polluted environment. Front. Biol. 2017, 12, 210–218. [Google Scholar] [CrossRef]
- Zimmerling, J.; Oelschlägel, M.; Großmann, C.; Voitel, M.; Schlömann, M.; Tischler, D. Biochemical characterization of phenylacetaldehyde dehydrogenases from styrene-degrading soil bacteria. Appl. Biochem. Biotechnol. 2021, 193, 650–667. [Google Scholar] [CrossRef]
- Karlsson, E.; Mapelli, V.; Olsson, L. Adipic acid tolerance screening for potential adipic acid production hosts. Microb. Cell Factories 2017, 16, 20. [Google Scholar] [CrossRef]
- Ackermann, Y.S.; Li, W.J.; Op de Hipt, L.; Niehoff, P.J.; Casey, W.; Polen, T.; Köbbing, S.; Ballerstedt, H.; Wynands, B.; O’Connor, K.; et al. Engineering adipic acid metabolism in Pseudomonas putida. Metab. Eng. 2021, 67, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, G.; Li, A.; Deng, Y. Directed synthesis of biobased 1, 6-diaminohexane from adipic acid by rational regulation of a functional enzyme cascade in Escherichia coli. ACS Sustain. Chem. Eng. 2023, 11, 6011–6020. [Google Scholar] [CrossRef]
- Chia, Y.C.; Rajbanshi, R.; Calhoun, C.; Chiu, R.H. Anti-neoplastic effects of gallic acid, a major component of Toona sinensis leaf extract, on oral squamous carcinoma cells. Molecules 2010, 15, 8377–8389. [Google Scholar] [CrossRef]
- Badhani, B.; Sharma, N.; Kakkar, R. Gallic acid: A versatile antioxidant with promising therapeutic and industrial applications. Rsc Adv. 2015, 5, 27540–27557. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.K.; Cha, H.G.; Kang, M.J.; Lee, H.S.; Khang, T.U.; Yun, E.J.; Lee, D.H.; Song, B.K.; Park, S.J. Biological valorization of poly(ethylene terephthalate) monomers for upcycling waste PET. ACS Sustain. Chem. Eng. 2019, 7, 19396–19406. [Google Scholar] [CrossRef]
- Luo, Z.W.; Choi, K.R.; Lee, S.Y. Improved terephthalic acid production from p-xylene using metabolically engineered Pseudomonas Putida. Metab. Eng. 2023, 76, 75–86. [Google Scholar] [CrossRef]
- Tang, H.; Wang, A.; Salley, S.O.; Ng, K.S. The effect of natural and synthetic antioxidants on the oxidative stability of biodiesel. J. Am. Oil Chem. Soc. 2008, 85, 373–382. [Google Scholar] [CrossRef]
- Johnson, C.W.; Salvachúa, D.; Khanna, P.; Smith, H.; Peterson, D.J.; Beckham, G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016, 3, 111–119. [Google Scholar] [CrossRef]
- Wang, J.; Shen, X.; Wang, J.; Yang, Y.; Yuan, Q.; Yan, Y. Exploring the promiscuity of phenol hydroxylase from Pseudomonas stutzeri OX1 for the biosynthesis of phenolic compounds. ACS Synth. Biol. 2018, 7, 1238–1243. [Google Scholar] [CrossRef]
- Gao, R.; Pan, H.; Kai, L.; Han, K.; Lian, J. Microbial degradation and valorization of poly(ethylene terephthalate) (PET) monomers. World J. Microbiol. Biotechnol. 2022, 38, 89. [Google Scholar] [CrossRef]
- Werner, A.Z.; Clare, R.; Mand, T.D.; Pardo, I.; Ramirez, K.J.; Haugen, S.J.; Bratti, F.; Dexter, G.N.; Elmore, J.R.; Huenemann, J.D. Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 2021, 67, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Rorrer, N.A.; Notonier, S.F.; Knott, B.C.; Black, B.A.; Singh, A.; Nicholson, S.R.; Kinchin, C.P.; Schmidt, G.P.; Carpenter, A.C.; Ramirez, K.J.; et al. Production of β-ketoadipic acid from glucose in Pseudomonas putida KT2440 for use in performance-advantaged nylons. Cell Rep. Phys. Sci. 2022, 3, 100840. [Google Scholar] [CrossRef]
- Wu, J.H.; Wu, F.Y.; Chuang, H.P.; Chen, W.Y.; Huang, H.J.; Chen, S.H.; Liu, W.T. Community and proteomic analysis of methanogenic consortia degrading terephthalate. Appl. Environ. Microbiol. 2013, 79, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef]
- Kumar, A.; Wang, L.; Ng, C.Y.; Maranas, C.D. Pathway design using de novo steps through uncharted biochemical spaces. Nat. Commun. 2018, 9, 184. [Google Scholar] [CrossRef]
- Delépine, B.; Duigou, T.; Carbonell, P.; Faulon, J.L. RetroPath2.0: A retrosynthesis workflow for metabolic engineers. Metab. Eng. 2018, 45, 158–170. [Google Scholar] [CrossRef]
- Duigou, T.; Du Lac, M.; Carbonell, P.; Faulon, J.L. RetroRules: A database of reaction rules for engineering biology. Nucleic Acids Res. 2019, 47, D1229–D1235. [Google Scholar] [CrossRef]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Schwaneberg, U.; Roccatano, D. Computer-aided protein directed evolution: A review of web servers, databases and other computational tools for protein engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209008. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.K.; Wu, Z.; Arnold, F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Bennett, N.R.; Coventry, B.; Goreshnik, I.; Huang, B.; Allen, A.; Vafeados, D.; Peng, Y.P.; Dauparas, J.; Baek, M.; Stewart, L. Improving de novo protein binder design with deep learning. Nat. Commun. 2023, 14, 2625. [Google Scholar] [CrossRef]
| Enzyme 1 | Microorganism | Localization | Degradation Condition 2 | Results | Ref. |
|---|---|---|---|---|---|
| Polyethylene and Polypropylene | |||||
| Cutinase | Thermobifida fusca WSH03-11 | Extracellular | 60 °C for 7 days using pretreated PE films |
| [14] |
| Lipase | Halomonas sp. | Extracellular | 30 °C for 10 days using LDPE films |
| [15] |
| Esterase | Exiguobacterium sp. Halomonas sp. Ochrobactrum sp. | Extracellular Extracellular Extracellular | 30 °C for 24 h using PE films |
| [16] |
| P450 | Bacillus thuringiensis JNU01 | Cytoplasm | 37 °C for 18 h using PE powder |
| [10] |
| Alkane monooxygenase | Pseudomonas aeruginosa E7 | Membrane-bound | 37 °C for 80 days using low-molecular-weight PE powder |
| [17] |
| Laccase | Botrytis aclada Bacillus subtilis | Extracellular Membrane-bound | 30 °C for 5 days using mediator and pretreated PE films |
| [18] |
| Polyurethane | |||||
| Laccase | Trametes versicolor | Extracellular | 37 °C for 18 days using PCL-PU cubes and coatings |
| [19] |
| Cutinase (TfCut2) | Thermobifida fusca KW3 | Extracellular | 70 °C for 200 h using PU cubes |
| [20] |
| Esterase | Comamonas acidovorans TB-35 | Extracellular | 30 °C for 24 h using PU cubes |
| [21] |
| Polyethylene terephthalate | |||||
| PETase | Ideonella sakaiensis 201-F6 | Extracellular | 37 °C for 72 h using PET films |
| [22] |
| Cutinase (LCCWCCG) | Leaf-branch compost metagenome | Extracellular | 72 °C for 10.5 h using pretreated PET |
| [23] |
| Cutinase (PE-HY250S) | Pseudomonas aestusnigri VGXO14T | Extracellular | 30 °C for 48 h using PET films |
| [24] |
| Cutinase (TfCaWA) | Thermobifida fusca KW3 | Extracellular | 50 °C for 1 h using 1 mM of cyclic PET trimer |
| [25] |
| BHETase (ΔBsEst) | Bacillus subtilis PET-86 | Extracellular | 30 °C for 3 h using 5 mM of BHET |
| [26] |
| BHETase (ΔChryBHETase) | Chryseobacterium sp. PET-29 | Extracellular | 30 °C for 9 h using 5 mM of BHET |
| [26] |
| MHETase (MHETaseR411K/S416A/F424I) | Ideonella sakaiensis 201-F6 | Extracellular | 30 °C for 1 h using 2.5 μM of PET pentamer |
| [27] |
| Polystyrene | |||||
| Lipase and esterase | Pseudomonas spp. Bacillus spp. | Extracellular | Treated with culture supernatant, 30 °C for 4 days, using 2 mg/mL of emulsified HIPS |
| [28] |
| Hydroquinone peroxidase | Azotobacter beijerinckii HM121 | Extracellular | 30 °C for 10 min using 2 g/L of PS |
| [29] |
| Polylactic acid | |||||
| PLA depolymerase | Paenibacillus amylolyticus strain TB-13 | Extracellular | 37 °C for 6 h using 0.5% of emulsified PLA |
| [30] |
| Proteinase K | Tritirachium album | Extracellular | 37 °C for 6 h using 0.5% of emulsified PLA |
| [30] |
| Lipase | Pseudomonas sp. strain DS04-T | Extracellular | 50 °C for 3 h using 1 g/L of emulsified PLA |
| [31] |
| Polybutylene succinate | |||||
| Cutinase | Fusarium solani | Extracellular | 37 °C for 26 h, pretreated PBS films |
| [32] |
| Lipase (Lipase Asahi) | Chromobacterium viscosum | Extracellular | 37 °C for 17 days using PBS films |
| [33] |
| Lipase (Lipase PS®) | Pseudomonas cepacia | Extracellular | 50 °C for 7 days using PBS films |
| [34] |
| Microorganism | Degradation Condition | Results | Ref. |
|---|---|---|---|
| Polyethylene and Polypropylene | |||
| Pseudomonas aeruginosa WGH-6 | Modified mineral salt medium containing PP particles at 30 °C for 40 days |
| [35] |
| Microbacterium paraoxydans | Minimal broth containing 0.25 g of pretreated LDPE powder at room temperature for 2 months |
| [36] |
| Pseudomonas aeruginosa | Minimal medium containing 0.25 g of pretreated LDPE powder at RT for 2 months |
| [36] |
| Alternaria alternata FB1 | Basic medium containing PE films for 28 days |
| [37] |
| Aneurinibacillus spp. Brevibacillus spp. | Minimal medium containing LDPE, HDPE, and PP films and pellets at 50 °C for 140 days |
| [38] |
| Pseudomonas Vibrio Aspergillus niger | B7 medium supplemented with starch containing PE strips at 30 °C for 175 days |
| [39] |
| Polyvinyl chloride | |||
| Klebsiella sp. EMBL-1 | Mineral salt medium containing PVC films at 30 °C for 90 days |
| [40] |
| Polyurethane | |||
| Comamonas acidovorans | Basal medium containing PU cubes at 30 °C for 7 days |
| [41] |
| Rhodococcus equi TB-60 | Mineral medium containing TDCB at 30 °C for 10 days |
| [42] |
| Polyethylene terephthalate | |||
| Ideonella sakaiensis 201-F6 | Yeast extract–sodium carbonate– vitamins medium containing PET films at 30 °C for 13 days |
| [43] |
| Polystyrene | |||
| Pseudomonas aeruginosa DSM 50071 | Liquid carbon-free basal medium containing PS films at 25 °C for 60 days |
| [44] |
| Acinetobacter johnsoniii JNU01 | Basal medium containing PS powder at 28 °C for 7 days |
| [45] |
| Bacillus paralicheniformis G1 | Mineral salt medium containing PS films at 30 °C for 60 days |
| [46] |
| Polylactic acid | |||
| Saccharothrix waywayandensis | Basal medium supplemented with 0.1% gelatin containing 100 mg of PLA films at 30 °C for 7 days |
| [47] |
| Amycolatopsis HT-32 | Basal medium containing 100 mg of pretreated PLA films at 30 °C for 14 days |
| [48] |
| Tritirachium album ATCC 22563 | Basal medium supplemented with 0.1% gelatin containing 100 mg of PLA films at 30 °C for 14 days |
| [49] |
| Kibdelosporangium aridum | Basal medium supplemented with 0.1% gelatin containing 100 mg of high-molecular-weight PLA films at 30 °C for 10 days |
| [50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, G.H.; Kim, D.-W.; Jin, Y.H.; Kim, S.M.; Lim, E.S.; Cha, M.J.; Ko, J.K.; Gong, G.; Lee, S.-M.; Um, Y.; et al. Biotechnological Plastic Degradation and Valorization Using Systems Metabolic Engineering. Int. J. Mol. Sci. 2023, 24, 15181. https://doi.org/10.3390/ijms242015181
Lee GH, Kim D-W, Jin YH, Kim SM, Lim ES, Cha MJ, Ko JK, Gong G, Lee S-M, Um Y, et al. Biotechnological Plastic Degradation and Valorization Using Systems Metabolic Engineering. International Journal of Molecular Sciences. 2023; 24(20):15181. https://doi.org/10.3390/ijms242015181
Chicago/Turabian StyleLee, Ga Hyun, Do-Wook Kim, Yun Hui Jin, Sang Min Kim, Eui Seok Lim, Min Ji Cha, Ja Kyong Ko, Gyeongtaek Gong, Sun-Mi Lee, Youngsoon Um, and et al. 2023. "Biotechnological Plastic Degradation and Valorization Using Systems Metabolic Engineering" International Journal of Molecular Sciences 24, no. 20: 15181. https://doi.org/10.3390/ijms242015181
APA StyleLee, G. H., Kim, D.-W., Jin, Y. H., Kim, S. M., Lim, E. S., Cha, M. J., Ko, J. K., Gong, G., Lee, S.-M., Um, Y., Han, S. O., & Ahn, J. H. (2023). Biotechnological Plastic Degradation and Valorization Using Systems Metabolic Engineering. International Journal of Molecular Sciences, 24(20), 15181. https://doi.org/10.3390/ijms242015181





