Engineering and Expression Strategies for Optimization of L-Asparaginase Development and Production
Abstract
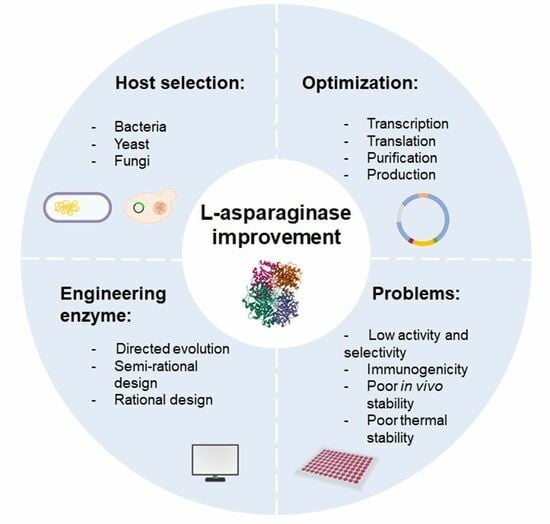
:1. Introduction
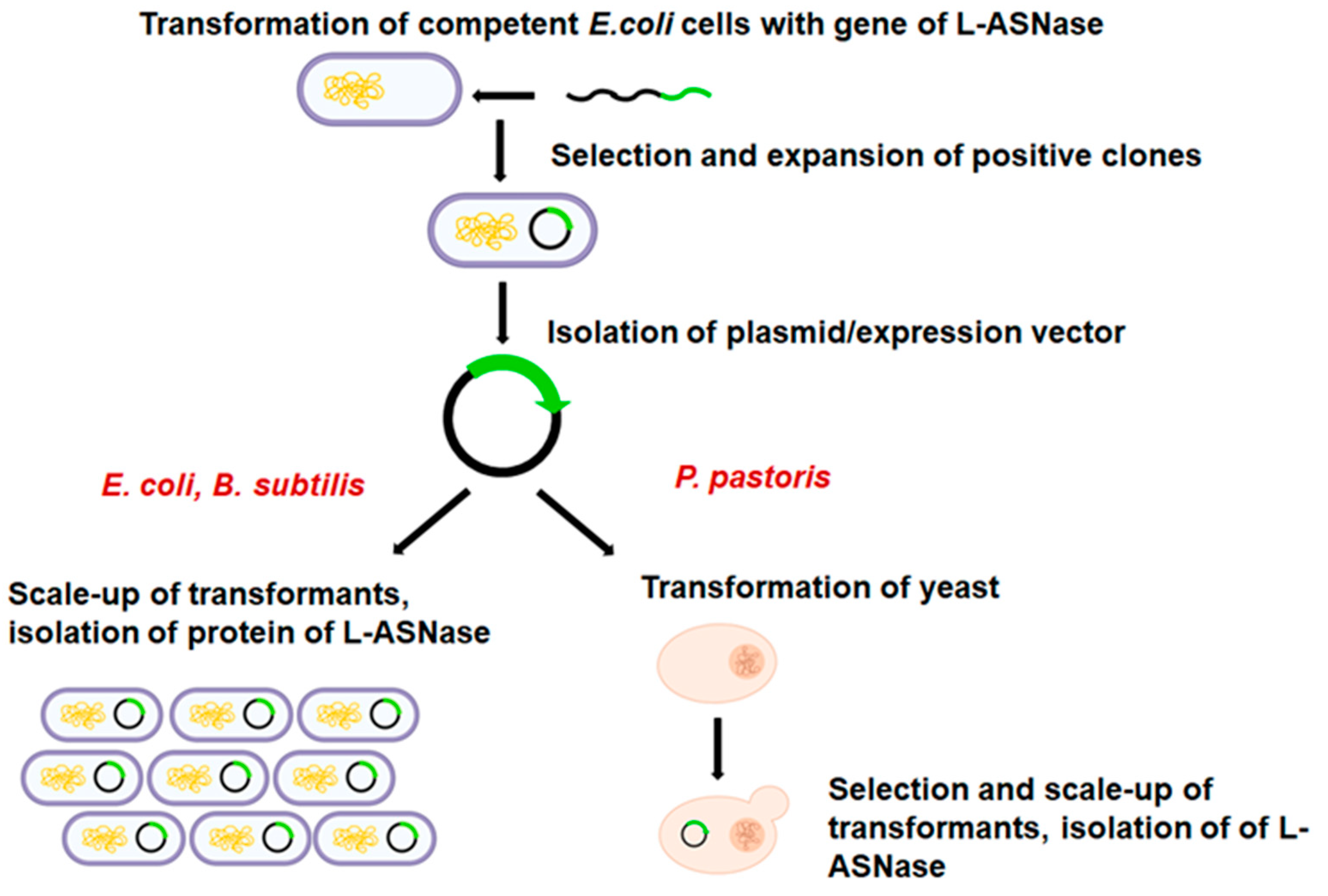
2. Host Systems for Expression of L-Asparaginase
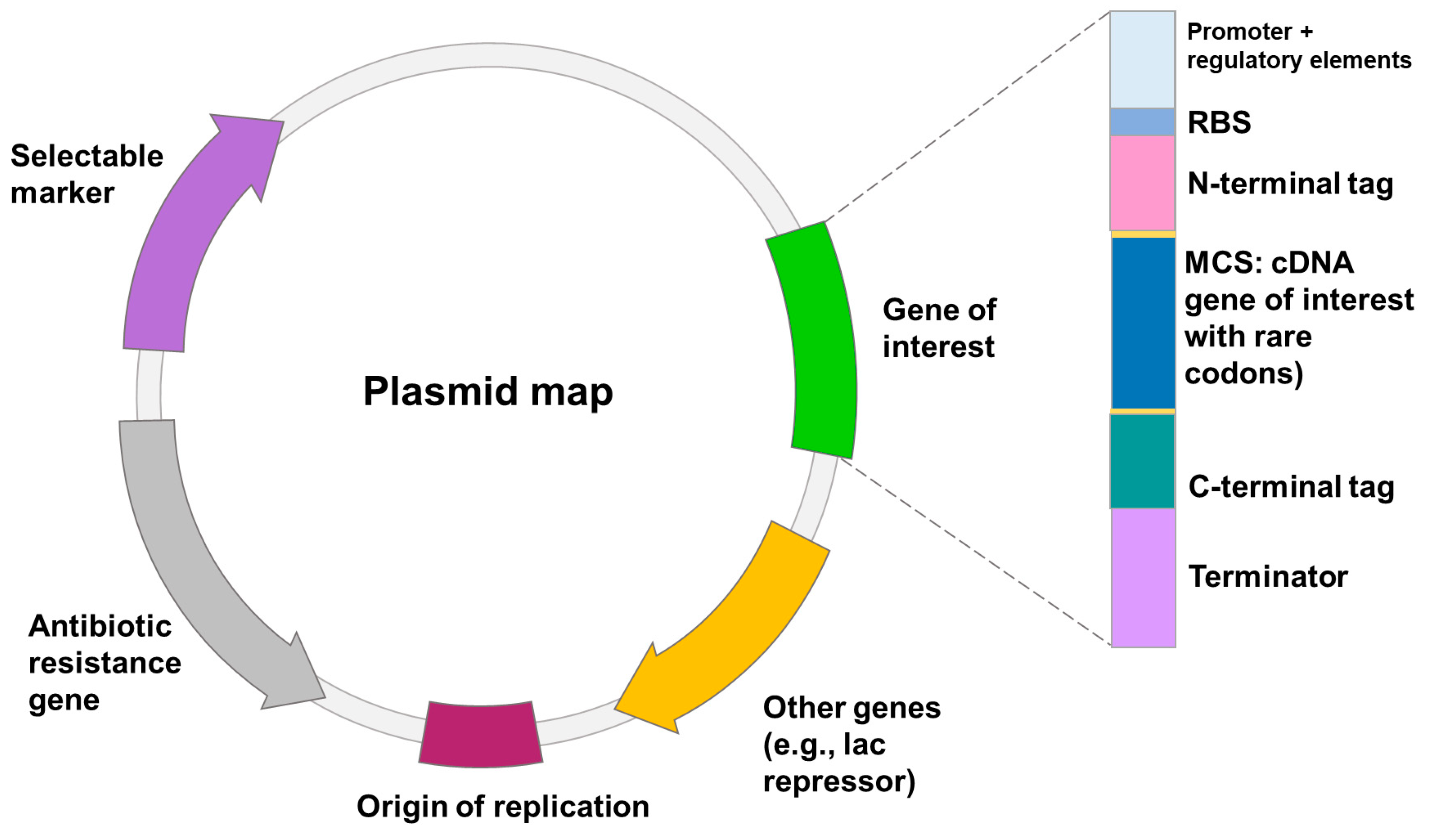
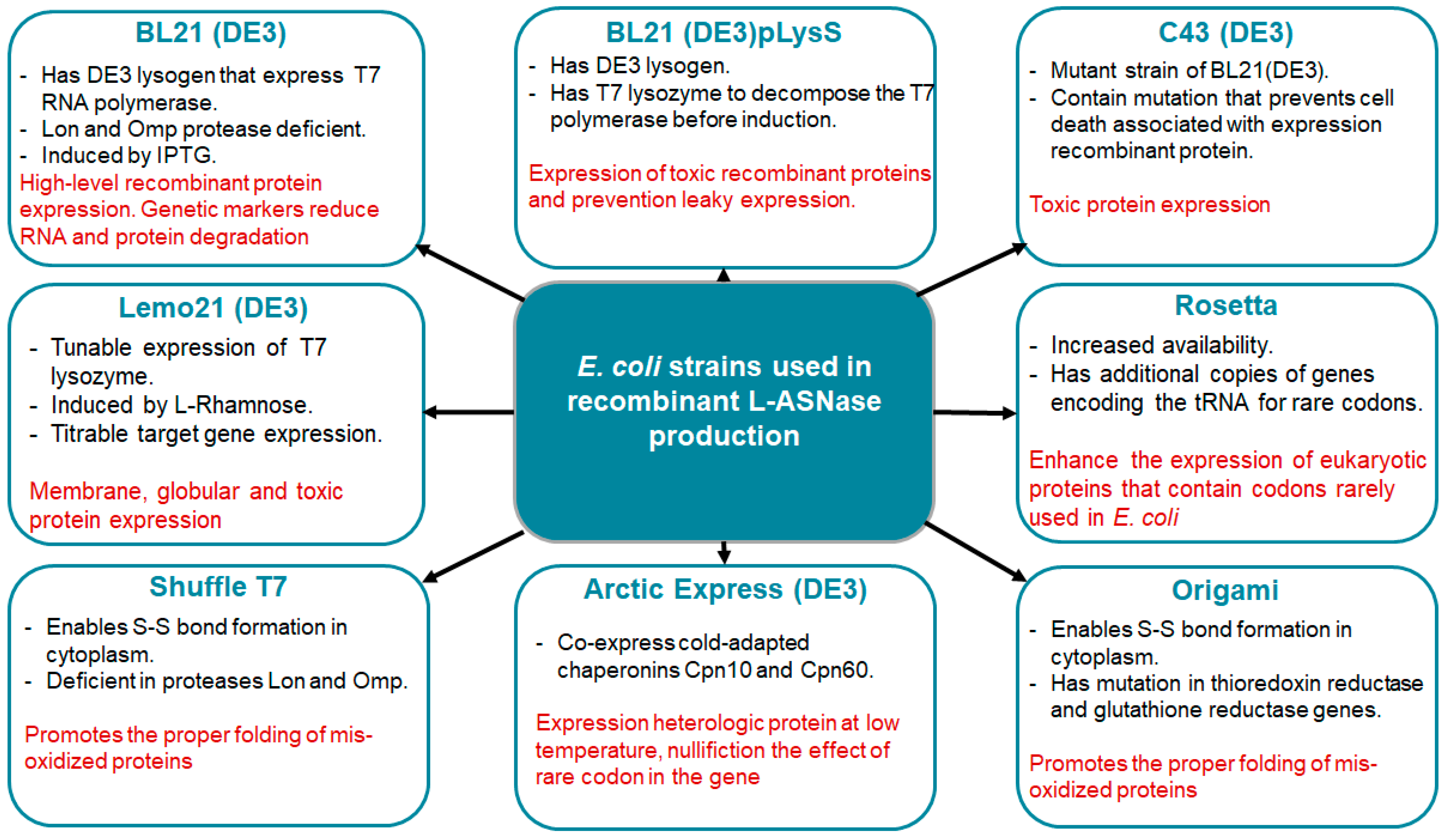
2.1. Escherichia coli Expression System
2.1.1. Optimization of Promoter Strength
2.1.2. Signal Sequence
2.1.3. Codon Optimization
2.1.4. Fusion Tags
2.1.5. Co-Expression with Chaperones
2.1.6. Optimization of L-Asparaginase Production in Escherichia coli
2.2. Bacillus subtilis Expression System
2.2.1. Promoter Engineering
2.2.2. Plasmid Engineering
2.2.3. Enhancement of Protein Secretion Level
2.2.4. Optimization of L-Asparaginase Production in Bacillus subtilis
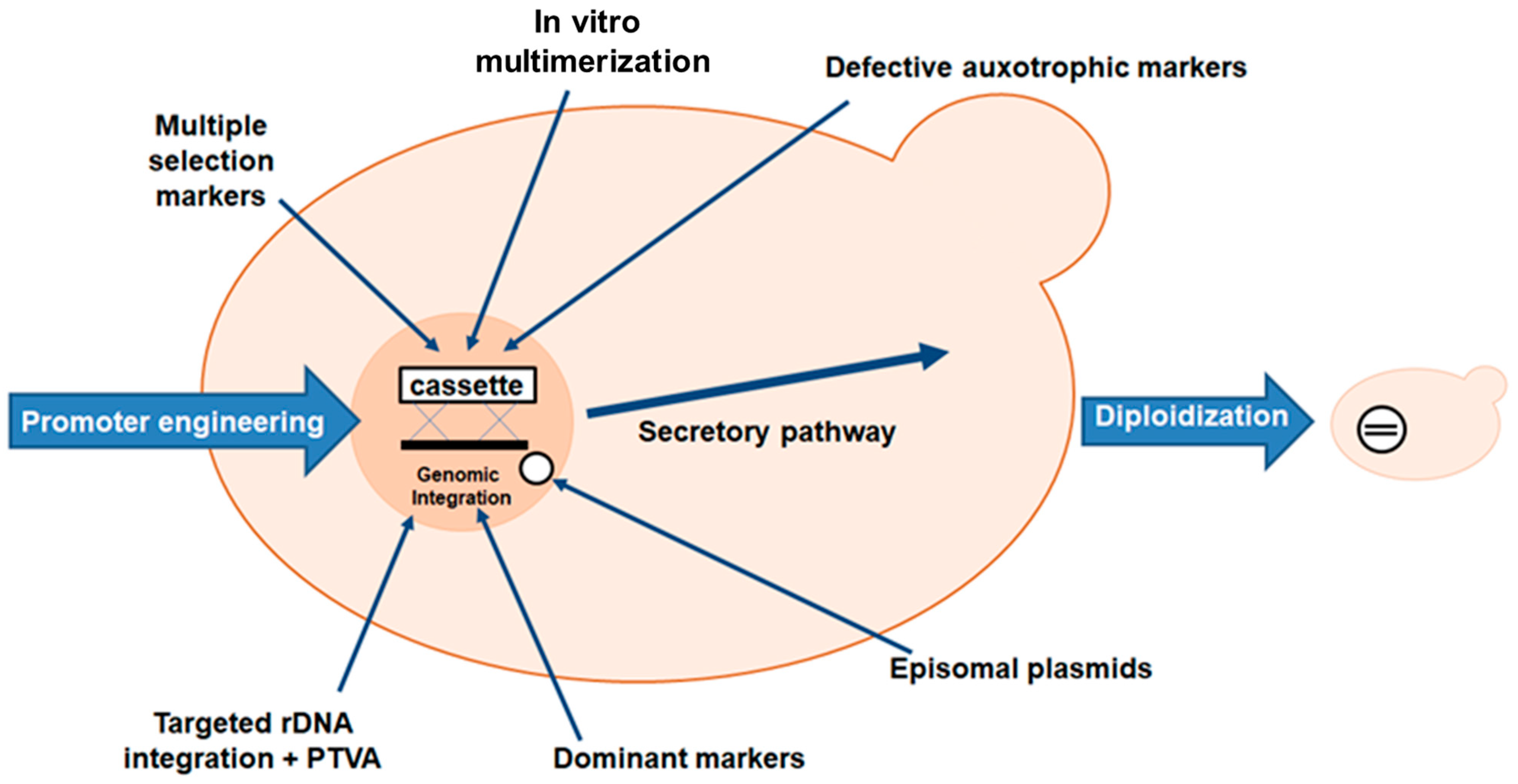
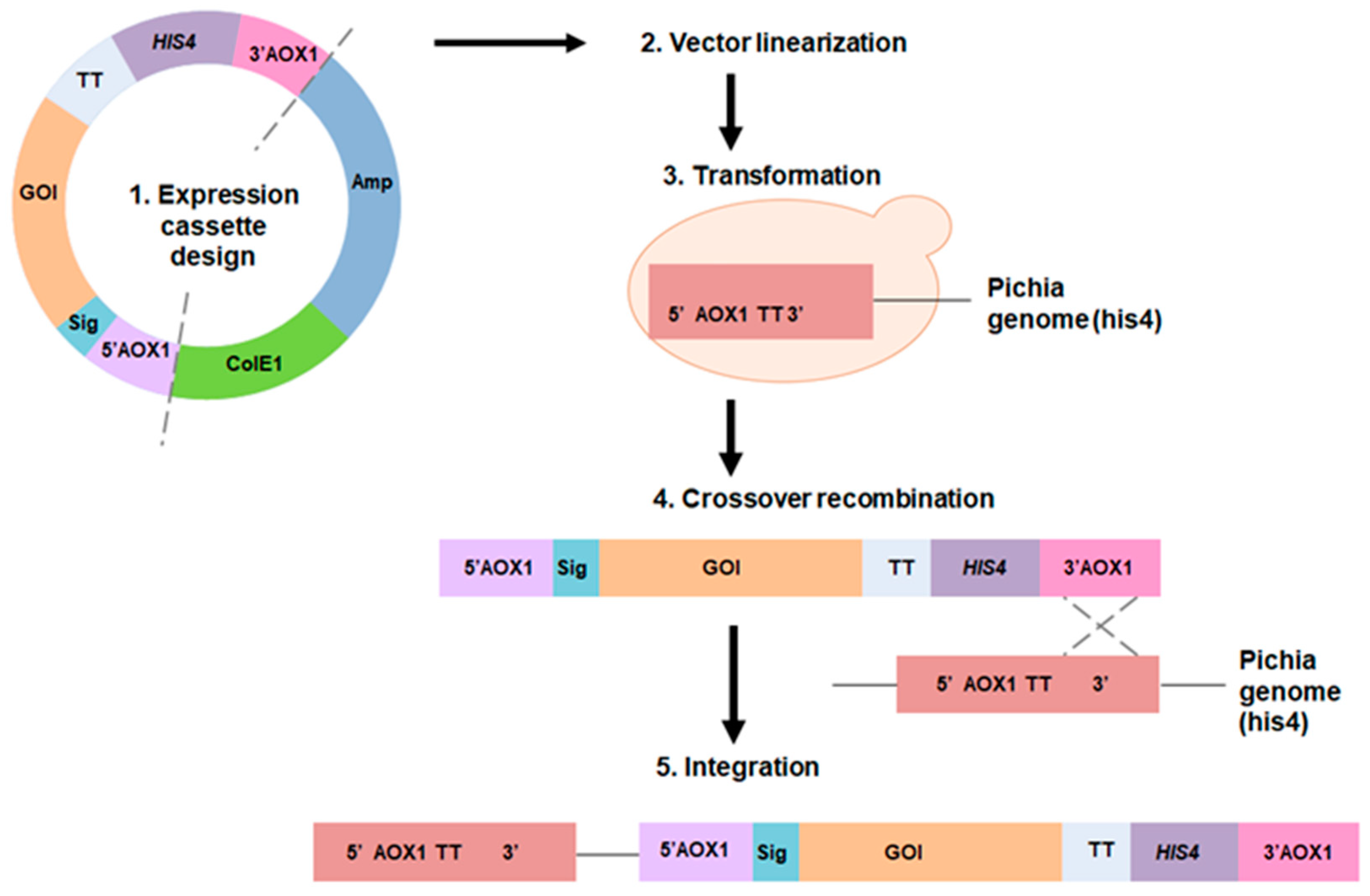
2.3. Pichia pastoris Expression System
2.3.1. Promoter and Vector Engineering
2.3.2. Affinity Tags
2.3.3. Signal Sequence and Post-Translational Modifications
2.3.4. Optimization of L-Asparaginase Production in Pichia pastoris
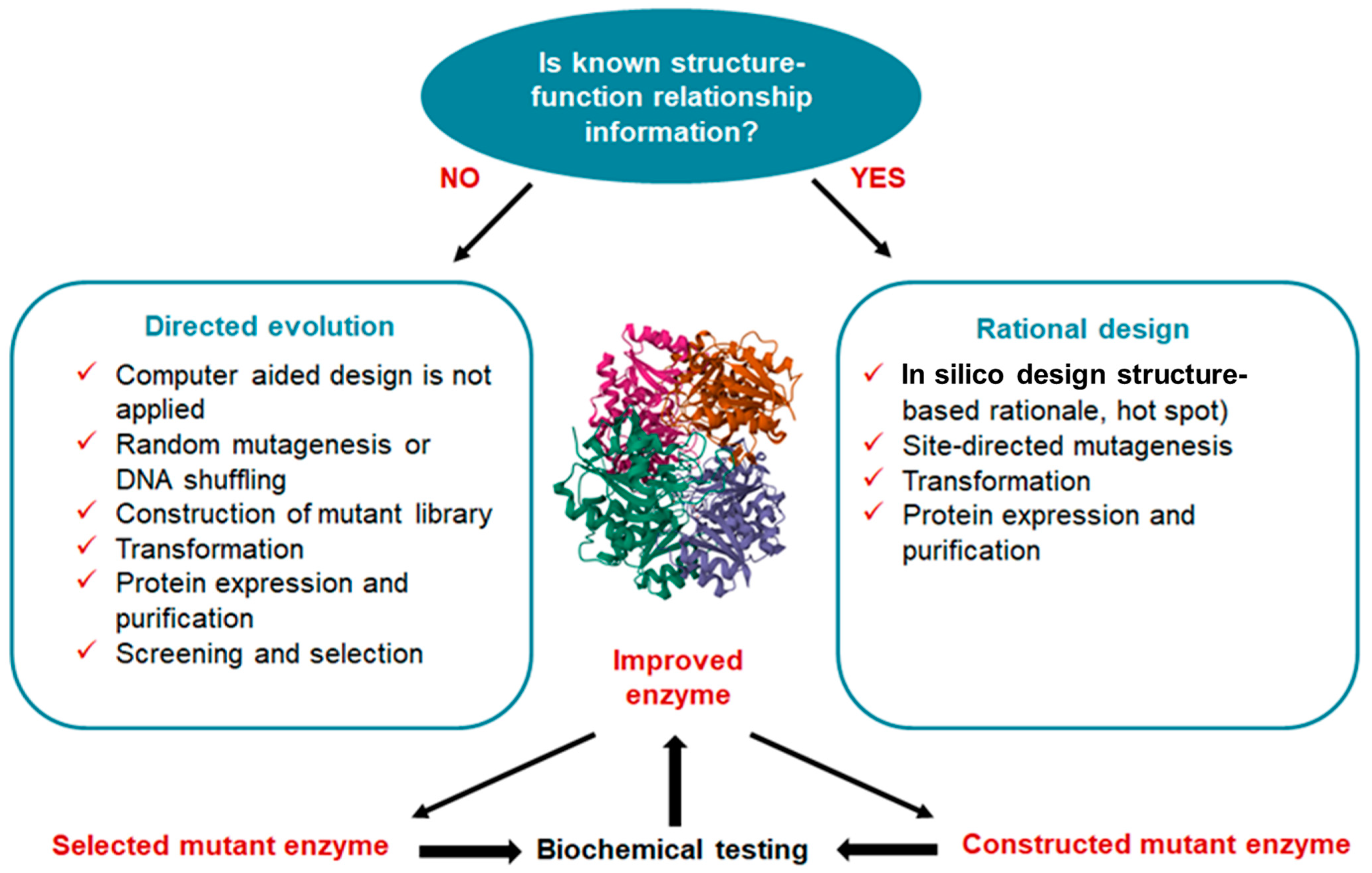
3. Approaches for Engineering Novel L-ASNases
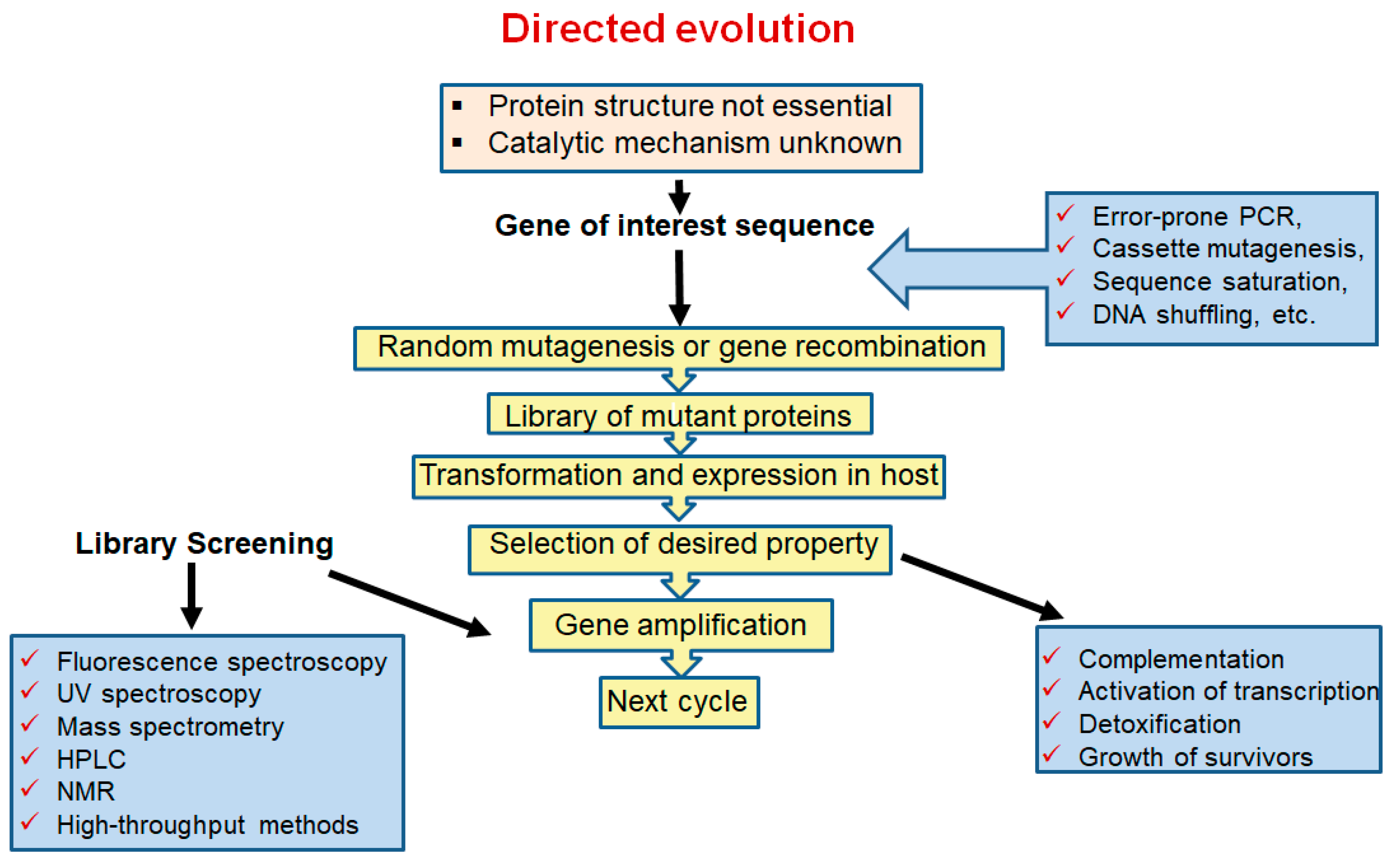
3.1. Directed Evolution for Designing L-ASNases
High-Throughput Screening
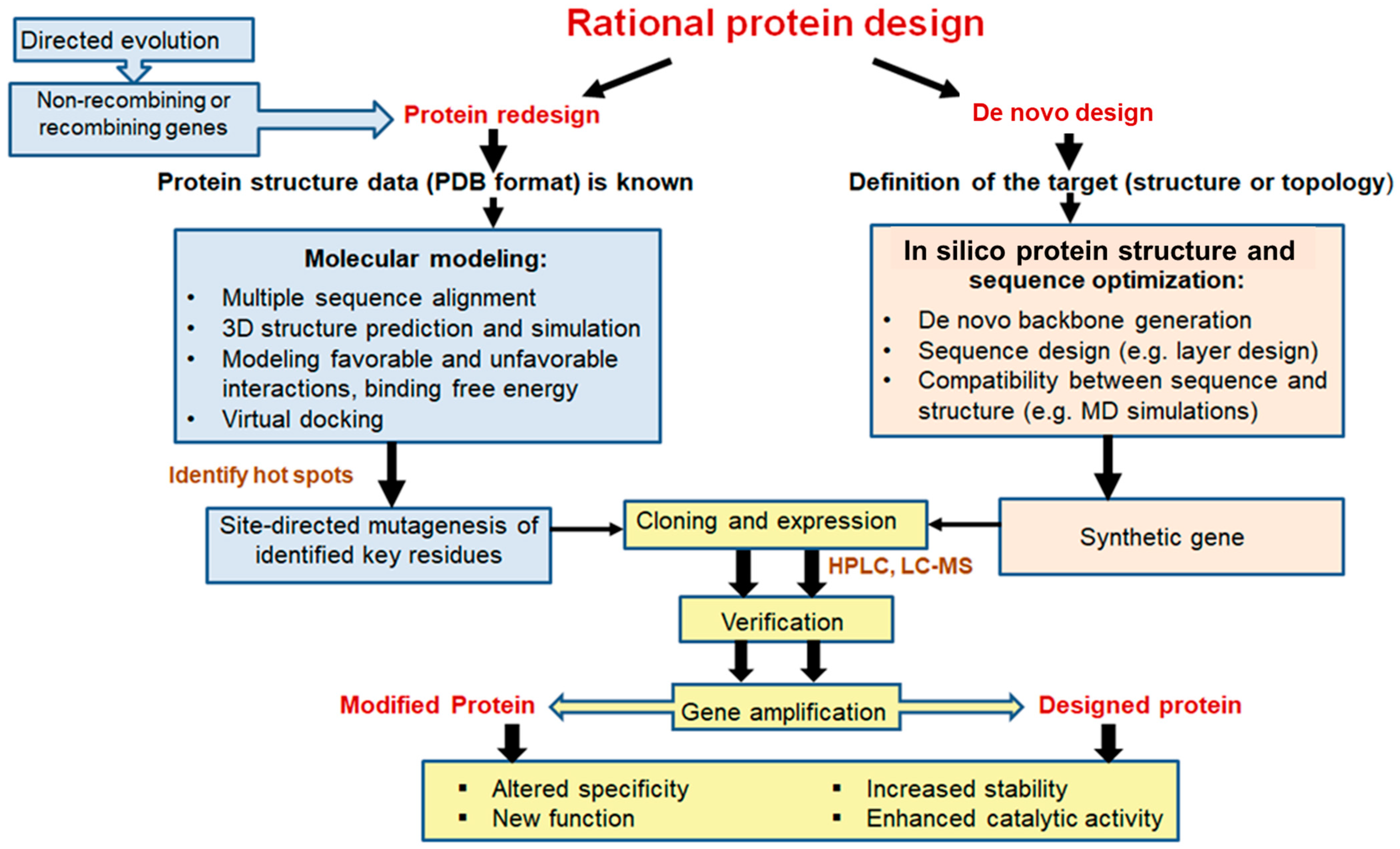
3.2. Rational Design Strategies and Bioinformatic Tools to Improve the Structural and Functional Properties of L-ASNases
3.2.1. Multiple Sequence Alignment
3.2.2. Homology Modeling
3.2.3. Molecular Dynamics
3.2.4. Molecular Docking of Ligand Binding
3.2.5. Algorithms for Predicting Protein Stability
3.2.6. De Novo Computational Design
3.2.7. Machine Learning Approaches
4. Combining Computer Design Techniques to Obtain L-Asparaginases with Improved Properties
4.1. Modifications to Increase the Expression
4.2. Modifications to Decrease Glutaminase Activity
4.3. Modifications to Increase Catalytic Activity and Specificity
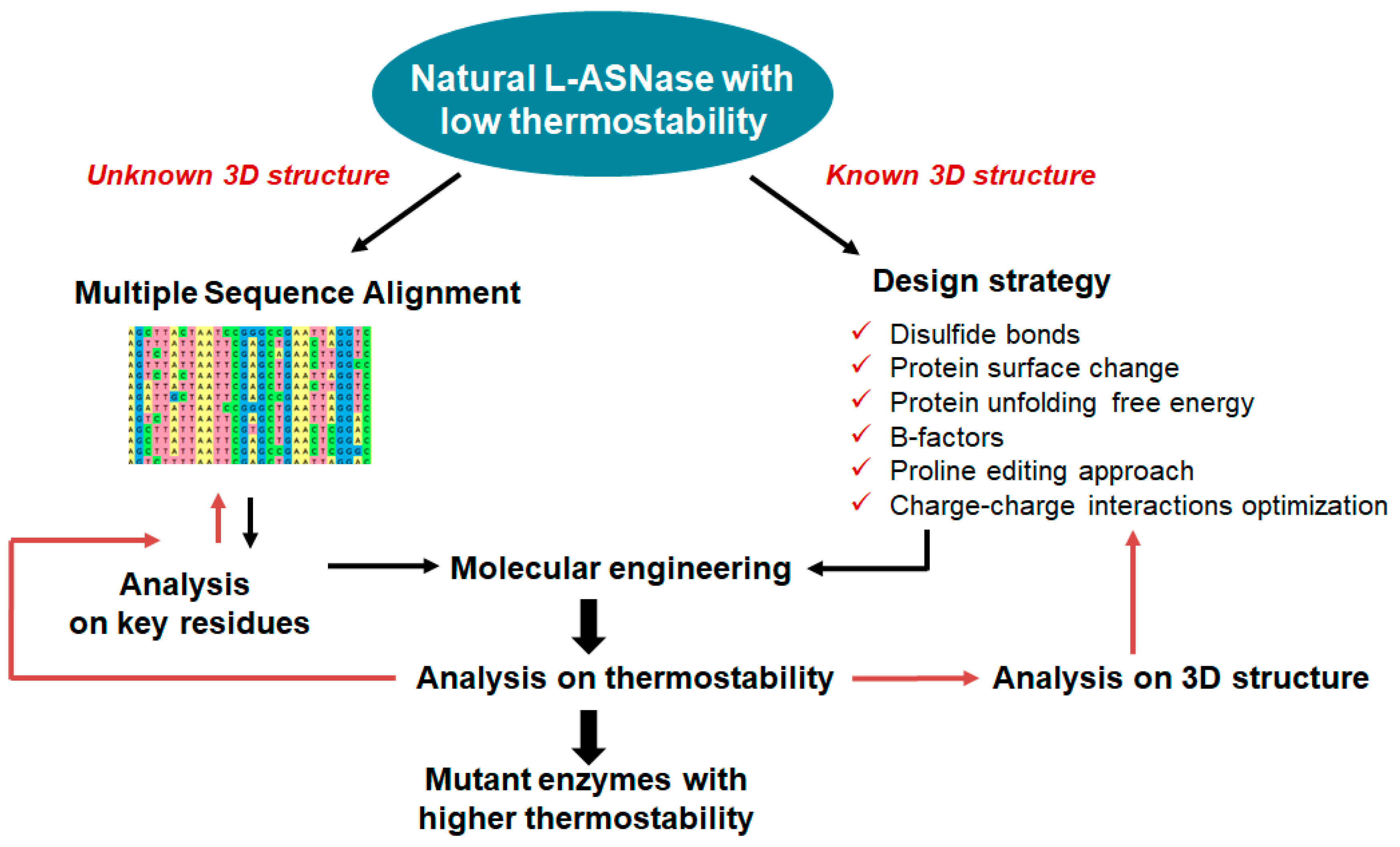
4.4. Modifications to Increase Thermostability
4.5. Modifications to Decrease Immunogenicity
4.5.1. T-Cell Epitope Prediction and Allergenicity Prediction
4.5.2. Prediction of the Toxicity of the Protein
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pokrovskaya, M.V.; Pokrovsky, V.S.; Aleksandrova, S.S.; Sokolov, N.N.; Zhdanov, D.D. Molecular Analysis of L-Asparaginases for Clarification of the Mechanism of Action and Optimization of Pharmacological Functions. Pharmaceutics 2022, 14, 599. [Google Scholar] [CrossRef]
- Sankaran, H.; Sengupta, S.; Purohit, V.; Kotagere, A.; Moulik, N.R.; Prasad, M.; Dhamne, C.; Narula, G.; Banavali, S.; Gota, V. A Comparison of Asparaginase Activity in Generic Formulations of E. coli Derived L-Asparaginase: In-vitro Study and Retrospective Analysis of Asparaginase Monitoring in Pediatric Patients with Leukemia. Br. J. Clin. Pharmacol. 2020, 86, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.C.; McCorkle, J.R.; Barnett, K.R.; Bonten, E.J.; Bergeron, B.P.; Bhattarai, K.R.; Yang, W.; Smith, C.; Hansen, B.S.; Bajpai, R.; et al. Amino Acid Stress Response Genes Promote L-Asparaginase Resistance in Pediatric Acute Lymphoblastic leukemia. Blood Adv. 2022, 6, 3386–3397. [Google Scholar] [CrossRef] [PubMed]
- Vimal, A.; Kumar, A. L-Asparaginase: A Feasible Therapeutic Molecule for Multiple Diseases. 3 Biotech 2018, 8, 278. [Google Scholar] [CrossRef]
- Völler, S.; Pichlmeier, U.; Zens, A.; Hempel, G. Pharmacokinetics of Recombinant Asparaginase in Children with Acute Lymphoblastic leukemia. Cancer Chemother. Pharmacol. 2018, 81, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Van Trimpont, M.; Peeters, E.; De Visser, Y.; Schalk, A.M.; Mondelaers, V.; De Moerloose, B.; Lavie, A.; Lammens, T.; Goossens, S.; Van Vlierberghe, P. Novel Insights on the Use of L-Asparaginase as an Efficient and Safe Anti-Cancer Therapy. Cancers 2022, 14, 902. [Google Scholar] [CrossRef]
- Mazloum-Ravasan, S.; Madadi, E.; Mohammadi, A.; Mansoori, B.; Amini, M.; Mokhtarzadeh, A.; Baradaran, B.; Darvishi, F. Yarrowia Lipolytica L-Asparaginase Inhibits the Growth and Migration of Lung (A549) and Breast (MCF7) Cancer Cells. Int. J. Biol. Macromol. 2021, 170, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, F.; Jahanafrooz, Z.; Mokhtarzadeh, A. Microbial L-Asparaginase as a Promising Enzyme for Treatment of Various Cancers. Appl. Microbiol. Biotechnol. 2022, 106, 5335–5347. [Google Scholar] [CrossRef]
- Ravi, A.; Gurunathan, B. Acrylamide Mitigation in Fried Kochchi Kesel Chips Using Free and Immobilized Fungal Asparaginase. Food Technol. Biotechnol. 2018, 56, 51–57. [Google Scholar] [CrossRef]
- Gazi, S.; Göncüoğlu Taş, N.; Görgülü, A.; Gökmen, V. Effectiveness of Asparaginase on Reducing Acrylamide Formation in Bakery Products according to Their Dough Type and Properties. Food Chem. 2023, 402, 134224. [Google Scholar] [CrossRef]
- Sajed, M.; Ahmad, N.; Rashid, N. Temperature Dependent Autocleavage and Applications of Recombinant L-Asparaginase from Thermococcus Kodakarensis for Acrylamide Mitigation. 3 Biotech 2022, 12, 129. [Google Scholar] [CrossRef] [PubMed]
- Bosmann, H.B.; Kessel, D. Inhibition of Glycoprotein Synthesis in L5178Y Mouse Lukaemic Cells by L-Asparaginase in Vitro. Nature 1970, 226, 850–851. [Google Scholar] [CrossRef] [PubMed]
- Plyasova, A.A.; Pokrovskaya, M.V.; Lisitsyna, O.M.; Pokrovsky, V.S.; Alexandrova, S.S.; Hilal, A.; Sokolov, N.N.; Zhdanov, D.D. Penetration into Cancer Cells via Clathrin-dependent Mechanism Allows L-asparaginase from Rhodospirillum rubrum to Inhibit Telomerase. Pharmaceuticals 2020, 13, 286. [Google Scholar] [CrossRef]
- Ankel, E.G.; Zirneski, J.; Ring, B.J.; Holcenberg, J.S. Effect of Asparaginase on Cell Membranes of Sensitive and Resistants Mouse Lymphoma Cells. Vitr. Cell. Dev. Biol.-Plant 1984, 20, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Castro, D.; Marques, A.S.C.; Almeida, M.R.; de Paiva, G.B.; Bento, H.B.S.; Pedrolli, D.B.; Freire, M.G.; Tavares, A.P.M.; Santos-Ebinuma, V.C. L-Asparaginase Production Review: Bioprocess Design and Biochemical Characteristics. Appl. Microbiol. Biotechnol. 2021, 105, 4515–4534. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Wang, Y.; Li, X.; Zhu, M.; Zhang, H.; Xu, M.; Yang, T.; Rao, Z. Heterologous Expression and Rational Design of L-Asparaginase from Rhizomucor miehei to Improve Thermostability. Biology 2021, 10, 1346. [Google Scholar] [CrossRef]
- Ratuchne, A.; Izidoro, S.C.; Beitel, S.M.; Lacerda, L.T.; Knob, A. A New Extracellular Glutaminase and Urease-Free l-Asparaginase from Meyerozyma guilliermondii. Braz. J. Microbiol. 2023, 54, 715–723. [Google Scholar] [CrossRef]
- Wang, N.; Ji, W.; Wang, L.; Wu, W.; Zhang, W.; Wu, Q.; Du, W.; Bai, H.; Peng, B.; Ma, B.; et al. Overview of the Structure, Side Effects, and Activity Assays of L-Asparaginase as a Therapy Drug of Acute Lymphoblastic leukemia. RSC Med. Chem. 2022, 13, 117–128. [Google Scholar] [CrossRef]
- LiverTox: Clinical and Research Information on Drug-Induced Liver Injury; National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2023; Volume 2012.
- Patel, P.G.; Panseriya, H.Z.; Vala, A.K.; Dave, B.P.; Gosai, H.B. Exploring Current Scenario and Developments in the Field of Microbial L-Asparaginase Production and Applications: A Review. Process Biochem. 2022, 121, 529–541. [Google Scholar] [CrossRef]
- Xu, F.; Oruna-Concha, M.-J.; Elmore, J.S. The Use of Asparaginase to Reduce Acrylamide Levels in Cooked Food. Food Chem. 2016, 210, 163–171. [Google Scholar] [CrossRef]
- Vimal, A.; Kumar, A. L-Asparaginase: Need for an Expedition from an Enzymatic Molecule to Antimicrobial Drug. Int. J. Pept. Res. Ther. 2022, 28, 9. [Google Scholar] [CrossRef]
- Ali, M.; Ishqi, H.M.; Husain, Q. Enzyme Engineering: Reshaping the Biocatalytic Functions. Biotechnol. Bioeng. 2020, 117, 1877–1894. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.; Lefin, N.; Beltran, J.F.; Belén, L.H.; Tsipa, A.; Farias, J.G.; Zamorano, M. Enzyme Engineering Strategies for the Bioenhancement of L-Asparaginase Used as a Biopharmaceutical. BioDrugs 2023, 1–19. [Google Scholar] [CrossRef]
- Yang, X.; Rao, Y.; Zhang, M.; Liu, W.; Cai, D.; Chen, S. Efficient Production of L-Asparaginase in Bacillus licheniformis by Optimizing Expression Elements and Host. Sheng Wu Gong Cheng Xue Bao 2023, 39, 1096–1106. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, S.; Zhang, X.; Xu, M.; Yang, T.; Wang, L.; Zhang, H.; Fang, H.; Osire, T.; Yang, S.; et al. Design of a High-Efficiency Synthetic System for L-Asparaginase Production in Bacillus subtilis. Eng. Life Sci. 2019, 19, 229–239. [Google Scholar] [CrossRef]
- Andjelkovic, M.; Zinovjev, K.; Ramos-Guzmán, C.A.; Ruiz-Pernía, J.J.; Tuñón, I. Elucidation of the Active Form and Reaction Mechanism in Human Asparaginase Type III Using Multiscale Simulations. J. Chem. Inf. Model. 2023, 63, 5676–5688. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Darnal, S.; Patial, V.; Kumar, V.; Kumar, V.; Kumar, S.; Singh, D. Molecular Cloning, Characterization, and in-Silico Analysis of l-Asparaginase from Himalayan pseudomonas sp. PCH44. 3 Biotech 2022, 12, 162. [Google Scholar] [CrossRef]
- Modi, T.; Regufe da Mota, S.; Gervais, D. L-Asparaginase and HCP Quantification by SWATH LC-MS/MS for New and Improved Purification Step in Erwinia Chrysanthemi l-Asparaginase Manufacture. J. Pharm. Biomed. Anal. 2022, 209, 114537. [Google Scholar] [CrossRef]
- Lefin, N.; Miranda, J.; Beltrán, J.F.; Belén, L.H.; Effer, B.; Pessoa, A.; Farias, J.G.; Zamorano, M. Current State of Molecular and Metabolic Strategies for the Improvement of L-Asparaginase Expression in Heterologous Systems. Front. Pharmacol. 2023, 14, 1208277. [Google Scholar] [CrossRef]
- Pourhassan, H.; Douer, D.; Pullarkat, V.; Aldoss, I. Asparaginase: How to Better Manage Toxicities in Adults. Curr. Oncol. Rep. 2023, 25, 51–61. [Google Scholar] [CrossRef]
- Mahboobi, M.; Salmanian, A.-H.; Sedighian, H.; Bambai, B. Molecular Modeling and Optimization of Type II E. coli l-Asparginase Activity by In Silico Design and In Vitro Site-Directed Mutagenesis. Protein J. 2023, 1–11. [Google Scholar] [CrossRef]
- Agrawal, S.; Jana, U.K.; Kango, N. Heterologous Expression and Molecular Modelling of L-Asparaginase from Bacillus subtilis ETMC-2. Int. J. Biol. Macromol. 2021, 192, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Golbabaie, A.; Nouri, H.; Moghimi, H.; Khaleghian, A. L-Asparaginase Production and Enhancement by Sarocladium strictum: In Vitro Evaluation of Anti-Cancerous Properties. J. Appl. Microbiol. 2020, 129, 356–366. [Google Scholar] [CrossRef]
- Costa-Silva, T.A.; Camacho-Córdova, D.I.; Agamez-Montalvo, G.S.; Parizotto, L.A.; Sánchez-Moguel, I.; Pessoa-Jr, A. Optimization of Culture Conditions and Bench-Scale Production of Anticancer Enzyme L-Asparaginase by Submerged Fermentation from Aspergillus Terreus CCT 7693. Prep. Biochem. Biotechnol. 2019, 49, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, S.L.; de Freitas, M.M.; de Souza, P.M.; Homem-de-Mello, M.; Silveira, D.; Fonseca-Bazzo, Y.M.; Filho, E.X.; Junior, A.P.; Magalhães, P.O. Optimization of Aqueous Two-Phase Micellar System for Partial Purification of L-Asparaginase from Penicillium sp. Grown in Wheat Bran as Agro-Industrial Residue. Braz. J. Microbiol. 2020, 51, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Mishra, A. Synergistic Effects of Ternary Mixture Formulation and Process Parameters Optimization in a Sequential Approach for Enhanced L-Asparaginase Production Using Agro-Industrial Wastes. Environ. Sci. Pollut. Res. 2023, 1–16. [Google Scholar] [CrossRef]
- Brumano, L.P.; da Silva, F.V.S.; Costa-Silva, T.A.; Apolinário, A.C.; Santos, J.H.P.M.; Kleingesinds, E.K.; Monteiro, G.; Rangel-Yagui, C.d.O.; Benyahia, B.; Junior, A.P. Development of L-Asparaginase Biobetters: Current Research Status and Review of the Desirable Quality Profiles. Front. Bioeng. Biotechnol. 2018, 6, 212. [Google Scholar] [CrossRef]
- da Cunha, M.C.; dos Santos Aguilar, J.G.; de Melo, R.R.; Nagamatsu, S.T.; Ali, F.; de Castro, R.J.S.; Sato, H.H. Fungal L-Asparaginase: Strategies for Production and Food Applications. Food Res. Int. 2019, 126, 108658. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, W.; Wu, H.; Zhang, W.; Guang, C.; Mu, W. Microbial Production, Molecular Modification, and Practical Application of l-Asparaginase: A Review. Int. J. Biol. Macromol. 2021, 186, 975–983. [Google Scholar] [CrossRef]
- Chakravarty, N.; Priyanka; Singh, J.; Singh, R.P. A Potential Type-II L-Asparaginase from Marine Isolate Bacillus australimaris NJB19: Statistical Optimization, in Silico Analysis and Structural Modeling. Int. J. Biol. Macromol. 2021, 174, 527–539. [Google Scholar] [CrossRef]
- Mahajan, R.V.; Kumar, V.; Rajendran, V.; Saran, S.; Ghosh, P.C.; Saxena, R.K. Purification and Characterization of a Novel and Robust L-Asparaginase Having Low-Glutaminase Activity from Bacillus licheniformis: In Vitro Evaluation of Anti-Cancerous Properties. PLoS ONE 2014, 9, e99037. [Google Scholar] [CrossRef] [PubMed]
- Cachumba, J.J.M.; Antunes, F.A.F.; Peres, G.F.D.; Brumano, L.P.; Dos Santos, J.C.; Da Silva, S.S. Current Applications and Different Approaches for Microbial L-Asparaginase Production. Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2016, 47 (Suppl. S1), 77–85. [Google Scholar] [CrossRef]
- Rieder, L.; Teuschler, N.; Ebner, K.; Glieder, A. Eukaryotic Expression Systems for Industrial Enzymes. In Industrial Enzyme Applications; Wiley: Hoboken, NJ, USA, 2019; pp. 47–69. [Google Scholar]
- Al-Hazmi, N.E.; Naguib, D.M. Plant Asparaginase versus Microbial Asparaginase as Anticancer Agent. Environ. Sci. Pollut. Res. 2022, 29, 27283–27293. [Google Scholar] [CrossRef]
- Lopes, A.M.; de Oliveira-Nascimento, L.; Ribeiro, A.; Tairum, C.A.J.; Breyer, C.A.; de Oliveira, M.A.; Monteiro, G.; de Souza-Motta, C.M.; Magalhães, P.d.O.; Avendaño, J.G.F.; et al. Therapeutic L-Asparaginase: Upstream, Downstream and Beyond. Crit. Rev. Biotechnol. 2017, 37, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, F.; Faraji, N.; Shamsi, F. Production and Structural Modeling of a Novel Asparaginase in Yarrowia lipolytica. Int. J. Biol. Macromol. 2019, 125, 955–961. [Google Scholar] [CrossRef]
- Chand, S.; Mahajan, R.V.; Prasad, J.P.; Sahoo, D.K.; Mihooliya, K.N.; Dhar, M.S.; Sharma, G. A Comprehensive Review on Microbial L-Asparaginase: Bioprocessing, Characterization, and Industrial Applications. Biotechnol. Appl. Biochem. 2020, 67, 619–647. [Google Scholar] [CrossRef]
- Orabi, H.M.; El-Fakharany, E.M.; Abdelkhalek, E.S.; Sidkey, N.M. L-Asparaginase And L-Glutaminase: Sources, Production, and Applications In Medicine and Industry. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 179–190. [Google Scholar] [CrossRef]
- Dantas, R.C.; Caetano, L.F.; Torres, A.L.S.; Alves, M.S.; Silva, E.T.M.F.; Teixeira, L.P.R.; Teixeira, D.C.; de Azevedo Moreira, R.; Fonseca, M.H.G.; Gaudêncio Neto, S.; et al. Expression of a Recombinant Bacterial L-Asparaginase in Human Cells. BMC Res. Notes 2019, 12, 794. [Google Scholar] [CrossRef] [PubMed]
- L-Asparaginase (ASRGL1) Recombinant Protein. Available online: https://www.mybiosource.com/recombinant-protein/l-asparaginase-asrgl1/1420071 (accessed on 11 October 2023).
- Izadpanah Qeshmi, F.; Homaei, A.; Fernandes, P.; Javadpour, S. Marine Microbial L-Asparaginase: Biochemistry, Molecular Approaches and Applications in Tumor Therapy and in Food Industry. Microbiol. Res. 2018, 208, 99–112. [Google Scholar] [CrossRef]
- Loch, J.I.; Jaskolski, M. Structural and Biophysical Aspects of L-Asparaginases: A Growing Family with Amazing Diversity. IUCrJ 2021, 8, 514–531. [Google Scholar] [CrossRef]
- Michalska, K.; Jaskolski, M. Structural Aspects of L-Asparaginases, Their Friends and Relations. Acta Biochim. Pol. 2006, 53, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Bejger, M.; Imiolczyk, B.; Clavel, D.; Gilski, M.; Pajak, A.; Marsolais, F.; Jaskolski, M. Na+/K+ Exchange Switches the Catalytic Apparatus of Potassium-Dependent Plant L-Asparaginase. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1854–1872. [Google Scholar] [CrossRef] [PubMed]
- Dumina, M.; Zhgun, A. Thermo-L-Asparaginases: From the Role in the Viability of Thermophiles and Hyperthermophiles at High Temperatures to a Molecular Understanding of Their Thermoactivity and Thermostability. Int. J. Mol. Sci. 2023, 24, 2674. [Google Scholar] [CrossRef]
- Lubkowski, J.; Wlodawer, A. Geometric Considerations Support the Double-displacement Catalytic Mechanism of l-asparaginase. Protein Sci. 2019, 28, 1850–1864. [Google Scholar] [CrossRef]
- Kumar, S.; Darnal, S.; Patial, V.; Kumar, V.; Singh, D. Molecular Characterization of a Stable and Robust L-Asparaginase from Pseudomonas sp. PCH199: Evaluation of Cytotoxicity and Acrylamide Mitigation Potential. Fermentation 2022, 8, 568. [Google Scholar] [CrossRef]
- Zielezinski, A.; Loch, J.I.; Karlowski, W.M.; Jaskolski, M. Massive Annotation of Bacterial L-Asparaginases Reveals Their Puzzling Distribution and Frequent Gene Transfer Events. Sci. Rep. 2022, 12, 15797. [Google Scholar] [CrossRef] [PubMed]
- Lubkowski, J.; Wlodawer, A. Structural and Biochemical Properties of L-Asparaginase. FEBS J. 2021, 288, 4183–4209. [Google Scholar] [CrossRef]
- Kotzia, G.A.; Lappa, K.; Labrou, N.E. Tailoring Structure-Function Properties of L-Asparaginase: Engineering Resistance to Trypsin Cleavage. Biochem. J. 2007, 404, 337–343. [Google Scholar] [CrossRef]
- Gesto, D.S.; Cerqueira, N.M.F.S.A.; Fernandes, P.A.; Ramos, M.J. Unraveling the Enigmatic Mechanism of L-Asparaginase II with QM/QM Calculations. J. Am. Chem. Soc. 2013, 135, 7146–7158. [Google Scholar] [CrossRef]
- Fonseca, M.H.G.; Fiúza, T.d.S.; de Morais, S.B.; de Souza, T.d.A.C.B.; Trevizani, R. Circumventing the Side Effects of L-Asparaginase. Biomed. Pharmacother. Biomed. Pharmacother. 2021, 139, 111616. [Google Scholar] [CrossRef]
- Aghaiypour, K.; Wlodawer, A.; Lubkowski, J. Structural Basis for the Activity and Substrate Specificity of Erwinia chrysanthemi L-Asparaginase. Biochemistry 2001, 40, 5655–5664. [Google Scholar] [CrossRef] [PubMed]
- Jaskólski, M.; Kozak, M.; Lubkowski, J.; Palm, G.; Wlodawer, A. Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001, 57, 369–377. [Google Scholar] [CrossRef]
- Offman, M.N.; Krol, M.; Patel, N.; Krishnan, S.; Liu, J.; Saha, V.; Bates, P.A. Rational Engineering of L-Asparaginase Reveals Importance of Dual Activity for Cancer Cell Toxicity. Blood 2011, 117, 1614–1621. [Google Scholar] [CrossRef]
- Alexandrova, S.S.; Gladilina, Y.A.; Pokrovskaya, M.V.; Sokolov, N.N.; Zhdanov, D.D. Mechanisms of Development of Side Effects and Drug Resistance to Asparaginase and Ways to Overcome Them. Biochem. (Mosc.) Suppl. Ser. B Biomed. Chem. 2022, 16, 175–186. [Google Scholar] [CrossRef]
- Gupta, S.K.; Shukla, P. Advanced Technologies for Improved Expression of Recombinant Proteins in Bacteria: Perspectives and Applications. Crit. Rev. Biotechnol. 2016, 36, 1089–1098. [Google Scholar] [CrossRef]
- Asitok, A.; Ekpenyong, M.; Amenaghawon, A.; Akwagiobe, E.; Asuquo, M.; Rao, A.; Ubi, D.; Iheanacho, J.; Etiosa, J.; Antai, A.; et al. Production, Characterization and Techno-Economic Evaluation of Aspergillus Fusant l-Asparaginase. AMB Express 2023, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.E.; Martí, J.M.; Radivojevic, T.; Jonnalagadda, S.V.R.; Gentz, R.; Hillson, N.J.; Peisert, S.; Kim, J.; Simmons, B.A.; Petzold, C.J.; et al. Machine Learning for Metabolic Engineering: A Review. Metab. Eng. 2021, 63, 34–60. [Google Scholar] [CrossRef]
- Shakambari, G.; Sameer Kumar, R.; Ashokkumar, B.; Ganesh, V.; Vasantha, V.S.; Varalakshmi, P. Cloning and Expression of L-Asparaginase from Bacillus tequilensis PV9W and Therapeutic Efficacy of Solid Lipid Particle Formulations against Cancer. Sci. Rep. 2018, 8, 18013. [Google Scholar] [CrossRef]
- Kim, S.-K.; Min, W.-K.; Park, Y.-C.; Seo, J.-H. Application of Repeated Aspartate Tags to Improving Extracellular Production of Escherichia coli L-Asparaginase Isozyme II. Enzym. Microb. Technol. 2015, 79–80, 49–54. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant Protein Expression in Escherichia coli: Advances and Challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef]
- Baeshen, N.A.; Baeshen, M.N.; Sheikh, A.; Bora, R.S.; Ahmed, M.M.M.; Ramadan, H.A.I.; Saini, K.S.; Redwan, E.M. Cell Factories for Insulin Production. Microb. Cell Factories 2014, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Plomp, P.J.A.M.; Boer, L.D.; Rooijen, R.J.V.; Meima, R.B. Asparaginase and Its Use in Food Production. Available online: https://patents.google.com/patent/US8105815B2/en (accessed on 11 October 2023).
- Pouresmaeil, M.; Azizi-Dargahlou, S. Factors Involved in Heterologous Expression of Proteins in E. coli Host. Arch. Microbiol. 2023, 205, 212. [Google Scholar] [CrossRef] [PubMed]
- Heyde, S.A.H.; Nørholm, M.H.H. Tailoring the Evolution of BL21(DE3) Uncovers a Key Role for RNA Stability in Gene Expression Toxicity. Commun. Biol. 2021, 4, 963. [Google Scholar] [CrossRef] [PubMed]
- Radha, R.; Arumugam, N.; Gummadi, S.N. Glutaminase Free L-Asparaginase from Vibrio Cholerae: Heterologous Expression, Purification and Biochemical Characterization. Int. J. Biol. Macromol. 2018, 111, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Einsfeldt, K.; Baptista, I.C.; Pereira, J.C.C.V.; Costa-Amaral, I.C.; da Costa, E.S.; Ribeiro, M.C.M.; Land, M.G.P.; Alves, T.L.M.; Larentis, A.L.; Almeida, R.V. Recombinant L-Asparaginase from Zymomonas Mobilis: A Potential New Antileukemic Agent Produced in Escherichia coli. PLoS ONE 2016, 11, e0156692. [Google Scholar] [CrossRef]
- Studier, F.W.; Daegelen, P.; Lenski, R.E.; Maslov, S.; Kim, J.F. Understanding the Differences between Genome Sequences of Escherichia coli B Strains REL606 and BL21(DE3) and Comparison of the E. coli B and K-12 Genomes. J. Mol. Biol. 2009, 394, 653–680. [Google Scholar] [CrossRef]
- Pourhossein, M.; Korbekandi, H. Cloning, Expression, Purification and Characterisation of Erwinia Carotovora L-Asparaginase in Escherichia coli. Adv. Biomed. Res. 2014, 3, 82. [Google Scholar] [CrossRef]
- de Moura, W.A.F.; Schultz, L.; Breyer, C.A.; de Oliveira, A.L.P.; Tairum, C.A.; Fernandes, G.C.; Toyama, M.H.; Pessoa, A., Jr.; Monteiro, G.; de Oliveira, M.A. Functional and Structural Evaluation of the Antileukaemic Enzyme L-Asparaginase II Expressed at Low Temperature by Different Escherichia coli Strains. Biotechnol. Lett. 2020, 42, 2333–2344. [Google Scholar] [CrossRef]
- Firat Duzenli, O.; Okay, S. Promoter Engineering for the Recombinant Protein Production in Prokaryotic Systems. AIMS Bioeng. 2020, 7, 62–81. [Google Scholar] [CrossRef]
- Alba Burbano, D.; Cardiff, R.A.L.; Tickman, B.I.; Kiattisewee, C.; Maranas, C.J.; Zalatan, J.G.; Carothers, J.M. Engineering Activatable Promoters for Scalable and Multi-Input CRISPRa/i Circuits. Proc. Natl. Acad. Sci. USA 2023, 120, e2220358120. [Google Scholar] [CrossRef]
- Lozano Terol, G.; Gallego-Jara, J.; Sola Martínez, R.A.; Martínez Vivancos, A.; Cánovas Díaz, M.; de Diego Puente, T. Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef] [PubMed]
- Hyvönen, M.E. Coli Expression and Fusion Proteins. Available online: https://hyvonen.bioc.cam.ac.uk/wp-content/uploads/2017/10/ecoli_expression_and_fusion_proteins_2017_web.pdf (accessed on 2 September 2023).
- Hayat, S.M.G.; Farahani, N.; Golichenari, B.; Sahebkar, A. Recombinant Protein Expression in Escherichia coli (E. coli): What We Need to Know. Curr. Pharm. Des. 2018, 24, 718–725. [Google Scholar] [CrossRef]
- Zhang, Z.-X.; Nong, F.-T.; Wang, Y.-Z.; Yan, C.-X.; Gu, Y.; Song, P.; Sun, X.-M. Strategies for Efficient Production of Recombinant Proteins in Escherichia coli: Alleviating the Host Burden and Enhancing Protein Activity. Microb. Cell Factories 2022, 21, 191. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, Z.; Wang, L.; Wang, J.; Liang, Y.; Yang, H.; Tao, R.; Jiang, Y.; Yang, J.; Yang, S. Downregulation of T7 RNA Polymerase Transcription Enhances PET-based Recombinant Protein Production in Escherichia coli BL21 (DE3) by Suppressing Autolysis. Biotechnol. Bioeng. 2021, 118, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Gileadi, O. Recombinant Protein Expression in E. coli: A Historical Perspective. In Heterologous Gene Expression in E.coli; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1586, pp. 3–10. [Google Scholar]
- Dumina, M.V.; Zhgun, A.A.; Pokrovskay, M.V.; Aleksandrova, S.S.; Zhdanov, D.D.; Sokolov, N.N.; El’darov, M.A. Comparison of Enzymatic Activity of Novel Recombinant L-Asparaginases of Extremophiles. Appl. Biochem. Microbiol. 2021, 57, 594–602. [Google Scholar] [CrossRef]
- Roth, G.; Nunes, J.E.S.; Rosado, L.A.; Bizarro, C.V.; Volpato, G.; Nunes, C.P.; Renard, G.; Basso, L.A.; Santos, D.S.; Chies, J.M. Recombinant Erwinia Carotovora L-Asparaginase II Production in Escherichia coli Fed-Batch Cultures. Braz. J. Chem. Eng. 2013, 30, 245–256. [Google Scholar] [CrossRef]
- Khushoo, A.; Pal, Y.; Mukherjee, K.J. Optimization of Extracellular Production of Recombinant Asparaginase in Escherichia coli in Shake-Flask and Bioreactor. Appl. Microbiol. Biotechnol. 2005, 68, 189–197. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Singh, A.; Mukherjee, K.J.; Panda, A.K. Refolding and Purification of Recombinant L-Asparaginase from Inclusion Bodies of E. coli into Active Tetrameric Protein. Front. Microbiol. 2014, 5, 486. [Google Scholar] [CrossRef]
- Huang, L.; Liu, Y.; Sun, Y.; Yan, Q.; Jiang, Z. Biochemical Characterization of a Novel L-Asparaginase with Low Glutaminase Activity from Rhizomucor Miehei and Its Application in Food Safety and Leukemia Treatment. Appl. Environ. Microbiol. 2014, 80, 1561–1569. [Google Scholar] [CrossRef]
- Costa, I.M.; Schultz, L.; de Araujo Bianchi Pedra, B.; Leite, M.S.M.; Farsky, S.H.P.; de Oliveira, M.A.; Pessoa, A.; Monteiro, G. Recombinant L-Asparaginase 1 from Saccharomyces cerevisiae: An Allosteric Enzyme with Antineoplastic Activity. Sci. Rep. 2016, 6, 36239. [Google Scholar] [CrossRef]
- Saeed, H.; Hemida, A.; El-Nikhely, N.; Abdel-Fattah, M.; Shalaby, M.; Hussein, A.; Eldoksh, A.; Ataya, F.; Aly, N.; Labrou, N.; et al. Highly Efficient Pyrococcus furiosus Recombinant L-Asparaginase with No Glutaminase Activity: Expression, Purification, Functional Characterization, and Cytotoxicity on THP-1, A549 and Caco-2 Cell Lines. Int. J. Biol. Macromol. 2020, 156, 812–828. [Google Scholar] [CrossRef] [PubMed]
- Pokrovskaya, M.V.; Aleksandrova, S.S.; Pokrovsky, V.S.; Omeljanjuk, N.M.; Borisova, A.A.; Anisimova, N.Y.; Sokolov, N.N. Cloning, Expression and Characterization of the Recombinant Yersinia Pseudotuberculosis l-Asparaginase. Protein Expr. Purif. 2012, 82, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Chohan, S.M.; Rashid, N.; Sajed, M.; Imanaka, T. Pcal_0970: An Extremely Thermostable L-Asparaginase from Pyrobaculum calidifontis with No Detectable Glutaminase Activity. Folia Microbiol. 2019, 64, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Q.; Weng, H.; Shi, Y.; Chen, J.; Du, G.; Kang, Z. Construction of Synthetic Promoters by Assembling the Sigma Factor Binding-35 and -10 Boxes. Biotechnol. J. 2019, 14, e1800298. [Google Scholar] [CrossRef]
- Lei, S.P.; Lin, H.C.; Wang, S.S.; Callaway, J.; Wilcox, G. Characterization of the Erwinia Carotovora PelB Gene and Its Product Pectate Lyase. J. Bacteriol. 1987, 169, 4379–4383. [Google Scholar] [CrossRef]
- Khushoo, A.; Pal, Y.; Singh, B.N.; Mukherjee, K.J. Extracellular Expression and Single Step Purification of Recombinant Escherichia coli L-Asparaginase II. Protein Expr. Purif. 2004, 38, 29–36. [Google Scholar] [CrossRef]
- Chan, W.K.; Lorenzi, P.L.; Anishkin, A.; Purwaha, P.; Rogers, D.M.; Sukharev, S.; Rempe, S.B.; Weinstein, J.N. The Glutaminase Activity of L-Asparaginase Is Not Required for Anticancer Activity against ASNS-Negative Cells. Blood 2014, 123, 3596–3606. [Google Scholar] [CrossRef]
- Aghaeepoor, M.; Mozafari, S.; Shahraki, M.K.; Shahraki, M.K.; Bambai, B. High Level of Extracellular Fermentation and Alternative Purification of Escherichia coli Asparaginase II. Biharean Biol. 2011, 5, 96–101. [Google Scholar]
- Yari, M.; Ghoshoon, M.B.; Nezafat, N.; Ghasemi, Y. Experimental Evaluation of In Silico Selected Signal Peptides for Secretory Expression of Erwinia Asparaginase in Escherichia coli. Int. J. Pept. Res. Ther. 2020, 26, 1583–1591. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, K.; Han, Y.; Xing, Y.; Zhang, Y.; Yang, Q.; Zhou, M. Codon Usage Bias Regulates Gene Expression and Protein Conformation in Yeast Expression System, P. Pastoris. Microb. Cell Factories 2021, 20, 91. [Google Scholar] [CrossRef]
- Dumina, M.; Zhgun, A.; Pokrovskaya, M.; Aleksandrova, S.; Zhdanov, D.; Sokolov, N.; El’darov, M.; El’darov, M.; El’darov, M. A Novel L-Asparaginase from Hyperthermophilic Archaeon Thermococcus sibiricus: Heterologous Expression and Characterization for Biotechnology Application. Int. J. Mol. Sci. 2021, 22, 9894. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, V.; Falcone, F.H. Polyionic Tags as Enhancers of Protein Solubility in Recombinant Protein Expression. Microorganisms 2018, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.A.; Su, Y.; Lavie, A. Structural Insight into Substrate Selectivity of Erwinia Chrysanthemi L-Asparaginase. Biochemistry 2016, 55, 1246–1253. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, D.C.; Landwehr, M.; Reissner, C.; Smalla, K.-H.; Richter, K.; Wolf, G.; Böckers, T.M.; Gundelfinger, E.D.; Kreutz, M.R. Gliap—A Novel Untypical L-Asparaginase Localized to Rat Brain Astrocytes. J. Neurochem. 2003, 85, 1117–1125. [Google Scholar] [CrossRef]
- Sindhu, R.; Manonmani, H.K. Expression and Characterization of Recombinant l-Asparaginase from Pseudomonas fluorescens. Protein Expr. Purif. 2018, 143, 83–91. [Google Scholar] [CrossRef]
- Darwesh, D.B.; Al-Awthan, Y.S.; Elfaki, I.; Habib, S.A.; Alnour, T.M.; Darwish, A.B.; Youssef, M.M. Anticancer Activity of Extremely Effective Recombinant L-Asparaginase from Burkholderia Pseudomallei. J. Microbiol. Biotechnol. 2022, 32, 551–563. [Google Scholar] [CrossRef]
- Ki, M.-R.; Pack, S.P. Fusion Tags to Enhance Heterologous Protein Expression. Appl. Microbiol. Biotechnol. 2020, 104, 2411–2425. [Google Scholar] [CrossRef]
- Saeed, H.; Hemida, A.; Abdel-Fattah, M.; Eldoksh, A.; Shalaby, M.; Nematalla, H.; El-Nikhely, N.; Elkewedi, M. Pseudomonas aeruginosa Recombinant L-Asparaginase: Large Scale Production, Purification, and Cytotoxicity on THP-1, MDA-MB-231, A549, Caco2 and HCT-116 Cell Lines. Protein Expr. Purif. 2021, 181, 105820. [Google Scholar] [CrossRef]
- Lopes, W.; dos Santos, B.A.F.; Sampaio, A.L.F.; Gregório Alves Fontão, A.P.; Nascimento, H.J.; Jurgilas, P.B.; Torres, F.A.G.; Bon, E.P.d.S.; Almeida, R.V.; Ferrara, M.A. Expression, Purification, and Characterization of Asparaginase II from Saccharomyces cerevisiae in Escherichia coli. Protein Expr. Purif. 2019, 159, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.H.P.M.; Costa, I.M.; Molino, J.V.D.; Leite, M.S.M.; Pimenta, M.V.; Coutinho, J.A.P.; Pessoa, A.; Ventura, S.P.M.; Lopes, A.M.; Monteiro, G. Heterologous Expression and Purification of Active L-Asparaginase I of Saccharomyces cerevisiae in Escherichia coli Host. Biotechnol. Prog. 2017, 33, 416–424. [Google Scholar] [CrossRef]
- Nguyen, H.A.; Su, Y.; Lavie, A. Design and Characterization of Erwinia Chrysanthemi L-Asparaginase Variants with Diminished l-Glutaminase Activity. J. Biol. Chem. 2016, 291, 17664–17676. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.A.; Su, Y.; Zhang, J.Y.; Antanasijevic, A.; Caffrey, M.; Schalk, A.M.; Liu, L.; Rondelli, D.; Oh, A.; Mahmud, D.L.; et al. A Novel L-Asparaginase with Low l-Glutaminase Coactivity Is Highly Efficacious against Both T- and B-Cell Acute Lymphoblastic leukemias In Vivo. Cancer Res. 2018, 78, 1549–1560. [Google Scholar] [CrossRef]
- Van Trimpont, M.; Schalk, A.M.; De Visser, Y.; Nguyen, H.A.; Reunes, L.; Vandemeulebroecke, K.; Peeters, E.; Su, Y.; Lee, H.; Lorenzi, P.L.; et al. In Vivo Stabilization of a Less Toxic Asparaginase Variant Leads to a Durable Antitumor Response in Acute Leukemia. Haematologica 2022, 108, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Yang, S.J.; Son, B.; Lee, H.; Lee, J.; Joo, J.; Park, H.H.; Park, T.H. Enhanced Anti-Cancer Effect Using MMP-Responsive L-Asparaginase Fused with Cell-Penetrating 30Kc19 Protein. Artif. Cells Nanomed. Biotechnol. 2022, 50, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, J.; Qian, S.; Chen, R.; Meng, G. Construction and Structural Modeling of a Single-Chain Fv-Asparaginase Fusion Protein Resistant to Proteolysis. Biotechnol. Bioeng. 2000, 70, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. Recent Advances in Understanding Catalysis of Protein Folding by Molecular Chaperones. FEBS Lett. 2020, 594, 2770–2781. [Google Scholar] [CrossRef] [PubMed]
- Ruan, A.; Ren, C.; Quan, S. Conversion of the Molecular Chaperone Spy into a Novel Fusion Tag to Enhance Recombinant Protein Expression. J. Biotechnol. 2020, 307, 131–138. [Google Scholar] [CrossRef]
- Fatima, K.; Naqvi, F.; Younas, H. A Review: Molecular Chaperone-Mediated Folding, Unfolding and Disaggregation of Expressed Recombinant Proteins. Cell Biochem. Biophys. 2021, 79, 153–174. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, M.; Lv, X.; Fan, J.; Zhang, J.; Sun, J.; Shen, Y. GroEL/ES Mediated the in Vivo Recovery of TRAIL Inclusion Bodies in Escherichia coli. Sci. Rep. 2018, 8, 15766. [Google Scholar] [CrossRef] [PubMed]
- Utami, D.F.; Azizah, M.I.; Sriwidodo, S.; Haryanto, R.A. Review Article: Effect of Co-Expression Chaperones on the Expression of Intracellular Recombinant Proteins in Escherichia coli. Chim. Nat. Acta 2023, 11, 25–33. [Google Scholar]
- Sajed, M.; Falak, S.; Muhammad, M.A.; Ahmad, N.; Rashid, N. A Plant-Type L-Asparaginase from Pyrobaculum calidifontis Undergoes Temperature Dependent Autocleavage. Biologia 2022, 77, 3623–3631. [Google Scholar] [CrossRef]
- Seyed Hosseini Fin, N.A.; Barshan-tashnizi, M.; Sajjadi, S.M.; Asgari, S.; Mohajerani, N.; Mirzahoseini, H. The Effects of Overexpression of Cytoplasmic Chaperones on Secretory Production of Hirudin-PA in E. coli. Protein Expr. Purif. 2019, 157, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Biglari Goliloo, E.; Tollabi, A.; Jaliani, H.Z. Soluble Expression and Purification of Q59L Mutant L-Asparaginase in the Presence of Chaperones in SHuffleTM T7 Straine. Int. J. Med. Lab. 2021, 8, 138–146. [Google Scholar] [CrossRef]
- Bhatwa, A.; Wang, W.; Hassan, Y.I.; Abraham, N.; Li, X.-Z.; Zhou, T. Challenges Associated with the Formation of Recombinant Protein Inclusion Bodies in Escherichia coli and Strategies to Address Them for Industrial Applications. Front. Bioeng. Biotechnol. 2021, 9, 630551. [Google Scholar] [CrossRef] [PubMed]
- Vernet, E.; Kotzsch, A.; Voldborg, B.; Sundström, M. Screening of Genetic Parameters for Soluble Protein Expression in Escherichia coli. Protein Expr. Purif. 2011, 77, 104–111. [Google Scholar] [CrossRef]
- Tomar, R.; Garg, D.K.; Mishra, R.; Thakur, A.K.; Kundu, B. N-Terminal Domain of Pyrococcus Furiosus L-Asparaginase Functions as a Non-Specific, Stable, Molecular Chaperone. FEBS J. 2013, 280, 2688–2699. [Google Scholar] [CrossRef]
- Jena, R.; Garg, D.K.; Choudhury, L.; Saini, A.; Kundu, B. Heterologous Expression of an Engineered Protein Domain Acts as Chaperone and Enhances Thermotolerance of Escherichia coli. Int. J. Biol. Macromol. 2018, 107, 2086–2093. [Google Scholar] [CrossRef]
- Looser, V.; Bruhlmann, B.; Bumbak, F.; Stenger, C.; Costa, M.; Camattari, A.; Fotiadis, D.; Kovar, K. Cultivation Strategies to Enhance Productivity of Pichia pastoris: A Review. Biotechnol. Adv. 2015, 33, 1177–1193. [Google Scholar] [CrossRef]
- Hymavathi, M.; Sathish, T.; Rao, C.S.; Prakasham, R.S. Enhancement of L-Asparaginase Production by Isolated Bacillus circulans (MTCC 8574) Using Response Surface Methodology. Appl. Biochem. Biotechnol. 2009, 159, 191–198. [Google Scholar] [CrossRef]
- Darnal, S.; Patial, V.; Kumar, V.; Kumar, S.; Kumar, V.; Padwad, Y.S.; Singh, D. Biochemical Characterization of Extremozyme L-Asparaginase from Pseudomonas sp. PCH199 for Therapeutics. AMB Express 2023, 13, 22. [Google Scholar] [CrossRef]
- Ghosh, S.; Murthy, S.; Govindasamy, S.; Chandrasekaran, M. Optimization of L-Asparaginase Production by Serratia Marcescens (NCIM 2919) under Solid State Fermentation Using Coconut Oil Cake. Sustain. Chem. Process. 2013, 1, 9. [Google Scholar] [CrossRef]
- Trang, T.H.N.; Cuong, T.N.; Thanh, S.L.N.; Tuyen, T. Do Optimization, Purification and Characterization of Recombinant L-Asparaginase II in Escherichia coli. Afr. J. Biotechnol. 2016, 15, 1681–1691. [Google Scholar] [CrossRef]
- Borah, D.; Yadav, R.N.S.; Sangra, A.; Sangra, L. Production, Purification and Process Optimization of L-Asparagines. Int. J. Pharm. Pharm. Sci. 2012, 4, 560–563. [Google Scholar]
- Goswami, R.; Veeranki, V.D.; Mishra, V.K. Optimization of Process Conditions and Evaluation of PH & Thermal Stability of Recombinant l-Asparaginase II of Erwinia carotovora Subsp. Atroseptica SCRI 1043 in E. coli. Biocatal. Agric. Biotechnol. 2019, 22, 101377. [Google Scholar] [CrossRef]
- Doriya, K.; Jose, N.; Gowda, M.; Kumar, D.S. Solid-State Fermentation vs. Submerged Fermentation for the Production of l-Asparaginase. Adv. Food Nutr. Res. 2016, 78, 115–135. [Google Scholar]
- Barros, T.; Brumano, L.; Freitas, M.; Pessoa, A.; Parachin, N.; Magalhães, P.O. Development of Processes for Recombinant L-Asparaginase II Production by Escherichia coli Bl21 (De3): From Shaker to Bioreactors. Pharmaceutics 2020, 13, 14. [Google Scholar] [CrossRef]
- Ukkonen, K.; Neubauer, A.; Pereira, V.J.; Vasala, A. High Yield of Recombinant Protein in Shaken E. coli Cultures with Enzymatic Glucose Release Medium EnPresso B. Methods Mol. Biol. 2017, 1586, 127–137. [Google Scholar]
- Mihooliya, K.N.; Nandal, J.; Kumari, A.; Nanda, S.; Verma, H.; Sahoo, D.K. Studies on Efficient Production of a Novel L-Asparaginase by a Newly Isolated Pseudomonas resinovorans IGS-131 and Its Heterologous Expression in Escherichia coli. 3 Biotech 2020, 10, 148. [Google Scholar] [CrossRef]
- Abhini, K.N.; Rajan, A.B.; Fathimathu Zuhara, K.; Sebastian, D. Response Surface Methodological Optimization of L-Asparaginase Production from the Medicinal Plant Endophyte Acinetobacter Baumannii ZAS1. J. Genet. Eng. Biotechnol. 2022, 20, 22. [Google Scholar] [CrossRef] [PubMed]
- Ghoshoon, M.B.; Berenjian, A.; Hemmati, S.; Dabbagh, F.; Karimi, Z.; Negahdaripour, M.; Ghasemi, Y. Extracellular Production of Recombinant L-Asparaginase II in Escherichia coli: Medium Optimization Using Response Surface Methodology. Int. J. Pept. Res. Ther. 2015, 21, 487–495. [Google Scholar] [CrossRef]
- Chergui, A.; Kecha, M.; Tighrine, A.; Adrar, N.; Adrar, S.; Titouche, Y.; Boughani, L.; Kadri, N.; Houali, K. Characterization and Optimization of Extracellular L-Asparaginase Production by Selected Actinomycete Strain Isolated from an Algerian Wheat Bran. Cell. Mol. Biol. 2018, 64, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Mahalik, S.; Sharma, A.; Das, D.R.; Mittra, D.; Mukherjee, K.J. Co-Expressing Leucine Responsive Regulatory Protein (Lrp) Enhances Recombinant L-Asparaginase-II Production in Escherichia coli. J. Biotechnol. 2022, 351, 99–108. [Google Scholar] [CrossRef]
- Sharma, A.K.; Shukla, E.; Janoti, D.S.; Mukherjee, K.J.; Shiloach, J. A Novel Knock out Strategy to Enhance Recombinant Protein Expression in Escherichia coli. Microb. Cell Factories 2020, 19, 148. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.R.; Byregowda, S.M.; Veeregowda, B.M.; Balamurugan, V. An Overview of Heterologous Expression Host Systems for the Production of Recombinant Proteins. Adv. Anim. Vet. Sci. 2016, 4, 346–356. [Google Scholar] [CrossRef]
- de Souza, C.C.; Guimarães, J.M.; Pereira, S.d.S.; Mariúba, L.A.M. The Multifunctionality of Expression Systems in Bacillus subtilis: Emerging Devices for the Production of Recombinant Proteins. Exp. Biol. Med. 2021, 246, 2443–2453. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Qu, J.; Zou, W.; Shen, W.; Chen, X. An Overview and Future Prospects of Recombinant Protein Production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2021, 105, 6607–6626. [Google Scholar] [CrossRef]
- Cui, W.; Han, L.; Suo, F.; Liu, Z.; Zhou, L.; Zhou, Z. Exploitation of Bacillus subtilis as a Robust Workhorse for Production of Heterologous Proteins and Beyond. World J. Microbiol. Biotechnol. 2018, 34, 145. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, J.; Tan, M.; Zhen, J.; Shu, W.; Yang, S.; Ma, Y.; Zheng, H.; Song, H. High Copy Number and Highly Stable Escherichia coli–Bacillus subtilis Shuttle Plasmids Based on PWB980. Microb. Cell Factories 2020, 19, 25. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Nguyen, Q.A.; Ferreira, R.C.; Ferreira, L.C.S.; Tran, L.T.; Schumann, W. Construction of Plasmid-Based Expression Vectors for Bacillus subtilis Exhibiting Full Structural Stability. Plasmid 2005, 54, 241–248. [Google Scholar] [CrossRef]
- Schumann, W. Production of Recombinant Proteins in Bacillus subtilis. Adv. Appl. Microbiol. 2007, 62, 137–189. [Google Scholar]
- Krüger, A.; Welsch, N.; Dürwald, A.; Brundiek, H.; Wardenga, R.; Piascheck, H.; Mengers, H.G.; Krabbe, J.; Beyer, S.; Kabisch, J.F.; et al. A Host-Vector Toolbox for Improved Secretory Protein Overproduction in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2022, 106, 5137–5151. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Cai, D.; Wang, H.; Xu, Y.; Xiong, S.; Gao, L.; Xiong, M.; Wang, Z.; Chen, S.; Ma, X. Construction and Application of a Dual Promoter System for Efficient Protein Production and Metabolic Pathway Enhancement in Bacillus licheniformis. J. Biotechnol. 2020, 312, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Yan, R.; Shen, J.; Zhu, X.; Meng, F.; Lu, Z.; Lu, F. Cis-Element Engineering Promotes the Expression of Bacillus subtilis Type I L-Asparaginase and Its Application in Food. Int. J. Mol. Sci. 2022, 23, 6588. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, S.; Jiao, Y.; Gao, H.; Wang, M.; Du, G.; Chen, J. Enhanced Extracellular Production of L-Asparaginase from Bacillus subtilis 168 by B. Subtilis WB600 through a Combined Strategy. Appl. Microbiol. Biotechnol. 2017, 101, 1509–1520. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Xu, M.; He, B.; Rao, Z. Cloning, Expression, and Characterization of L-Asparaginase from a Newly Isolated Bacillus subtilis B11–06. J. Agric. Food Chem. 2013, 61, 9428–9434. [Google Scholar] [CrossRef]
- Bento, H.B.S.; Paiva, G.B.; Almeida, M.R.; Silva, C.G.; Carvalho, P.J.; Tavares, A.P.M.; Pedrolli, D.B.; Santos-Ebinuma, V.C. Aliivibrio Fischeri L-Asparaginase Production by Engineered Bacillus subtilis: A Potential New Biopharmaceutical. Bioprocess Biosyst. Eng. 2022, 45, 1635–1644. [Google Scholar] [CrossRef]
- Chityala, S.; Venkata Dasu, V.; Ahmad, J.; Prakasham, R.S. High Yield Expression of Novel Glutaminase Free L-Asparaginase II of Pectobacterium carotovorum MTCC 1428 in Bacillus subtilis WB800N. Bioprocess Biosyst. Eng. 2015, 38, 2271–2284. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Meng, F.; Zhou, Y.; Zhang, C.; Lu, Z.; Lu, F.; Chen, M. Non-Classical Secretion of a Type I L-Asparaginase in Bacillus subtilis. Int. J. Biol. Macromol. 2021, 180, 677–683. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, S.; Jiao, Y.; Wang, Y.; Wang, M.; Du, G. Gene Cloning and Expression of the L-Asparaginase from Bacillus cereus BDRD-ST26 in Bacillus subtilis WB600. J. Biosci. Bioeng. 2019, 127, 418–424. [Google Scholar] [CrossRef]
- Moorthy, V.; Ramalingam, A.; Alagarsamy, S.; Shankaranaya, R.T. Production, Purification and Characterisation of Extracellular L-Asparaginase from a Soil Isolate of Bacillus sp. Afr. J. Microbiol. Res. 2010, 4, 1862–1867. [Google Scholar]
- Pan, Y.; Yang, J.; Wu, J.; Yang, L.; Fang, H. Current Advances of Pichia pastoris as Cell Factories for Production of Recombinant Proteins. Front. Microbiol. 2022, 13, 1059777. [Google Scholar] [CrossRef]
- Daly, R.; Hearn, M.T.W. Expression of Heterologous Proteins in Pichia pastoris: A Useful Experimental Tool in Protein Engineering and Production. J. Mol. Recognit. 2005, 18, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A Highly Successful Expression System for Optimal Synthesis of Heterologous Proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, R.; Karbalaei, M.; Soleimanpour, S.; Mosavat, A.; Rezaee, S.A.; Ghazvini, K.; Farsiani, H. Practical Methods for Expression of Recombinant Protein in the Pichia pastoris System. Curr. Protoc. 2021, 1, e155. [Google Scholar] [CrossRef] [PubMed]
- Macauley-Patrick, S.; Fazenda, M.L.; McNeil, B.; Harvey, L.M. Heterologous Protein Production Using the Pichia pastoris Expression System. Yeast 2005, 22, 249–270. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Anumanthan, A.; Gao, X.-G.; Ilangovan, K.; Suzara, V.V.; Düzgüneş, N.; Renugopalakrishnan, V. Expression of Recombinant Proteins in Pichia pastoris. Appl. Biochem. Biotechnol. 2007, 142, 105–124. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Heterologous Protein Expression in Pichia pastoris: Latest Research Progress and Applications. Chembiochem A Eur. J. Chem. Biol. 2018, 19, 7–21. [Google Scholar] [CrossRef]
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein Expression in Pichia pastoris: Recent Achievements and Perspectives for Heterologous Protein Production. Appl. Microbiol. Biotechnol. 2014, 98, 5301–5317. [Google Scholar] [CrossRef]
- Safder, I.; Islam, I.-U.; Kazim, M.; Khan, S. Pichia pastoris Expression System: A Potential Candidate to Express Protein in Industrial and Biopharmaceutical Domains. Biomed. Lett. 2018, 4, 1–14. [Google Scholar]
- Gao, J.; Jiang, L.; Lian, J. Development of Synthetic Biology Tools to Engineer Pichia pastoris as a Chassis for the Production of Natural Products. Synth. Syst. Biotechnol. 2021, 6, 110–119. [Google Scholar] [CrossRef]
- Thor, D.; Xiong, S.; Orazem, C.; Kwan, A.; Cregg, J.; Lincereghino, J.; Lincereghino, G. Cloning and Characterization of the Gene as a Selectable Marker. FEMS Yeast Res. 2005, 5, 935–942. [Google Scholar] [CrossRef]
- Piva, L.C.; Bentacur, M.O.; Reis, V.C.B.; De Marco, J.L.; de Moraes, L.M.P.; Torres, F.A.G. Molecular Strategies to Increase the Levels of Heterologous Transcripts in Komagataella Phaffii for Protein Production. Bioengineered 2017, 8, 441–445. [Google Scholar] [CrossRef]
- Erden-Karaoğlan, F.; Karaoğlan, M. Improvement of Recombinant L-Asparaginase Production in Pichia pastoris. 3 Biotech 2023, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Cai, H.; Liu, J.; Zeng, M.; Chen, J.; Cheng, Q.; Zhang, L. Controlling AOX1 Promoter Strength in Pichia pastoris by Manipulating Poly (DA:DT) Tracts. Sci. Rep. 2018, 8, 1401. [Google Scholar] [CrossRef] [PubMed]
- Türkanoğlu Özçelik, A.; Yılmaz, S.; Inan, M. Pichia pastoris Promoters. Methods Mol. Biol. 2019, 1923, 97–112. [Google Scholar] [PubMed]
- Waterham, H.R.; Digan, M.E.; Koutz, P.J.; Lair, S.V.; Cregg, J.M. Isolation of the Pichia pastoris Glyceraldehyde-3-Phosphate Dehydrogenase Gene and Regulation and Use of Its Promoter. Gene 1997, 186, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T. Engineering of Promoters for Gene Expression in Pichia pastoris. Methods Mol. Biol. 2022, 2513, 153–177. [Google Scholar]
- Ferrara, M.A.; Severino, N.M.B.B.; Mansure, J.J.; Martins, A.S.; Oliveira, E.M.M.M.; Siani, A.C.; Pereira, N.; Torres, F.A.G.G.; Bon, E.P.S.S. Asparaginase Production by a Recombinant Pichia pastoris Strain Harbouring Saccharomyces cerevisiae ASP3 Gene. Enzym. Microb. Technol. 2006, 39, 1457–1463. [Google Scholar] [CrossRef]
- Nguyen, T.H.T.T.C.; Do, T.T.; Nguyen, T.H.T.T.C.; Quyen, D.T. Expression, Purification and Evaluation of Recombinant L-Asparaginase in Mehthylotrophic Yeast Pichia pastoris. J. Vietnam. Environ. 2014, 6, 288–292. [Google Scholar] [CrossRef]
- Jacobs, P.P.; Geysens, S.; Vervecken, W.; Contreras, R.; Callewaert, N. Engineering Complex-Type N-Glycosylation in Pichia pastoris Using GlycoSwitch Technology. Nat. Protoc. 2009, 4, 58–70. [Google Scholar] [CrossRef]
- Effer, B.; Lima, G.M.; Cabarca, S.; Pessoa, A.; Farías, J.G.; Monteiro, G. L-Asparaginase from E. Chrysanthemi Expressed in Glycoswitch®: Effect of His-Tag Fusion on the Extracellular Expression. Prep. Biochem. Biotechnol. 2019, 49, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Sajitha, S.; Vidya, J.; Varsha, K.; Binod, P.; Pandey, A. Cloning and Expression of L-Asparaginase from E. coli in Eukaryotic Expression System. Biochem. Eng. J. 2015, 102, 14–17. [Google Scholar] [CrossRef]
- Biasoto, H.P.; Hebeda, C.B.; Farsky, S.H.P.; Pessoa, A.; Costa-Silva, T.A.; Monteiro, G. Extracellular Expression of Saccharomyces cerevisiae’s L-Asparaginase II in Pichia pastoris Results in Novel Enzyme with Better Parameters. Prep. Biochem. Biotechnol. 2023, 53, 511–522. [Google Scholar] [CrossRef]
- Viswanath, V. A Short Communication on Pichia pastoris vs. E. coli: Efficient Expression System. Ann. Proteom. Bioinform. 2021, 5, 49–50. [Google Scholar] [CrossRef]
- de Almeida Parizotto, L.; Krebs Kleingesinds, E.; Manfrinato Pedrotti da Rosa, L.; Effer, B.; Meira Lima, G.; Herkenhoff, M.E.; Li, Z.; Rinas, U.; Monteiro, G.; Pessoa, A.; et al. Increased Glycosylated L-Asparaginase Production through Selection of Pichia pastoris Platform and Oxygen-Methanol Control in Fed-Batches. Biochem. Eng. J. 2021, 173, 108083. [Google Scholar] [CrossRef]
- Lima, G.M.; Effer, B.; Biasoto, H.P.; Feijoli, V.; Pessoa, A.; Palmisano, G.; Monteiro, G. Glycosylation of L-Asparaginase from E. coli through Yeast Expression and Site-Directed Mutagenesis. Biochem. Eng. J. 2020, 156, 107516. [Google Scholar] [CrossRef]
- Effer, B.; Kleingesinds, E.K.; Lima, G.M.; Costa, I.M.; Sánchez-Moguel, I.; Pessoa, A.; Santiago, V.F.; Palmisano, G.; Farías, J.G.; Monteiro, G. Glycosylation of Erwinase Results in Active Protein Less Recognized by Antibodies. Biochem. Eng. J. 2020, 163, 107750. [Google Scholar] [CrossRef]
- Çalık, P.; Ata, Ö.; Güneş, H.; Massahi, A.; Boy, E.; Keskin, A.; Öztürk, S.; Zerze, G.H.; Özdamar, T.H. Recombinant Protein Production in Pichia pastoris under Glyceraldehyde-3-Phosphate Dehydrogenase Promoter: From Carbon Source Metabolism to Bioreactor Operation Parameters. Biochem. Eng. J. 2015, 95, 20–36. [Google Scholar] [CrossRef]
- Rodrigues, D.; Pillaca-Pullo, O.; Torres-Obreque, K.; Flores-Santos, J.; Sánchez-Moguel, I.; Pimenta, M.V.; Basi, T.; Converti, A.; Lopes, A.M.; Monteiro, G.; et al. Fed-Batch Production of Saccharomyces cerevisiae L-Asparaginase II by Recombinant Pichia pastoris MUTs Strain. Front. Bioeng. Biotechnol. 2019, 7, 16. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, Q.; Wu, W.; Pu, Z.; Yu, H. Rational Design of Enzyme Activity and Enantioselectivity. Front. Bioeng. Biotechnol. 2023, 11, 1129149. [Google Scholar] [CrossRef]
- Korendovych, I.V. Rational and Semirational Protein Design. Methods Mol. Biol. 2018, 1685, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Miton, C.M.; Tokuriki, N. A Mechanistic View of Enzyme Evolution. Protein Sci. 2020, 29, 1724–1747. [Google Scholar] [CrossRef]
- Zeymer, C.; Hilvert, D. Directed Evolution of Protein Catalysts. Annu. Rev. Biochem. 2018, 87, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Basener, W.F.; Sanford, J.C. The Fundamental Theorem of Natural Selection with Mutations. J. Math. Biol. 2018, 76, 1589–1622. [Google Scholar] [CrossRef]
- Reetz, M.T. Directed Evolution of Selective Enzymes: Catalysts for Organic Chemistry and Biotechnology; Wiley: Hoboken, NJ, USA, 2016; ISBN 9783527316601. [Google Scholar]
- Moore, G.L.; Maranas, C.D. Modeling DNA Mutation and Recombination for Directed Evolution Experiments. J. Theor. Biol. 2000, 205, 483–503. [Google Scholar] [CrossRef] [PubMed]
- Sellés Vidal, L.; Isalan, M.; Heap, J.T.; Ledesma-Amaro, R. A Primer to Directed Evolution: Current Methodologies and Future Directions. RSC Chem. Biol. 2023, 4, 271–291. [Google Scholar] [CrossRef]
- Cirino, P.C.; Mayer, K.M.; Umeno, D. Generating Mutant Libraries Using Error-Prone PCR. In Directed Evolution Library Creation; Humana Press: Totowa, NJ, USA, 2003; Volume 231, pp. 3–10. [Google Scholar]
- Sen, S.; Venkata Dasu, V.; Mandal, B. Developments in Directed Evolution for Improving Enzyme Functions. Appl. Biochem. Biotechnol. 2007, 143, 212–223. [Google Scholar] [CrossRef]
- Worall, A.F. Site-Directed Mutagenesis by the Cassette Method. In DNA-Protein Interactions; Humana Press: Totowa, NJ, USA, 1994; Volume 30, pp. 199–210. [Google Scholar]
- Williams, E.M.; Copp, J.N.; Ackerley, D.F. Site-Saturation Mutagenesis by Overlap Extension PCR. In Directed Evolution Library Creation; Methods in Molecular Biology; Springer: New York, NY, USA, 2014; Volume 1179, pp. 83–101. [Google Scholar]
- Chembath, A.; Wagstaffe, B.P.G.; Ashraf, M.; Amaral, M.M.F.; Frigotto, L.; Hine, A.V. Nondegenerate Saturation Mutagenesis: Library Construction and Analysis via MAX and ProxiMAX Randomization. In Directed Evolution; Humana: New York, NY, USA, 2022; Volume 2461, pp. 19–41. [Google Scholar]
- Handal-Marquez, P.; Koch, M.; Kestemont, D.; Arangundy-Franklin, S.; Pinheiro, V.B. Antha-Guided Automation of Darwin Assembly for the Construction of Bespoke Gene Libraries. In Directed Evolution; Humana: New York, NY, USA, 2022; Volume 2461, pp. 43–66. [Google Scholar]
- Levi, T.; Sloutskin, A.; Kalifa, R.; Juven-Gershon, T.; Gerlitz, O. Efficient In Vivo Introduction of Point Mutations Using SsODN and a Co-CRISPR Approach. Biol. Proced. Online 2020, 22, 14. [Google Scholar] [CrossRef]
- Zhao, H.; Zha, W. In Vitro “sexual” Evolution through the PCR-Based Staggered Extension Process (StEP). Nat. Protoc. 2006, 1, 1865–1871. [Google Scholar] [CrossRef]
- Patrick, W.M.; Gerth, M.L. ITCHY: Incremental Truncation for the Creation of Hybrid Enzymes. In Directed Evolution Library Creation; Springer: New York, NY, USA, 2014; Volume 1179, pp. 225–244. [Google Scholar]
- Coco, W.M. RACHITT Gene Family Shuffling by Random Chimeragenesis on Transient Templates. In Directed Evolution Library Creation; Humana Press: Totowa, NJ, USA, 2003; Volume 231, pp. 111–128. [Google Scholar]
- McClellan, M.J. In Vitro Site Directed Mutagenesis. In DNA Manipulation and Analysis; Humana: New York, NY, USA, 2023; Volume 2633, pp. 87–95. [Google Scholar]
- Butt, H.; Zaidi, S.S.-A.; Hassan, N.; Mahfouz, M. CRISPR-Based Directed Evolution for Crop Improvement. Trends Biotechnol. 2020, 38, 236–240. [Google Scholar] [CrossRef]
- Cui, H.; Davari, M.D.; Schwaneberg, U. Recombination of Single Beneficial Substitutions Obtained from Protein Engineering by Computer-Assisted Recombination (CompassR). Methods Mol. Biol. 2022, 2461, 9–18. [Google Scholar] [CrossRef]
- Lu, X.; Chen, J.; Jiao, L.; Zhong, L.; Lu, Z.; Zhang, C.; Lu, F. Improvement of the Activity of L-Asparaginase I Improvement of the Catalytic Activity of l-Asparaginase I from Bacillus megaterium H-1 by In Vitro Directed Evolution. J. Biosci. Bioeng. 2019, 128, 683–689. [Google Scholar] [CrossRef]
- Rigouin, C.; Nguyen, H.A.; Schalk, A.M.; Lavie, A. Discovery of Human-like L-Asparaginases with Potential Clinical Use by Directed Evolution. Sci. Rep. 2017, 7, 10224. [Google Scholar] [CrossRef] [PubMed]
- Kotzia, G.A.; Labrou, N.E. Engineering Thermal Stability of L-Asparaginase by in Vitro Directed Evolution. FEBS J. 2009, 276, 1750–1761. [Google Scholar] [CrossRef] [PubMed]
- Kotzia, G.A.; Labrou, N.E. Engineering Substrate Specificity of E. carotovora l-Asparaginase for the Development of Biosensor. J. Mol. Catal. B Enzym. 2011, 72, 95–101. [Google Scholar] [CrossRef]
- Karamitros, C.S.; Konrad, M. Fluorescence-Activated Cell Sorting of Human l-Asparaginase Mutant Libraries for Detecting Enzyme Variants with Enhanced Activity. ACS Chem. Biol. 2016, 11, 2596–2607. [Google Scholar] [CrossRef]
- Attene-Ramos, M.S.; Austin, C.P.; Xia, M. High Throughput Screening. In Encyclopedia of Toxicology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 916–917. [Google Scholar]
- Coleman, R.J.; Bruck, T. Method for Production of Recombinant Erwinia Asparaginase. Available online: https://patents.google.com/patent/US10787671B2/en2019 (accessed on 11 October 2023).
- Beckett, A.; Gervais, D. What Makes a Good New Therapeutic L-Asparaginase? World J. Microbiol. Biotechnol. 2019, 35, 152. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, N.; Lamminmäki, U.; Khanna, N.; Batra, G. Enhanced Cell Density Cultivation and Rapid Expression-Screening of Recombinant Pichia pastoris Clones in Microscale. Sci. Rep. 2020, 10, 7458. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Kickenweiz, T.; Pitzer, J.; Sturmberger, L.; Weninger, A.; Biggs, B.W.; Köhler, E.-M.; Baumschlager, A.; Fischer, J.E.; Hyden, P.; et al. Publisher Correction: Engineered Bidirectional Promoters Enable Rapid Multi-Gene Co-Expression Optimization. Nat. Commun. 2021, 12, 1287. [Google Scholar] [CrossRef]
- Pongsupasa, V.; Anuwan, P.; Maenpuen, S.; Wongnate, T. Rational-Design Engineering to Improve Enzyme Thermostability. Methods Mol. Biol. 2022, 2397, 159–178. [Google Scholar]
- Poluri, K.M.; Gulati, K. Rational Designing of Novel Proteins Through Computational Approaches. In Protein Engineering Techniques: Gateways to Synthetic Protein Universe; Springer: Singapore, 2017; pp. 61–83. [Google Scholar]
- Xie, W.J.; Asadi, M.; Warshel, A. Enhancing Computational Enzyme Design by a Maximum Entropy Strategy. Proc. Natl. Acad. Sci. USA 2022, 119, e2122355119. [Google Scholar] [CrossRef]
- Yin, C.; Yau, S.S.-T. A Coevolution Analysis for Identifying Protein-Protein Interactions by Fourier Transform. PLoS ONE 2017, 12, e0174862. [Google Scholar] [CrossRef] [PubMed]
- Lladós, J.; Cores, F.; Guirado, F.; Lérida, J.L. Accurate Consistency-Based MSA Reducing the Memory Footprint. Comput. Methods Programs Biomed. 2021, 208, 106237. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kaushik, V.; Goel, M. Insights into the Distribution and Functional Properties of L-Asparaginase in the Archaeal Domain and Characterization of Picrophilus torridus Asparaginase Belonging to the Novel Family Asp2like1. ACS Omega 2022, 7, 40750–40765. [Google Scholar] [CrossRef] [PubMed]
- Safrhansova, L.; Hlozkova, K.; Starkova, J. Targeting Amino Acid Metabolism in Cancer. In International Review of Cell and Molecular Biology; Elsevier Inc.: Alpharetta, GA, USA, 2022; Volume 373, pp. 37–79. [Google Scholar] [CrossRef]
- Hobbs, J.K.; Prentice, E.J.; Groussin, M.; Arcus, V.L. Reconstructed Ancestral Enzymes Impose a Fitness Cost upon Modern Bacteria Despite Exhibiting Favourable Biochemical Properties. J. Mol. Evol. 2015, 81, 110–120. [Google Scholar] [CrossRef]
- Ayuso-Fernández, I.; Molpeceres, G.; Camarero, S.; Ruiz-Dueñas, F.J.; Martínez, A.T. Ancestral Sequence Reconstruction as a Tool to Study the Evolution of Wood Decaying Fungi. Front. Fungal Biol. 2022, 3, 1003489. [Google Scholar] [CrossRef]
- Planas-Iglesias, J.; Marques, S.M.; Pinto, G.P.; Musil, M.; Stourac, J.; Damborsky, J.; Bednar, D. Computational Design of Enzymes for Biotechnological Applications. Biotechnol. Adv. 2021, 47, 107696. [Google Scholar] [CrossRef]
- Faber, M.S.; Whitehead, T.A. Data-Driven Engineering of Protein Therapeutics. Curr. Opin. Biotechnol. 2019, 60, 104–110. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.; Song, J. Computational Enzyme Design Approaches with Significant Biological Outcomes: Progress and Challenges. Comput. Struct. Biotechnol. J. 2012, 2, e201209007. [Google Scholar] [CrossRef]
- Ju, F.; Zhu, J.; Shao, B.; Kong, L.; Liu, T.-Y.; Zheng, W.-M.; Bu, D. Learning Residue Co-Evolution Directly from Multiple Sequence Alignment for Protein Structure Prediction. Nat. Commun. 2021, 12, 2535. [Google Scholar] [CrossRef]
- Safary, A.; Moniri, R.; Hamzeh-Mivehroud, M.; Dastmalchi, S. Highly Efficient Novel Recombinant L-Asparaginase with No Glutaminase Activity from a New Halo-Thermotolerant Bacillus Strain. BioImpacts BI 2019, 9, 15–23. [Google Scholar] [CrossRef]
- Chi, H.; Chen, M.; Jiao, L.; Lu, Z.; Bie, X.; Zhao, H.; Lu, F. Characterization of a Novel L-Asparaginase from Mycobacterium gordonae with Acrylamide Mitigation Potential. Foods 2021, 10, 2819. [Google Scholar] [CrossRef] [PubMed]
- Dumina, M.; Zhgun, A.; Pokrovskaya, M.; Aleksandrova, S.; Zhdanov, D.; Sokolov, N.; El’darov, M. Highly Active Thermophilic L-Asparaginase from Melioribacter roseus Represents a Novel Large Group of Type II Bacterial L-Asparaginases from Chlorobi-Ignavibacteriae-Bacteroidetes Clade. Int. J. Mol. Sci. 2021, 22, 13632. [Google Scholar] [CrossRef]
- Agnihotry, S.; Pathak, R.K.; Singh, D.B.; Tiwari, A.; Hussain, I. Protein Structure Prediction. In Bioinformatics; Elsevier: Amsterdam, The Netherlands, 2022; pp. 177–188. [Google Scholar]
- Praveen, R.P. Modeling and Validation of L-Asparaginase Enzyme, an Anticancer Agent Using the Tools of Computational Biology. Int. J. Res. Med. Sci. 2019, 8, 211–214. [Google Scholar] [CrossRef]
- Farahat, M.G.; Amr, D.; Galal, A. Molecular Cloning, Structural Modeling and Characterization of a Novel Glutaminase-Free L-Asparaginase from Cobetia amphilecti AMI6. Int. J. Biol. Macromol. 2020, 143, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Saeed, H.; Ali, H.; Soudan, H.; Embaby, A.; El-Sharkawy, A.; Farag, A.; Hussein, A.; Ataya, F. Molecular Cloning, Structural Modeling and Production of Recombinant Aspergillus terreus L. Asparaginase in Escherichia coli. Int. J. Biol. Macromol. 2018, 106, 1041–1051. [Google Scholar] [CrossRef]
- Vasina, M.; Velecký, J.; Planas-Iglesias, J.; Marques, S.M.; Skarupova, J.; Damborsky, J.; Bednar, D.; Mazurenko, S.; Prokop, Z. Tools for Computational Design and High-Throughput Screening of Therapeutic Enzymes. Adv. Drug Deliv. Rev. 2022, 183, 114143. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Khosrow, A.; Elham, B.; Shiva, J. Bioinformatic Analysis of L-Asparaginase II from Citrobacter Freundii 1101, Erwinia Chrysanthemi DSM 4610, E. coli BL21 and Klebsiella pneumoniae ATCC 10031. Int. J. Med. Lab. 2017, 4, 123–134. [Google Scholar]
- Long, S.; Zhang, X.; Rao, Z.; Chen, K.; Xu, M.; Yang, T.; Yang, S. Amino Acid Residues Adjacent to the Catalytic Cavity of Tetramer L-Asparaginase II Contribute Significantly to Its Catalytic Efficiency and Thermostability. Enzym. Microb. Technol. 2016, 82, 15–22. [Google Scholar] [CrossRef]
- Rizzuti, B. Molecular Simulations of Proteins: From Simplified Physical Interactions to Complex Biological Phenomena. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2022, 1870, 140757. [Google Scholar] [CrossRef] [PubMed]
- Malinverni, D.; Barducci, A. Coevolutionary Analysis of Protein Sequences for Molecular Modeling. Methods Mol. Biol. 2019, 2022, 379–397. [Google Scholar] [PubMed]
- Pola, M.; Rajulapati, S.B.; Potla Durthi, C.; Erva, R.R.; Bhatia, M. In Silico Modelling and Molecular Dynamics Simulation Studies on L-Asparaginase Isolated from Bacterial Endophyte of Ocimum tenuiflorum. Enzym. Microb. Technol. 2018, 117, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Srivastava, A.; Mukherjee, G.; Pandey, R.; Verma, A.K.; Mishra, P.; Kundu, B. Hyperthermophilic asparaginase Mutants with Enhanced Substrate Affinity and Antineoplastic Activity: Structural Insights on Their Mechanism of Action. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012, 26, 1161–1171. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, H.; Yao, X.; Li, D.; Xu, L.; Li, Y.; Tian, S.; Hou, T. Comprehensive Evaluation of Ten Docking Programs on a Diverse Set of Protein–Ligand Complexes: The Prediction Accuracy of Sampling Power and Scoring Power. Phys. Chem. Chem. Phys. 2016, 18, 12964–12975. [Google Scholar] [CrossRef]
- Erva, R.R.; Rajulapati, S.B.; Potla Durthi, C.; Bhatia, M.; Pola, M. Molecular Dynamic Simulations of Escherichia coli L-Asparaginase to Illuminate Its Role in Deamination of Asparagine and Glutamine residues. 3 Biotech 2016, 6, 2. [Google Scholar] [CrossRef]
- Reddy, E.R.; Babu, R.S.; Chandrasai, P.D.; Madhuri, P. Exploration of the Binding Modes of L-Asparaginase Complexed with Its Amino Acid Substrates by Molecular Docking, Dynamics and Simulation. 3 Biotech 2016, 6, 105. [Google Scholar] [CrossRef]
- Hozoorbakhsh, F.; Ghiasian, M.; Ghandehari, F.; Emami-Karvani, Z.; Khademi Dehkordi, M. An Immunoinformatic Approach Employing Molecular Docking and Molecular Dynamics Simulation for Evaluation of L-Asparaginase Produced by Bacillus velezensis. J. Biomol. Struct. Dyn. 2022, 41, 9057–9071. [Google Scholar] [CrossRef]
- Buß, O.; Rudat, J.; Ochsenreither, K. FoldX as Protein Engineering Tool: Better than Random Based Approaches? Comput. Struct. Biotechnol. J. 2018, 16, 25–33. [Google Scholar] [CrossRef]
- Mahboobi, M.; Sedighian, H.; Hedayati CH, M.; Bambai, B.; Esmaeil Soofian, S.; Amani, J. Applying Bioinformatic Tools for Modeling and Modifying Type II E. coli l-Asparginase to Present a Better Therapeutic Agent/Drug for Acute Lymphoblastic leukemia. Int. J. Cancer Manag. 2017, 10, e5785. [Google Scholar] [CrossRef]
- Chi, H.; Wang, Y.; Xia, B.; Zhou, Y.; Lu, Z.; Lu, F.; Zhu, P. Enhanced Thermostability and Molecular Insights for L -Asparaginase from Bacillus licheniformis Structure- and Computation-Based Rational Design. J. Agric. Food Chem. 2022, 70, 14499–14509. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX Web Server: An Online Force Field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Dastmalchi, M.; Alizadeh, M.; Jamshidi-Kandjan, O.; Rezazadeh, H.; Hamzeh-Mivehroud, M.; Farajollahi, M.M.; Dastmalchi, S. Expression and Biological Evaluation of an Engineered Recombinant L-Asparaginase Designed by in Silico Method Based on Sequence of the Enzyme from E. coli. Adv. Pharm. Bull. 2023. [Google Scholar] [CrossRef]
- Marcos, E.; Silva, D. Essentials of de Novo Protein Design: Methods and Applications. WIREs Comput. Mol. Sci. 2018, 8, e1374. [Google Scholar] [CrossRef]
- Dhanjal, J.K.; Malik, V.; Radhakrishnan, N.; Sigar, M.; Kumari, A.; Sundar, D. Computational Protein Engineering Approaches for Effective Design of New Molecules. In Encyclopedia of Bioinformatics and Computational Biology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 631–643. [Google Scholar]
- Ferreira, P.; Fernandes, P.A.; Ramos, M.J. Modern Computational Methods for Rational Enzyme Engineering. Chem Catal. 2022, 2, 2481–2498. [Google Scholar] [CrossRef]
- King, C.; Garza, E.N.; Mazor, R.; Linehan, J.L.; Pastan, I.; Pepper, M.; Baker, D. Removing T-Cell Epitopes with Computational Protein Design. Proc. Natl. Acad. Sci. USA 2014, 111, 8577–8582. [Google Scholar] [CrossRef]
- Baskar, G.; Rajasekar, V.; Renganathan, S. Modeling and Optimization of L-Asparaginase Productionby Enterobacter Aerogenes Using Artificial Neural Network Linked Genetic Algorithm. Int. J. Chem. Eng. Appl. 2011, 2, 98–100. [Google Scholar] [CrossRef]
- Mortazavi, M.; Torkzadeh-Mahani, M.; Kargar, F.; Nezafat, N.; Ghasemi, Y. In Silico Analysis of Codon Usage and Rare Codon Clusters in the Halophilic Bacteria L-Asparaginase. Biologia 2020, 75, 151–160. [Google Scholar] [CrossRef]
- Orhan, H.; Aktaş Uygun, D. Immobilization of L-Asparaginase on Magnetic Nanoparticles for Cancer Treatment. Appl. Biochem. Biotechnol. 2020, 191, 1432–1443. [Google Scholar] [CrossRef]
- Kante, R.K.; Somavarapu, S.; Vemula, S.; Kethineni, C.; Mallu, M.R.; Ronda, S.R. Production of Recombinant Human Asparaginase from Escherichia coli under Optimized Fermentation Conditions: Effect of Physicochemical Properties on Enzyme Activity. Biotechnol. Bioprocess Eng. 2019, 24, 824–832. [Google Scholar] [CrossRef]
- González-Torres, I.; Perez-Rueda, E.; Evangelista-Martínez, Z.; Zárate-Romero, A.; Moreno-Enríquez, A.; Huerta-Saquero, A. Identification of L-Asparaginases from Streptomyces Strains with Competitive Activity and Immunogenic Profiles: A Bioinformatic Approach. PeerJ 2020, 8, e10276. [Google Scholar] [CrossRef] [PubMed]
- Ardalan, N.; Mirzaie, S.; Sepahi, A.A.; Khavari-Nejad, R.A. Novel Mutant of Escherichia coli Asparaginase II to Reduction of the Glutaminase Activity in Treatment of Acute Lymphocytic Leukemia by Molecular Dynamics Simulations and QM-MM Studies. Med. Hypotheses 2018, 112, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Derst, C.; Henseling, J.; Röhm, K.-H.H. Engineering the Substrate Specificity of Escherichia coli Asparaginase. II. Selective Reduction of Glutaminase Activity by Amino Acid Replacements at Position 248. Protein Sci. A Publ. Protein Soc. 2000, 9, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- LN, R.; Doble, M.; Rekha, V.P.B.; Pulicherla, K.K. In Silico Engineering of L-Asparaginase to Have Reduced Glutaminase Side Activity for Effective Treatment of Acute Lymphoblastic leukemia. J. Pediatr. Hematol./Oncol. 2011, 33, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Aghaeepoor, M.; Akbarzadeh, A.; Mirzaie, S.; Hadian, A.; Jamshidi Aval, S.; Dehnavi, E. Selective Reduction in Glutaminase Activity of L-Asparaginase by Asparagine 248 to Serine Mutation: A Combined Computational and Experimental Effort in Blood Cancer Treatment. Int. J. Biol. Macromol. 2018, 120, 2448–2457. [Google Scholar] [CrossRef] [PubMed]
- Baral, A.; Gorkhali, R.; Basnet, A.; Koirala, S.; Bhattarai, H.K. Selection of the Optimal L-Asparaginase II Against Acute Lymphoblastic leukemia: An In Silico Approach. JMIRx Med. 2021, 2, e29844. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jiao, L.; Shen, J.; Chi, H.; Lu, Z.; Liu, H.; Lu, F.; Zhu, P. Enhancing the Catalytic Activity of Type II L-Asparaginase from Bacillus licheniformis through Semi-Rational Design. Int. J. Mol. Sci. 2022, 23, 9663. [Google Scholar] [CrossRef] [PubMed]
- Sudhir, A.P.; Agarwaal, V.V.; Dave, B.R.; Patel, D.H.; Subramanian, R.B.B. Enhanced Catalysis of L-Asparaginase from Bacillus licheniformis by a Rational Redesign. Enzym. Microb. Technol. 2016, 86, 1–6. [Google Scholar] [CrossRef]
- Zhang, W.; Dai, Q.; Huang, Z.; Xu, W. Identification and Thermostability Modification of the Mesophilic L-Asparaginase from Limosilactobacillus secaliphilus. Appl. Biochem. Biotechnol. 2023, 1–15. [Google Scholar] [CrossRef]
- Jiao, L.; Chi, H.; Xia, B.; Lu, Z.; Bie, X.; Zhao, H.; Lu, F.; Chen, M. Thermostability Improvement of L-Asparaginase from Acinetobacter Soli via Consensus-Designed Cysteine Residue Substitution. Molecules 2022, 27, 6670. [Google Scholar] [CrossRef]
- Jia, R.; Wan, X.; Geng, X.; Xue, D.; Xie, Z.; Chen, C. Microbial L-Asparaginase for Application in Acrylamide Mitigation from Food: Current Research Status and Future Perspectives. Microorganisms 2021, 9, 1659. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Fang, X.; Su, F.; Chen, Y.; Xu, L.; Yan, Y. Enhancing the Thermostability of Rhizomucor miehei Lipase with a Limited Screening Library by Rational-Design Point Mutations and Disulfide Bonds. Appl. Environ. Microbiol. 2018, 84, e02129-17. [Google Scholar] [CrossRef] [PubMed]
- Cantor, J.R.; Panayiotou, V.; Agnello, G.; Georgiou, G.; Stone, E.M. Engineering Reduced-Immunogenicity Enzymes for Amino Acid Depletion Therapy in Cancer. Methods Enzymol. 2012, 502, 291–319. [Google Scholar] [CrossRef] [PubMed]
- Belén, L.H.; Lissabet, J.B.; de Oliveira Rangel-Yagui, C.; Effer, B.; Monteiro, G.; Pessoa, A.; Farías Avendaño, J.G. A Structural in Silico Analysis of the Immunogenicity of L-Asparaginase from Escherichia coli and Erwinia carotovora. Biologicals 2019, 59, 47–55. [Google Scholar] [CrossRef]
- Pedroso, A.; Herrera Belén, L.; Beltrán, J.F.; Castillo, R.L.; Pessoa, A.; Pedroso, E.; Farías, J.G. In Silico Design of a Chimeric Humanized L-Asparaginase. Int. J. Mol. Sci. 2023, 24, 7550. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Bootwala, A.; An, H.H.; Franklin, M.W.; Manning, B.J.; Xu, L.Y.; Panchal, S.; Garlick, J.D.; Baral, R.; Hudson, M.E.; Grigoryan, G.; et al. Protein Re-Surfacing of E. coli L-Asparaginase to Evade Pre-Existing Anti-Drug Antibodies and Hypersensitivity Responses. Front. Immunol. 2022, 13, 1016179. [Google Scholar] [CrossRef]
- Zhou, J.; Panaitiu, A.E.; Grigoryan, G. A General-Purpose Protein Design Framework Based on Mining Sequence–Structure Relationships in Known Protein Structures. Proc. Natl. Acad. Sci. USA 2020, 117, 1059–1068. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, Y.; Zhang, L.; Zhang, F.; Gao, W. Thermoresponsive polypeptide Fused L-Asparaginase with Mitigated Immunogenicity and Enhanced Efficacy in Treating Hematologic malignancies. Adv. Sci. 2023, 10, e2300469. [Google Scholar] [CrossRef]
- Chi, H.; Zhu, X.; Shen, J.; Lu, Z.; Lu, F.; Lyu, Y.; Zhu, P. Thermostability Enhancement and Insight of L-Asparaginase from Mycobacterium sp. via Consensus-Guided Engineering. Appl. Microbiol. Biotechnol. 2023, 107, 2321–2333. [Google Scholar] [CrossRef]
- Dadwal, A.; Sharma, S.; Satyanarayana, T. Progress in Ameliorating Beneficial Characteristics of Microbial Cellulases by Genetic Engineering Approaches for Cellulose saccharification. Front. Microbiol. 2020, 11, 1387. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Shukla, P. Current Trends in Protein Engineering: Updates and Progress. Curr. Protein Pept. Sci. 2019, 20, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Pardo, I.; Camarero, S. Laccase Engineering by Rational and Evolutionary Design. Cell. Mol. Life Sci. 2015, 72, 897–910. [Google Scholar] [CrossRef]
- Mate, D.M.; Alcalde, M. Laccase Engineering: From Rational Design to Directed Evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Seyedhosseini Ghaheh, H.; Sajjadi, S.; Shafiee, F.; Barzegari, E.; Moazen, F.; Mir Mohammad Sadeghi, H. Rational Design of a New Variant of Reteplase with Optimized Physicochemical Profile and Large-Scale Production in Escherichia coli. World J. Microbiol. Biotechnol. 2022, 38, 29. [Google Scholar] [CrossRef] [PubMed]

| Random Mutagenesis | References | |
|---|---|---|
| Error-prone PCR (epPCR) | A modification of the standard PCR method that is designed to alter and increase the natural error rate of the polymerase. Random mutations may be introduced into the DNA. | [204] |
| Sequence saturation mutagenesis (SeSaM) | Can be used to target one type of nucleotide in the selected sequence, and each type of nucleotide can be exchanged in a controlled manner. Completely independent of DNA polymerase mutation biases. | [205] |
| Cassette mutagenesis | The modification of a protein sequence at the level of the DNA by the replacement of a section of genetic information with an alternative sequence, usually provided by a synthetic duplex of DNA. Single or multiple amino acid changes to the protein sequence, as well as the insertion or deletion of sequences from the protein structure, can be achieved using this technique. | [206] |
| Site-saturation mutagenesis (SSM) | A strategy for the generation of high quality variant gene libraries of a defined size. Variation is introduced by creating a precise series of amino acid substitutions in the encoded protein through the incorporation of degenerate base combinations at specific codon positions. | [207] |
| Nondegenerate saturation mutagenesis | Contiguous (ProxiMAX) and noncontiguous (MAX) randomized codon generation techniques to create well-defined, diverse gene libraries in conjunction with other fully nondegenerate strategies. | [208] |
| The Darwin Assembly | Multiple site-saturation mutagenesis method capable of targeting up to 19 noncontiguous residues with less than 0.25% wild-type contamination. The assembly strategy has been designed with automation in mind, accelerating library construction and enabling the generation of highly complex libraries with minimal hands-on time and low potential for human error. | [209] |
| Recombination mutagenesis | ||
| DNA shuffling | Method for reassembling genes from their random DNA fragments, resulting in in vitro homologous recombination. Insertions and deletions result in nucleotide addition or removal. | [210] |
| Staggered extension protocol (StEP) | A modified PCR that generates staggered DNA fragments and promotes crossover events along the full length of the template sequence(s) using greatly shortened annealing and extension steps. Can be performed in a single tube and does not require DNA fragmentation. | [211] |
| Incremental truncation for the creation of hybrid enzymes (ITCHY) | Technique for the random recombination of two genes. The main advantage of ITCHY is that it does not require the two genes to share any sequence similarity. | [212] |
| Random chimeragenesis on transient templates (RACHITT) | Generates libraries with an average of 12 crossovers per gene in a single round of gene family shuffling. Generates chimeric genes by alignment of parental gene ‘donor’ fragments to a full-length DNA template | [213] |
| Rational mutagenesis | ||
| Site-directed mutagenesis | Specific point mutations can be introduced into plasmids by the use of primers (with the desired mutation) in a PCR protocol that will amplify the entire plasmid template. | [206,214] |
| Site-directed mutagenesis method mediated by Cas9 | Site-directed mutagenesis in a modern form. CRISPR-Cas9 systems can be programmed to target any locus in the genome and to carry out targeted CRISPR-mediated directed evolution. | [214,215] |
| Computer-assisted recombination (CompassR) strategy | Enables the effective and efficient recombination of identified beneficial substitutions to produce active enzymes with improved performance. By analyzing the relative free energy of folding, which has been used as a measure of protein stability and to assess the relationship between stability and function in several enzymes, the CompassR strategy guides the recombination of beneficial substitutions. | [216] |
| Source | Host | Method | Improvement | Reference |
|---|---|---|---|---|
| Directed evolution | ||||
| Bacillus megaterium H-1 | E. coli BL21(DE3) | epPCR, DNA shuffling | Catalytic activity | [217] |
| Guinea pig, human | E. coli BW5Δ strain | epPCR, DNA shuffling, CAST mutagenesis | Catalytic activity | [218] |
| Erwinia carotovora and E. chrysanthemi | E. coli BL21(DE3)pLysS | StEP, site-saturation mutagenesis | Increased thermostability | [219] |
| E. chrysanthemi and Erwinia carotovora | E. coli BL21(DE3)pLysS | StEP | Decreased glutaminase activity | [220] |
| Human | E. coli strain JC1(DE3), C41(DE3) | Site-saturation mutagenesis, epPCR, high-throughput screening | Increased catalytic activity | [221] |
| Rational design | ||||
| Halophilic bacteria | in silico design | Detection of rare codon cluster, molecular modeling and model evaluation, molecular docking, structural analysis of rare codon. | Increased expression | [269] |
| E. coli | in silico design | Genetic algorithm, molecular dynamics | Enhanced anticancer activity, decreased glutaminase activity | [66] |
| E. chrysantemi | E. coli BL21(DE3) C41 | Codon optimization, MSA, molecular dynamics | Decreased glutaminase activity | [109,117] |
| E. coli L-ASNase II | - | Saturation mutagenesis, molecular dynamics, enzymological screening | Decreased glutaminase activity | [103] |
| E. coli L-ASNase II | in silico redesign | Molecular dynamics, quantum mechanics molecular dynamics, molecular docking | Decreased glutaminase activity | [273] |
| E. coli L-ASNase II | E. coli CU1783 | Site-directed mutagenesis, MSA, molecular dynamics | Decreased glutaminase activity | [274] |
| Pectobacterium carotovorum | in silico | Molecular dynamics, molecular docking | Decreased glutaminase activity | [275] |
| Bacillus licheniformis | E. coli BL21 (DE3) | MSA, site-directed mutagenesis | Increased catalytic activity and half-life | [279] |
| E. coli and Erwinia species | in silico design | Phylogenetic analysis, homology modeling, active site prediction, molecular docking | Increased catalytic activity | [277] |
| E. coli L-ASNase II | in silico design | Phylogenetic analysis, homology modeling, active site prediction, molecular docking | Increased catalytic activity and low Km | [277] |
| Acinetobacter soli Y-3 | E. coli BL21 (DE3) | Consensus design, homology modeling, molecular dynamics | Improved thermostability | [281] |
| Bacillus licheniformis | E. coli BL21 (DE3) | Prediction important possible mutation hotspots, MSA, site-directed saturation mutagenesis, molecular dynamics | Improved thermostability | [261] |
| Rhizomucor miehei | B. subtilis 168 | Homology modeling, molecular dynamics, site-directed mutagenesis | Improved thermostability | [16] |
| Homo sapiens | in silico design chimeric humanized L-ASNase | T-cell epitope prediction and epitope density determination, prediction of allergenic epitopes, homology modeling, MSA, machine learning, molecular docking | Decreased immunogenicity | [286] |
| Streptomyces species | in silico search | MSA, phylogenetic analyses, homology modeling, molecular docking, antigenicity prediction, HLA class II binding prediction | Decreased immunogenicity | [272] |
| E. coli | in silico redesign | MSA, predicting protein stability, homology modeling, protein toxicity analysis, predicting antigens and allergens, prediction of B-cell epitopes | Reduced toxicity, increasing stability and half-life | [260] |
| E. coli WT | in silico redesign | MSA, homology modeling, anti-His-tag ELISA, de novo design, redesign of mCherry Surface | Decreased immunogenicity | [288,289] |
| In silico design | E. coli Rosetta (DE3) | Machine learning | Decreased immunogenicity, low toxicity | [290] |
| Semi-rational design | ||||
| Bacillus licheniformis | E. coli BL21 (DE3) | Coevolution analysis, screening of mutant libraries, saturation mutagenesis, homology modeling, molecular dynamics, molecular docking | Increased catalytic activity | [278] |
| Mycobacterium gordonae | E. coli BL21 (DE3) | Site-directed saturation and combinatorial mutagenesis, high-throughput screening, consensus design, homology modeling, molecular dynamics | Improved thermostability | [291] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shishparenok, A.N.; Gladilina, Y.A.; Zhdanov, D.D. Engineering and Expression Strategies for Optimization of L-Asparaginase Development and Production. Int. J. Mol. Sci. 2023, 24, 15220. https://doi.org/10.3390/ijms242015220
Shishparenok AN, Gladilina YA, Zhdanov DD. Engineering and Expression Strategies for Optimization of L-Asparaginase Development and Production. International Journal of Molecular Sciences. 2023; 24(20):15220. https://doi.org/10.3390/ijms242015220
Chicago/Turabian StyleShishparenok, Anastasiya N., Yulia A. Gladilina, and Dmitry D. Zhdanov. 2023. "Engineering and Expression Strategies for Optimization of L-Asparaginase Development and Production" International Journal of Molecular Sciences 24, no. 20: 15220. https://doi.org/10.3390/ijms242015220