Hypoplastic Left Heart Syndrome: Signaling & Molecular Perspectives, and the Road Ahead
Abstract
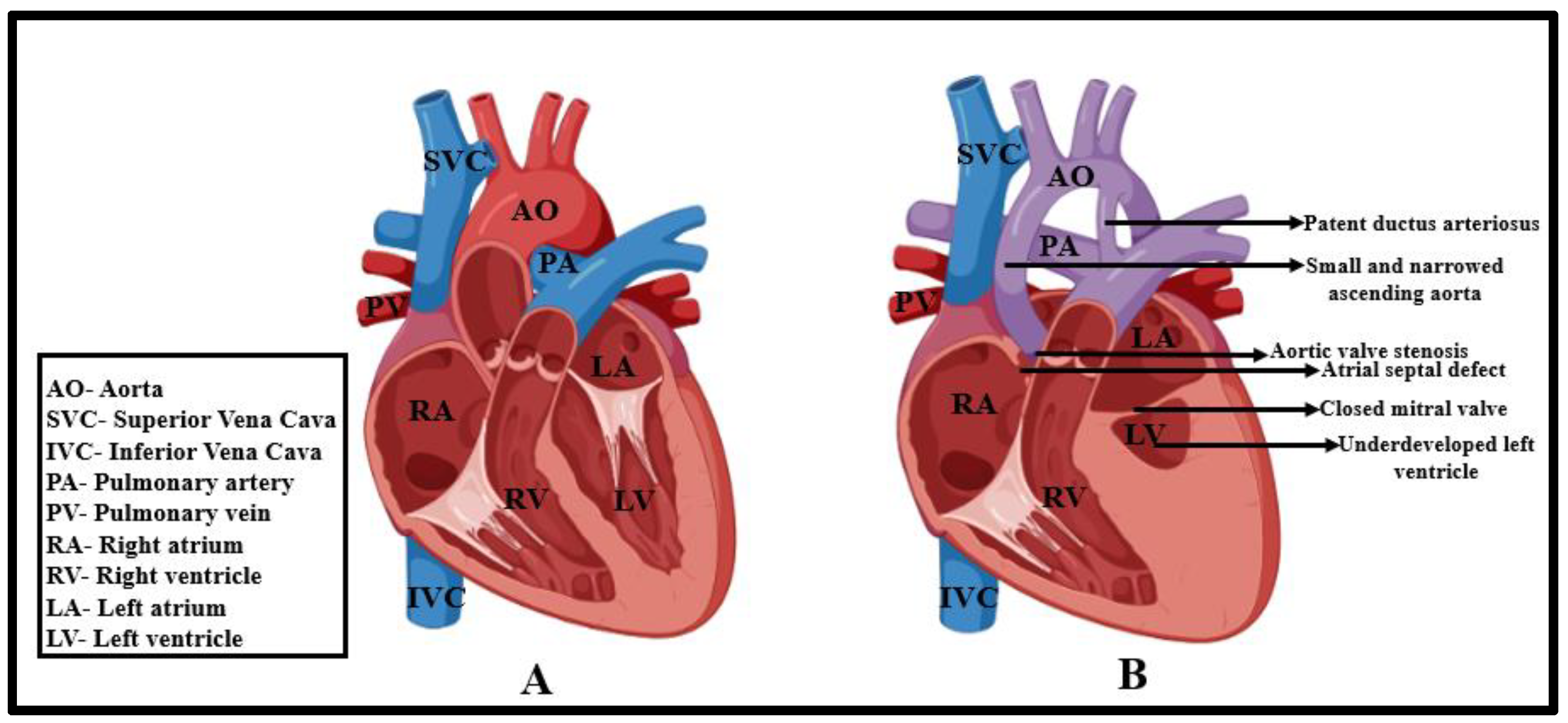
1. Introduction
2. Prenatal and Postnatal Risks
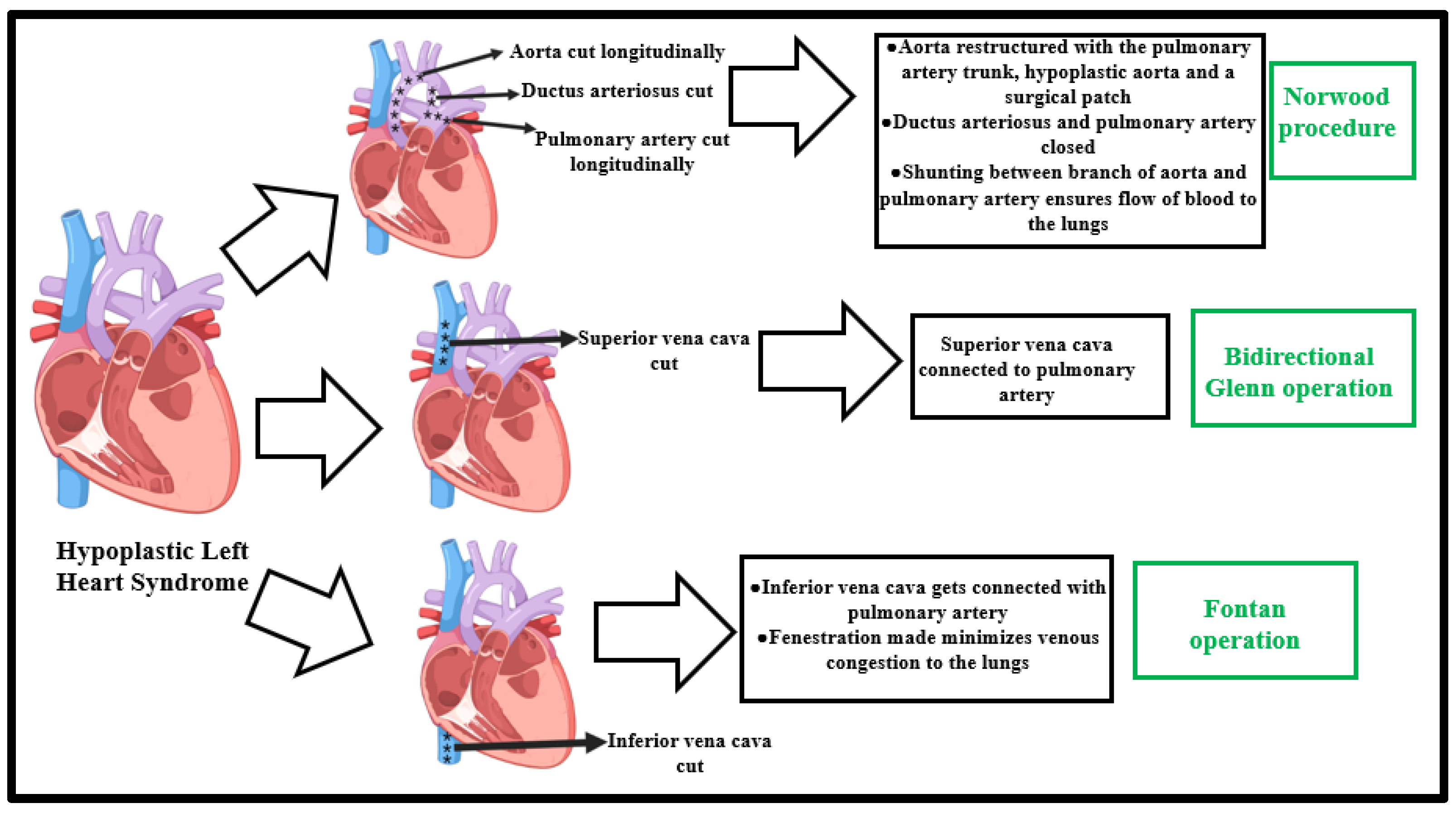
3. Existing Surgical Perspectives
3.1. Stage I—Norwood Procedure
3.2. Interstage Period
3.3. Stage II—Superior Cavopulmonary Connection Establishment
3.4. Stage III—Fontan Operation
4. Signaling and Molecular Mechanisms Outlining HLHS Incidence
4.1. Endocardial-Related Signaling Pathways
4.2. Notch Signaling
4.3. TGF-β/BMP Signaling
4.4. Wnt/SHH/p53 Signaling
4.5. Can Single Gene Mutation Attribute to HLHS?
4.5.1. RBFOX2
4.5.2. SAP130
4.5.3. PCDHA9
4.5.4. CONNEXIN43
4.5.5. HAND1
4.5.6. Myrf
5. Limitations in Current Understanding of HLHS Etiology
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Öhman, A.; El-Segaier, M.; Bergman, G.; Hanséus, K.; Malm, T.; Nilsson, B.; Pivodic, A.; Rydberg, A.; Sonesson, S.E.; Mellander, M. Changing epidemiology of hypoplastic left heart syndrome: Results of a national Swedish cohort study. J. Am. Heart Assoc. 2019, 8, e010893. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Congenital Heart Defects. 2021. Available online: https://www.cdc.gov/ncbddd/heartdefects/hlhs.html (accessed on 15 August 2023).
- Lev, M. Pathologic anatomy and interrelationship of the hypoplasia of the aortic tract complex. Lab Investig. 1952, 1, 61–70. [Google Scholar]
- Noonan, J.A.; Nadas, A.S. The hypoplastic left heart syndrome: An analysis of 101 cases. Pediatr. Clin. N. Am. 1958, 5, 1029–1056. [Google Scholar] [CrossRef]
- Aiello, V.D.; Ho, S.Y.; Anderson, R.H.; Thiene, G. Morphologic features of the hypoplastic left heart syndrome—A reappraisal. Pediatr. Pathol. 1990, 10, 931–943. [Google Scholar] [CrossRef]
- Bharati, S.; Lev, M.; Mills, B. The surgical anatomy of hypoplasia of aortic tract complex. J. Thorac. Cardiovasc. Surg. 1984, 88, 97–101. [Google Scholar] [CrossRef]
- Biorender. Create Professional Science Figures in Minutes. 2023. Available online: https://www.biorender.com/ (accessed on 10 September 2023).
- Miao, Y.; Tian, L.; Martin, M.; Paige, S.L.; Galdos, F.X.; Li, J.; Klein, A.; Zhang, H.; Ma, N.; Wei, Y. Intrinsic endocardial defects contribute to hypoplastic left heart syndrome. Cell Stem Cell 2020, 27, 574–589.e8. [Google Scholar] [CrossRef]
- Prendergast, C.; Nicholson, G. Left-Sided Obstructive Congenital Heart Lesions: Including Hypoplastic Left Heart; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Davis, C.K.; Pastuszko, P.; Lamberti, J.; Moore, J.; Hanley, F.; El Said, H. The hybrid procedure for the borderline left ventricle. Cardiol. Young 2011, 21, 26–30. [Google Scholar] [CrossRef]
- Hrstka, S.C.; Li, X.; Nelson, T.J.; Wanek Program Genetics Pipeline Group. NOTCH1-dependent nitric oxide signaling deficiency in hypoplastic left heart syndrome revealed through patient-specific phenotypes detected in bioengineered Cardiogenesis. Stem Cells 2017, 35, 1106–1119. [Google Scholar] [CrossRef]
- Liu, X.; Yagi, H.; Saeed, S.; Bais, A.S.; Gabriel, G.C.; Chen, Z.; Peterson, K.A.; Li, Y.; Schwartz, M.C.; Reynolds, W.T. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 2017, 49, 1152. [Google Scholar] [CrossRef]
- Yang, C.; Xu, Y.; Yu, M.; Lee, D.; Alharti, S.; Hellen, N.; Ahmad Shaik, N.; Banaganapalli, B.; Sheikh Ali Mohamoud, H.; Elango, R. Induced pluripotent stem cell modelling of HLHS underlines the contribution of dysfunctional NOTCH signalling to impaired cardiogenesis. Hum. Mol. Genet. 2017, 26, 3031–3045. [Google Scholar] [CrossRef]
- Hinton, R.B.; Martin, L.J.; Tabangin, M.E.; Mazwi, M.L.; Cripe, L.H.; Benson, D.W. Hypoplastic left heart syndrome is heritable. J. Am. Coll. Cardiol. 2007, 50, 1590–1595. [Google Scholar] [CrossRef]
- Shokeir, M. Hypoplastic left heart. Evidence for possible autosomal recessive inheritance. Birth Defects Orig. Artic. Ser. 1974, 10, 223–227. [Google Scholar]
- Ferencz, C.; Rubin, J.D.; Mccarter, R.J.; Brenner, J.I.; Neill, C.A.; Perry, L.W.; Hepner, S.I.; Downing, J.W. Congenital heart disease: Prevalence at livebirth: The Baltimore-Washington Infant Study. Am. J. Epidemiol. 1985, 121, 31–36. [Google Scholar] [CrossRef]
- Norwood, W.I.; Lang, P.; Castaneda, A.R.; Campbell, D.N. Experience with operations for hypoplastic left heart syndrome. J. Thorac. Cardiovasc. Surg. 1981, 82, 511–519. [Google Scholar] [CrossRef]
- Bharati, S.; Lev, M. The conduction system in hypoplasia of the aortic tract complex. Circulation 1979, 59, 1324–1332. [Google Scholar] [CrossRef]
- Lev, M. Some newer concepts of the pathology of congenital heart disease. Med. Clin. N. Am. 1966, 50, 3–14. [Google Scholar] [CrossRef]
- Lev, M.; Rimoldi, H.J.; Licata, R.H.; Gasul, B.M. Premature narrowing or closure of the foramen ovale. Am. Heart J. 1963, 65, 638–647. [Google Scholar] [CrossRef]
- Feinstein, J.A.; Benson, D.W.; Dubin, A.M.; Cohen, M.S.; Maxey, D.M.; Mahle, W.T.; Pahl, E.; Villafañe, J.; Bhatt, A.B.; Peng, L.F. Hypoplastic left heart syndrome: Current considerations and expectations. J. Am. Coll. Cardiol. 2012, 59, S1–S42. [Google Scholar] [CrossRef]
- Glatz, J.A.; Tabbutt, S.; Gaynor, J.W.; Rome, J.J.; Montenegro, L.; Spray, T.L.; Rychik, J. Hypoplastic left heart syndrome with atrial level restriction in the era of prenatal diagnosis. Ann. Thorac. Surg. 2007, 84, 1633–1638. [Google Scholar] [CrossRef]
- Rychik, J.; Rome, J.J.; Collins, M.H.; DeCampli, W.M.; Spray, T.L. The hypoplastic left heart syndrome with intact atrial septum: Atrial morphology, pulmonary vascular histopathology and outcome. J. Am. Coll. Cardiol. 1999, 34, 554–560. [Google Scholar] [CrossRef]
- Liu, M.Y.; Zielonka, B.; Snarr, B.S.; Zhang, X.; Gaynor, J.W.; Rychik, J. Longitudinal assessment of outcome from prenatal diagnosis through Fontan operation for over 500 fetuses with single ventricle-type congenital heart disease: The Philadelphia Fetus-to-Fontan Cohort Study. J. Am. Heart Assoc. 2018, 7, e009145. [Google Scholar] [CrossRef] [PubMed]
- Rychik, J.; Szwast, A.; Natarajan, S.; Quartermain, M.; Donaghue, D.; Combs, J.; Gaynor, J.; Gruber, P.; Spray, T.; Bebbington, M. Perinatal and early surgical outcome for the fetus with hypoplastic left heart syndrome: A 5-year single institutional experience. Ultrasound Obstet. Gynecol. 2010, 36, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Jantzen, D.W.; Moon-Grady, A.J.; Morris, S.A.; Armstrong, A.K.; Berg, C.; Dangel, J.; Fifer, C.G.; Frommelt, M.; Gembruch, U.; Herberg, U. Hypoplastic left heart syndrome with intact or restrictive atrial septum: A report from the International Fetal Cardiac Intervention Registry. Circulation 2017, 136, 1346–1349. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, M.K.; Rychik, J. Outcomes in Hypoplastic Left Heart Syndrome. Pediatr. Clin. 2020, 67, 945–962. [Google Scholar] [CrossRef]
- Ohye, R.G.; Sleeper, L.A.; Mahony, L.; Newburger, J.W.; Pearson, G.D.; Lu, M.; Goldberg, C.S.; Tabbutt, S.; Frommelt, P.C.; Ghanayem, N.S. Comparison of shunt types in the Norwood procedure for single-ventricle lesions. N. Engl. J. Med. 2010, 362, 1980–1992. [Google Scholar] [CrossRef]
- Norwood, W.I.; Kirklin, J.K.; Sanders, S.P. Hypoplastic left heart syndrome: Experience with palliative surgery. Am. J. Cardiol. 1980, 45, 87–91. [Google Scholar] [CrossRef]
- Hehir, D.A.; Dominguez, T.E.; Ballweg, J.A.; Ravishankar, C.; Marino, B.S.; Bird, G.L.; Nicolson, S.C.; Spray, T.L.; Gaynor, J.W.; Tabbutt, S. Risk factors for interstage death after stage 1 reconstruction of hypoplastic left heart syndrome and variants. J. Thorac. Cardiovasc. Surg. 2008, 136, 94–99.e93. [Google Scholar] [CrossRef]
- Anderson, J.B.; Beekman, R.H., III; Kugler, J.D.; Rosenthal, G.L.; Jenkins, K.J.; Klitzner, T.S.; Martin, G.R.; Neish, S.R.; Brown, D.W.; Mangeot, C. Improvement in interstage survival in a national pediatric cardiology learning network. Circ. Cardiovasc. Qual. Outcomes 2015, 8, 428–436. [Google Scholar] [CrossRef]
- Anderson, J.B.; Brown, D.W.; Lihn, S.; Mangeot, C.; Bates, K.E.; Van Bergen, A.H.; Rudd, N.A.; Hanke, S.; Tweddell, J.; Lannon, C. Power of a learning network in congenital heart disease. World J. Pediatr. Congenit. Heart Surg. 2019, 10, 66–71. [Google Scholar] [CrossRef]
- LaPar, D.J.; Mery, C.M.; Peeler, B.B.; Kron, I.L.; Gangemi, J.J. Short and long-term outcomes for bidirectional Glenn procedure performed with and without cardiopulmonary bypass. Ann. Thorac. Surg. 2012, 94, 164–171. [Google Scholar] [CrossRef]
- Brown, D.W.; Mangeot, C.; Anderson, J.B.; Peterson, L.E.; King, E.C.; Lihn, S.L.; Neish, S.R.; Fleishman, C.; Phelps, C.; Hanke, S. Digoxin use is associated with reduced interstage mortality in patients with no history of arrhythmia after stage I palliation for single ventricle heart disease. J. Am. Heart Assoc. 2016, 5, e002376. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.L.; Rychik, J.; Rome, J.J.; Apostolopoulou, S.; Pizarro, C.; Murphy, J.D.; Norwood, W.I., Jr. Early reduction of the volume work of the single ventricle: The hemi-Fontan operation. Ann. Thorac. Surg. 1996, 62, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Mavroudis, C.; Zales, V.R.; Backer, C.L.; Muster, A.J.; Latson, L.A. Fenestrated Fontan with delayed catheter closure. Effects of volume loading and baffle fenestration on cardiac index and oxygen delivery. Circulation 1992, 86 (Suppl. 5), II85–II92. [Google Scholar] [PubMed]
- Rychik, J.; Fogel, M.A.; Donofrio, M.T.; Goldmuntz, E.; Cohen, M.S.; Spray, T.L.; Jacobs, M.L. Comparison of patterns of pulmonary venous blood flow in the functional single ventricle heart after operative aortopulmonary shunt versus superior cavopulmonary shunt. Am. J. Cardiol. 1997, 80, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Gewillig, M.; Brown, S.C.; van de Bruaene, A.; Rychik, J. Providing a framework of principles for conceptualising the Fontan circulation. Acta Paediatr. 2020, 109, 651–658. [Google Scholar] [CrossRef]
- Downing, T.E.; Allen, K.Y.; Glatz, A.C.; Rogers, L.S.; Ravishankar, C.; Rychik, J.; Faerber, J.A.; Fuller, S.; Montenegro, L.M.; Steven, J.M. Long-term survival after the Fontan operation: Twenty years of experience at a single center. J. Thorac. Cardiovasc. Surg. 2017, 154, 243–253.e2. [Google Scholar] [CrossRef]
- Sharma, A.; Zhang, Y.; Buikema, J.W.; Serpooshan, V.; Chirikian, O.; Kosaric, N.; Churko, J.M.; Dzilic, E.; Shieh, A.; Burridge, P.W. Stage-specific effects of bioactive lipids on human iPSC cardiac differentiation and cardiomyocyte proliferation. Sci. Rep. 2018, 8, 6618. [Google Scholar] [CrossRef]
- Boselli, F.; Freund, J.B.; Vermot, J. Blood flow mechanics in cardiovascular development. Cell. Mol. Life Sci. 2015, 72, 2545–2559. [Google Scholar] [CrossRef]
- Yu, Z.; Liu, Z.; Ravichandran, V.; Lami, B.; Gu, M. Endocardium in Hypoplastic Left Heart Syndrome: Implications from In Vitro Study. J. Cardiovasc. Dev. Dis. 2022, 9, 442. [Google Scholar] [CrossRef]
- Miao, L.; Castillo, M.; Lu, Y.; Xiao, Y.; Liu, Y.; Burns, A.R.; Kumar, A.; Gunaratne, P.; Michael DiPersio, C.; Wu, M. beta1 integrins regulate cellular behaviors and cardiomyocyte organization during ventricular wall formation. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wu, M. Mechanisms of Trabecular Formation and Specification During Cardiogenesis. Pediatr. Cardiol. 2018, 39, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Miao, L.; Shieh, D.; Spiotto, E.; Li, J.; Zhou, B.; Paul, A.; Schwartz, R.J.; Firulli, A.B.; Singer, H.A.; et al. Single-Cell Lineage Tracing Reveals that Oriented Cell Division Contributes to Trabecular Morphogenesis and Regional Specification. Cell Rep. 2016, 15, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Li, J.; Li, J.; Lu, Y.; Shieh, D.; Mazurkiewicz, J.E.; Barroso, M.; Schwarz, J.J.; Xin, H.B.; Singer, H.A.; et al. Cardiomyocyte orientation modulated by the Numb family proteins-N-cadherin axis is essential for ventricular wall morphogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 15560–15569. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.C.; Pereira, C.; Fonseca, A.C.R.; Pinto-do-Ó, P.; Nascimento, D.S. Bearing my heart: The role of extracellular matrix on cardiac development, homeostasis, and injury response. Front. Cell Dev. Biol. 2021, 8, 621644. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Smith, C.L.; Hall, J.A.; Lee, I.; Luby-Phelps, K.; Tallquist, M.D. Epicardial spindle orientation controls cell entry into the myocardium. Dev. Cell 2010, 19, 114–125. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, X.; Liu, K.; Tang, J.; He, L.; Pu, W.; Liu, Q.; Li, Y.; Tian, X.; Wang, Y. Fibroblasts in an endocardial fibroelastosis disease model mainly originate from mesenchymal derivatives of epicardium. Cell Res. 2017, 27, 1157–1177. [Google Scholar] [CrossRef]
- Chaudhry, B.; Alqahtani, A.; Eley, L.; Coats, L.; Moldovan, C.; Annavarapu, S.R.; Henderson, D.J. The Left Ventricular Myocardium in Hypoplastic Left Heart Syndrome. J. Cardiovasc. Dev. Dis. 2022, 9, 279. [Google Scholar] [CrossRef]
- Li, J.; Miao, L.; Zhao, C.; Shaikh Qureshi, W.M.; Shieh, D.; Guo, H.; Lu, Y.; Hu, S.; Huang, A.; Zhang, L.; et al. CDC42 is required for epicardial and pro-epicardial development by mediating FGF receptor trafficking to the plasma membrane. Development 2017, 144, 1635–1647. [Google Scholar] [CrossRef]
- Cheng, L.; Xie, M.; Qiao, W.; Song, Y.; Zhang, Y.; Geng, Y.; Xu, W.; Wang, L.; Wang, Z.; Huang, K. Generation and characterization of cardiac valve endothelial-like cells from human pluripotent stem cells. Commun. Biol. 2021, 4, 1039. [Google Scholar] [CrossRef]
- de la Pompa, J.L.; Epstein, J.A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 2012, 22, 244–254. [Google Scholar] [CrossRef]
- Koenig, S.N.; Bosse, K.; Majumdar, U.; Bonachea, E.M.; Radtke, F.; Garg, V. Endothelial Notch1 is required for proper development of the semilunar valves and cardiac outflow tract. J. Am. Heart Assoc. 2016, 5, e003075. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.C.; Fu, Y.; Garside, V.C.; Niessen, K.; Chang, L.; Fuller, M.; Setiadi, A.; Smrz, J.; Kyle, A.; Minchinton, A. Notch initiates the endothelial-to-mesenchymal transition in the atrioventricular canal through autocrine activation of soluble guanylyl cyclase. Dev. Cell 2011, 21, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X.G. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Zhao, C.; Guo, H.; Li, J.; Myint, T.; Pittman, W.; Yang, L.; Zhong, W.; Schwartz, R.J.; Schwarz, J.J.; Singer, H.A.; et al. Numb family proteins are essential for cardiac morphogenesis and progenitor differentiation. Development 2014, 141, 281–295. [Google Scholar] [CrossRef]
- Miao, L.; Lu, Y.; Nusrat, A.; Abdelnasser, H.Y.; Datta, S.; Zhou, B.; Schwartz, R.J.; Wu, M. The Spatiotemporal Expression of Notch1 and Numb and Their Functional Interaction during Cardiac Morphogenesis. Cells 2021, 10, 2192. [Google Scholar] [CrossRef]
- Miao, L.; Li, J.; Li, J.; Tian, X.; Lu, Y.; Hu, S.; Shieh, D.; Kanai, R.; Zhou, B.Y.; Zhou, B.; et al. Notch signaling regulates Hey2 expression in a spatiotemporal dependent manner during cardiac morphogenesis and trabecular specification. Sci. Rep. 2018, 8, 2678. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.; Meier-Stiegen, F.; Schwanbeck, R.; Eilken, H.; Nishikawa, S.; Häsler, R.; Schreiber, S.; Bornkamm, G.W.; Nishikawa, S.-I.; Just, U. Activated Notch1 alters differentiation of embryonic stem cells into mesodermal cell lineages at multiple stages of development. Mech. Dev. 2006, 123, 570–579. [Google Scholar] [CrossRef]
- Liu, Y.; Li, P.; Liu, K.; He, Q.; Han, S.; Sun, X.; Li, T.; Shen, L. Timely inhibition of Notch signaling by DAPT promotes cardiac differentiation of murine pluripotent stem cells. PLoS ONE 2014, 9, e109588. [Google Scholar] [CrossRef]
- Kobayashi, J.; Yoshida, M.; Tarui, S.; Hirata, M.; Nagai, Y.; Kasahara, S.; Naruse, K.; Ito, H.; Sano, S.; Oh, H. Directed differentiation of patient-specific induced pluripotent stem cells identifies the transcriptional repression and epigenetic modification of NKX2-5, HAND1, and NOTCH1 in hypoplastic left heart syndrome. PLoS ONE 2014, 9, e102796. [Google Scholar] [CrossRef]
- Acharya, A.; Hans, C.P.; Koenig, S.N.; Nichols, H.A.; Galindo, C.L.; Garner, H.R.; Merrill, W.H.; Hinton, R.B.; Garg, V. Inhibitory role of Notch1 in calcific aortic valve disease. PLoS ONE 2011, 6, e27743. [Google Scholar] [CrossRef]
- Zeng, Q.; Jin, C.; Ao, L.; Cleveland, J.C., Jr.; Song, R.; Xu, D.; Fullerton, D.A.; Meng, X. Cross-talk between the Toll-like receptor 4 and Notch1 pathways augments the inflammatory response in the interstitial cells of stenotic human aortic valves. Circulation 2012, 126 (Suppl. 1), S222–S230. [Google Scholar] [CrossRef] [PubMed]
- Bosse, K.; Hans, C.P.; Zhao, N.; Koenig, S.N.; Huang, N.; Guggilam, A.; LaHaye, S.; Tao, G.; Lucchesi, P.A.; Lincoln, J. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J. Mol. Cell. Cardiol. 2013, 60, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Krebs, L.T.; Iwai, N.; Nonaka, S.; Welsh, I.C.; Lan, Y.; Jiang, R.; Saijoh, Y.; O’Brien, T.P.; Hamada, H.; Gridley, T. Notch signaling regulates left–right asymmetry determination by inducing Nodal expression. Genes Dev. 2003, 17, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Martinez-Fernandez, A.; Hartjes, K.A.; Kocher, J.-P.A.; Olson, T.M.; Terzic, A.; Nelson, T.J. Transcriptional atlas of cardiogenesis maps congenital heart disease interactome. Physiol. Genom. 2014, 46, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Drukker, M.; Tang, C.; Ardehali, R.; Rinkevich, Y.; Seita, J.; Lee, A.S.; Mosley, A.R.; Weissman, I.L.; Soen, Y. Isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells. Nat. Biotechnol. 2012, 30, 531–542. [Google Scholar] [CrossRef]
- Zhang, Y.; Jia, L.; Lee, S.J.; Wang, M.M. Conserved signal peptide of Notch3 inhibits interaction with proteasome. Biochem. Biophys. Res. Commun. 2007, 355, 245–251. [Google Scholar] [CrossRef][Green Version]
- Ricci, M.; Mohapatra, B.; Urbiztondo, A.; Birusingh, R.J.; Morgado, M.; Rodriguez, M.M.; Lincoln, J.; Vatta, M. Differential changes in TGF-β/BMP signaling pathway in the right ventricular myocardium of newborns with hypoplastic left heart syndrome. J. Card. Fail. 2010, 16, 628–634. [Google Scholar] [CrossRef]
- Bujak, M.; Frangogiannis, N.G. The role of TGF-β signaling in myocardial infarction and cardiac remodeling. Cardiovasc. Res. 2007, 74, 184–195. [Google Scholar] [CrossRef]
- Chaudhry, S.S.; Cain, S.A.; Morgan, A.; Dallas, S.L.; Shuttleworth, C.A.; Kielty, C.M. Fibrillin-1 regulates the bioavailability of TGFβ1. J. Cell Biol. 2007, 176, 355–367. [Google Scholar] [CrossRef]
- Frantz, S.; Hu, K.; Adamek, A.; Wolf, J.; Sallam, A.; Kg Maier, S.; Lonning, S.; Ling, H.; Ertl, G.; Bauersachs, J. Transforming growth factor beta inhibition increases mortality and left ventricular dilatation after myocardial infarction. Basic Res. Cardiol. 2008, 103, 485–492. [Google Scholar] [CrossRef]
- Xiao, H.; Zhang, Y.Y. Understanding the role of transforming growth factor-β signalling in the heart: Overview of studies using genetic mouse models. Clin. Exp. Pharmacol. Physiol. 2008, 35, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Kidani, T.; Miura, H. Molecular profiling of bone remodeling occurring in musculoskeletal tumors. J. Orthop. Res. 2021, 39, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
- Bayrak-Toydemir, P.; McDonald, J.; Markewitz, B.; Lewin, S.; Miller, F.; Chou, L.S.; Gedge, F.; Tang, W.; Coon, H.; Mao, R. Genotype-phenotype correlation in hereditary hemorrhagic telangiectasia: Mutations and manifestations. Am. J. Med. Genet. Part A 2006, 140, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.-B.; Wang, W.E.; Zeng, C.-Y. Wnt signaling pathways in myocardial infarction and the therapeutic effects of Wnt pathway inhibitors. Acta Pharmacol. Sin. 2019, 40, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Gessert, S.; Kuhl, M. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res. 2010, 107, 186–199. [Google Scholar] [CrossRef]
- Theis, J.L.; Vogler, G.; Missinato, M.A.; Li, X.; Nielsen, T.; Zeng, X.-X.I.; Martinez-Fernandez, A.; Walls, S.M.; Kervadec, A.; Kezos, J.N. Patient-specific genomics and cross-species functional analysis implicate LRP2 in hypoplastic left heart syndrome. Elife 2020, 9, e59554. [Google Scholar] [CrossRef]
- Christensen, E.I.; Birn, H. Megalin and cubilin: Multifunctional endocytic receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 258–267. [Google Scholar] [CrossRef]
- Marzolo, M.-P.; Farfán, P. New insights into the roles of megalin/LRP2 and the regulation of its functional expression. Biol. Res. 2011, 44, 89–105. [Google Scholar] [CrossRef]
- Kantarci, S.; Al-Gazali, L.; Hill, R.S.; Donnai, D.; Black, G.C.; Bieth, E.; Chassaing, N.; Lacombe, D.; Devriendt, K.; Teebi, A. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat. Genet. 2007, 39, 957–959. [Google Scholar] [CrossRef]
- Nykjaer, A.; Dragun, D.; Walther, D.; Vorum, H.; Jacobsen, C.; Herz, J.; Melsen, F.; Christensen, E.I.; Willnow, T.E. An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3. Cell 1999, 96, 507–515. [Google Scholar] [CrossRef]
- Briggs, L.E.; Burns, T.A.; Lockhart, M.M.; Phelps, A.L.; Van den Hoff, M.J.; Wessels, A. Wnt/β-catenin and sonic hedgehog pathways interact in the regulation of the development of the dorsal mesenchymal protrusion. Dev. Dyn. 2016, 245, 103–113. [Google Scholar] [CrossRef]
- Datta, M.W.; Hernandez, A.M.; Schlicht, M.J.; Kahler, A.J.; DeGueme, A.M.; Dhir, R.; Shah, R.B.; Farach-Carson, C.; Barrett, A.; Datta, S. Perlecan, a candidate gene for the CAPB locus, regulates prostate cancer cell growth via the Sonic Hedgehog pathway. Mol. Cancer 2006, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Christ, A.; Christa, A.; Klippert, J.; Eule, J.C.; Bachmann, S.; Wallace, V.A.; Hammes, A.; Willnow, T.E. LRP2 acts as SHH clearance receptor to protect the retinal margin from mitogenic stimuli. Dev. Cell 2015, 35, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Yeo, G.W.; Nostrand, E.L.V.; Liang, T.Y. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007, 3, e85. [Google Scholar]
- Minovitsky, S.; Gee, S.L.; Schokrpur, S.; Dubchak, I.; Conboy, J.G. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005, 33, 714–724. [Google Scholar] [CrossRef]
- Verma, S.K.; Deshmukh, V.; Nutter, C.A.; Jaworski, E.; Jin, W.; Wadhwa, L.; Abata, J.; Ricci, M.; Lincoln, J.; Martin, J.F. Rbfox2 function in RNA metabolism is impaired in hypoplastic left heart syndrome patient hearts. Sci. Rep. 2016, 6, 30896. [Google Scholar] [CrossRef]
- Venables, J.P.; Brosseau, J.-P.; Gadea, G.; Klinck, R.; Prinos, P.; Beaulieu, J.-F.; Lapointe, E.; Durand, M.; Thibault, P.; Tremblay, K. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol. Cell. Biol. 2013, 33, 396–405. [Google Scholar] [CrossRef]
- Braeutigam, C.; Rago, L.; Rolke, A.; Waldmeier, L.; Christofori, G.; Winter, J. The RNA-binding protein Rbfox2: An essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene 2014, 33, 1082–1092. [Google Scholar] [CrossRef]
- Gehman, L.T.; Meera, P.; Stoilov, P.; Shiue, L.; O’Brien, J.E.; Meisler, M.H.; Ares, M.; Otis, T.S.; Black, D.L. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012, 26, 445–460. [Google Scholar] [CrossRef]
- Gallagher, T.L.; Arribere, J.A.; Geurts, P.A.; Exner, C.R.; McDonald, K.L.; Dill, K.K.; Marr, H.L.; Adkar, S.S.; Garnett, A.T.; Amacher, S.L. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol. 2011, 359, 251–261. [Google Scholar] [CrossRef]
- Yeo, G.W.; Coufal, N.G.; Liang, T.Y.; Peng, G.E.; Fu, X.-D.; Gage, F.H. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009, 16, 130–137. [Google Scholar] [CrossRef]
- Jangi, M.; Boutz, P.L.; Paul, P.; Sharp, P.A. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014, 28, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Lovci, M.T.; Ghanem, D.; Marr, H.; Arnold, J.; Gee, S.; Parra, M.; Liang, T.Y.; Stark, T.J.; Gehman, L.T.; Hoon, S. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 2013, 20, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-A.; Tang, Z.-Z.; Black, D.L. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009, 23, 2284–2293. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, A.P.; Chen, J.R.; Garcia-Blanco, M.A. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell. Biol. 2006, 26, 1209–1222. [Google Scholar] [CrossRef]
- Mauger, D.M.; Lin, C.; Garcia-Blanco, M.A. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol. Cell. Biol. 2008, 28, 5403–5419. [Google Scholar] [CrossRef]
- Arya, A.D.; Wilson, D.I.; Baralle, D.; Raponi, M. RBFOX2 protein domains and cellular activities. Biochem. Soc. Trans. 2014, 42, 1180–1183. [Google Scholar] [CrossRef]
- Damianov, A.; Black, D.L. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA 2010, 16, 405–416. [Google Scholar] [CrossRef]
- Joo, J.-H.; Taxter, T.J.; Munguba, G.C.; Kim, Y.H.; Dhaduvai, K.; Dunn, N.W.; Degan, W.J.; Oh, S.P.; Sugrue, S.P. Pinin modulates expression of an intestinal homeobox gene, Cdx2, and plays an essential role for small intestinal morphogenesis. Dev. Biol. 2010, 345, 191–203. [Google Scholar] [CrossRef]
- Terashima, M.; Fujita, Y.; Togashi, Y.; Sakai, K.; De Velasco, M.A.; Tomida, S.; Nishio, K. KIAA1199 interacts with glycogen phosphorylase kinase β-subunit (PHKB) to promote glycogen breakdown and cancer cell survival. Oncotarget 2014, 5, 7040. [Google Scholar] [CrossRef]
- Wilson, J.M.; Martinez-De Luna, R.I.; El Hodiri, H.M.; Smith, R.; King, M.W.; Mescher, A.L.; Neff, A.W.; Belecky-Adams, T.L. RNA helicase Ddx39 is expressed in the developing central nervous system, limb, otic vesicle, branchial arches and facial mesenchyme of Xenopus laevis. Gene Expr. Patterns 2010, 10, 44–52. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information SAP130 Sin3A Associated Protein 130 [Homo Sapiens (Human)] 2023. Available online: https://www.ncbi.nlm.nih.gov/gene/79595 (accessed on 13 August 2023).
- Gaber, N.; Gagliardi, M.; Patel, P.; Kinnear, C.; Zhang, C.; Chitayat, D.; Shannon, P.; Jaeggi, E.; Tabori, U.; Keller, G. Fetal reprogramming and senescence in hypoplastic left heart syndrome and in human pluripotent stem cells during cardiac differentiation. Am. J. Pathol. 2013, 183, 720–734. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Yelon, D. Utilizing zebrafish to understand second heart field development. In Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology; Springer: Tokyo, Japan, 2016; pp. 193–199. [Google Scholar]
- Teekakirikul, P.; Zhu, W.; Gabriel, G.C.; Young, C.B.; Williams, K.; Martin, L.J.; Hill, J.C.; Richards, T.; Billaud, M.; Phillippi, J.A. Common deletion variants causing protocadherin-α deficiency contribute to the complex genetics of BAV and left-sided congenital heart disease. Hum. Genet. Genomics Adv. 2021, 2, 100037. [Google Scholar] [CrossRef]
- Etlioglu, H.E.; Sun, W.; Huang, Z.; Chen, W.; Schmucker, D. Characterization of a single genomic locus encoding the clustered protocadherin receptor diversity in Xenopus tropicalis. G3 Genes Genomes Genet. 2016, 6, 2309–2318. [Google Scholar] [CrossRef] [PubMed][Green Version]
- National Center for Biotechnology Information PCDHA9 Protocadherin Alpha 9 [Homo Sapiens (Human)] 2023. Available online: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=9752 (accessed on 13 August 2023).
- Strehl, S.; Glatt, K.; Liu, Q.M.; Glatt, H.; Lalande, M. Characterization of Two Novel Protocadherins (PCDH8andPCDH9) Localized on Human Chromosome 13 and Mouse Chromosome 14. Genomics 1998, 53, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhang, H.; Su, Y.; Wen, Z.; Zhu, Z.; Chen, G.; Peng, L.; Du, C.; Xie, H.; Li, H. Identification of two novel PCDHA9 mutations associated with Hirschsprung’s disease. Gene 2018, 658, 96–104. [Google Scholar] [CrossRef]
- Dasgupta, C.; Martinez, A.-M.; Zuppan, C.W.; Shah, M.M.; Bailey, L.L.; Fletcher, W.H. Identification of connexin43 (α1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE). Mutat. Res. Fundam. Mol. Mech. Mutagen. 2001, 479, 173–186. [Google Scholar] [CrossRef]
- Britz-Cunningham, S.H.; Shah, M.M.; Zuppan, C.W.; Fletcher, W.H. Mutations of the Connexin43 gap-junction gene in patients with heart malformations and defects of laterality. N. Engl. J. Med. 1995, 332, 1323–1330. [Google Scholar] [CrossRef]
- Dasgupta, C.; Escobar-Poni, B.; Shah, M.; Duncan, J.; Fletcher, W.H. Misregulation of connexin43 gap junction channels and congenital heart defects. In Novartis Foundation Symposium 219-Gap Junction-Mediated Intercellular Signalling in Health and Disease: Gap Junction-Mediated Intercellular Signalling in Health and Disease: Novartis Foundation Symposium 219; John Wiley & Sons, Ltd.: Chichester, UK, 2007; pp. 212–225. [Google Scholar]
- Stagg, R.B.; Fletcher, W.H. The hormone-induced regulation of contact-dependent cell-cell communication by phosphorylation. Endocr. Rev. 1990, 11, 302–325. [Google Scholar] [CrossRef]
- Godwin, A.J.; Green, L.M.; Walsh, M.P.; McDonald, J.R.; Walsh, D.A.; Fletcher, W.H. In situ regulation of cell-cell communication by the cAMP-dependent protein kinase and protein kinase C. In Reversible Protein Phosphorylation in Cell Regulation; Springer: Dordrecht, The Netherlands, 1993; pp. 293–307. [Google Scholar]
- Levin, M.; Mercola, M. Gap junctions are involved in the early generation of left–right asymmetry. Dev. Biol. 1998, 203, 90–105. [Google Scholar] [CrossRef]
- Levin, M.; Mercola, M. Gap junction-mediated transfer of left-right patterning signals in the early chick blastoderm is upstream of Shh asymmetry in the node. Development 1999, 126, 4703–4714. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Johnson, R.L.; Sterna, C.D.; Kuehn, M.; Tabin, C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell 1995, 82, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Reamon-Buettner, S.M.; Ciribilli, Y.; Inga, A.; Borlak, J. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum. Mol. Genet. 2008, 17, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Firulli, A.B. A HANDful of questions: The molecular biology of the heart and neural crest derivatives (HAND)-subclass of basic helix–loop–helix transcription factors. Gene 2003, 312, 27–40. [Google Scholar] [CrossRef] [PubMed]
- McFadden, D.G.; McAnally, J.; Richardson, J.A.; Charité, J.; Olson, E.N. Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development 2002, 129, 3077–3088. [Google Scholar] [CrossRef]
- Scott, I.C.; Anson-Cartwright, L.; Riley, P.; Reda, D.; Cross, J.C. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol. Cell. Biol. 2000, 20, 530–541. [Google Scholar] [CrossRef]
- Knöfler, M.; Meinhardt, G.; Bauer, S.; Loregger, T.; Vasicek, R.; Bloor, D.J.; Kimber, S.J.; Husslein, P. Human Hand1 basic helix-loop-helix (bHLH) protein: Extra-embryonic expression pattern, interaction partners and identification of its transcriptional repressor domains. Biochem. J. 2002, 361, 641–651. [Google Scholar] [CrossRef]
- Riley, P.; Anaon-Cartwight, L.; Cross, J.C. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 1998, 18, 271–275. [Google Scholar] [CrossRef]
- Firulli, A.B.; McFadden, D.G.; Lin, Q.; Srivastava, D.; Olson, E.N. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 1998, 18, 266–270. [Google Scholar] [CrossRef]
- Togi, K.; Kawamoto, T.; Yamauchi, R.; Yoshida, Y.; Kita, T.; Tanaka, M. Role of Hand1/eHAND in the dorso-ventral patterning and interventricular septum formation in the embryonic heart. Mol. Cell. Biol. 2004, 24, 4627–4635. [Google Scholar] [CrossRef]
- Doering, L.; Cornean, A.; Thumberger, T.; Benjaminsen, J.; Wittbrodt, B.; Kellner, T.; Hammouda, O.T.; Gorenflo, M.; Wittbrodt, J.; Gierten, J. CRISPR-based knockout and base editing confirm the role of MYRF in heart development and congenital heart disease. Dis. Models Mech. 2023, 16, dmm049811. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Sun, W.; Ouyang, J.; Li, S.; Jia, X.; Tan, Z.; Hejtmancik, J.F.; Zhang, Q. Novel truncation mutations in MYRF cause autosomal dominant high hyperopia mapped to 11p12-q13.3. Hum. Genet. 2019, 138, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Yu, L.; Zhou, X.; Wynn, J.; Zhao, H.; Guo, Y.; Zhu, N.; Kitaygorodsky, A.; Hernan, R.; Aspelund, G.; et al. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018, 14, e1007822. [Google Scholar] [CrossRef] [PubMed]
- Bujalka, H.; Koenning, M.; Jackson, S.; Perreau, V.M.; Pope, B.; Hay, C.M.; Mitew, S.; Hill, A.F.; Lu, Q.R.; Wegner, M.; et al. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol. 2013, 11, e1001625. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.D.; Stewart, B.; Prasov, L.; Pyle, L.C. MYRF-Related Cardiac Urogenital Syndrome. In GeneReviews((R)); University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Cross, S.H.; McKie, L.; Hurd, T.W.; Riley, S.; Wills, J.; Barnard, A.R.; Young, F.; MacLaren, R.E.; Jackson, I.J. The nanophthalmos protein TMEM98 inhibits MYRF self-cleavage and is required for eye size specification. PLoS Genet. 2020, 16, e1008583. [Google Scholar] [CrossRef]
- Kim, D.; Choi, J.O.; Fan, C.; Shearer, R.S.; Sharif, M.; Busch, P.; Park, Y. Homo-trimerization is essential for the transcription factor function of Myrf for oligodendrocyte differentiation. Nucleic Acids Res. 2017, 45, 5112–5125. [Google Scholar] [CrossRef]
- An, H.; Fan, C.; Sharif, M.; Kim, D.; Poitelon, Y.; Park, Y. Functional mechanism and pathogenic potential of MYRF ICA domain mutations implicated in birth defects. Sci. Rep. 2020, 10, 814. [Google Scholar] [CrossRef]
- Fan, C.; An, H.; Sharif, M.; Kim, D.; Park, Y. Functional mechanisms of MYRF DNA-binding domain mutations implicated in birth defects. J. Biol. Chem. 2021, 296, 100612. [Google Scholar] [CrossRef]
- Wu, P.; Zhen, X.; Li, B.; Yu, Q.; Huang, X.; Shi, N. Crystal structure of the MyRF ICA domain with its upstream beta-helical stalk reveals the molecular mechanisms underlying its trimerization and self-cleavage. Int. J. Biol. Sci. 2021, 17, 2931–2943. [Google Scholar] [CrossRef]
- Bove, E.L.; Lloyd, T.R. Staged reconstruction for hypoplastic left heart syndrome. Contemporary results. Ann. Surg. 1996, 224, 387. [Google Scholar] [CrossRef]
- Rossetti, L.Z.; Glinton, K.; Yuan, B.; Liu, P.; Pillai, N.; Mizerik, E.; Magoulas, P.; Rosenfeld, J.A.; Karaviti, L.; Sutton, V.R. Review of the phenotypic spectrum associated with haploinsufficiency of MYRF. Am. J. Med. Genet. Part A 2019, 179, 1376–1382. [Google Scholar] [CrossRef]
- Razzouk, A.J.; Chinnock, R.E.; Gundry, S.R.; Johnston, J.K.; Larsen, R.L.; Baum, M.F.; Mulla, N.F.; Bailey, L.L. Transplantation as a primary treatment for hypoplastic left heart syndrome: Intermediate-term results. Ann. Thorac. Surg. 1996, 62, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, X.-F. Signaling cross-talk between TGF-β/BMP and other pathways. Cell Res. 2009, 19, 71–88. [Google Scholar] [CrossRef]
- Bittle, G.J.; Morales, D.; Deatrick, K.B.; Parchment, N.; Saha, P.; Mishra, R.; Sharma, S.; Pietris, N.; Vasilenko, A.; Bor, C. Stem cell therapy for hypoplastic left heart syndrome: Mechanism, clinical application, and future directions. Circ. Res. 2018, 123, 288–300. [Google Scholar] [CrossRef]
- Pierpont, M.E.; Brueckner, M.; Chung, W.K.; Garg, V.; Lacro, R.V.; McGuire, A.L.; Mital, S.; Priest, J.R.; Pu, W.T.; Roberts, A. Genetic basis for congenital heart disease: Revisited: A scientific statement from the American Heart Association. Circulation 2018, 138, e653–e711. [Google Scholar] [CrossRef] [PubMed]
- Tinker, S.C.; Gilboa, S.M.; Moore, C.A.; Waller, D.K.; Simeone, R.M.; Kim, S.Y.; Jamieson, D.J.; Botto, L.D.; Fisher, S.C.; Reefhuis, J. Modification of the association between diabetes and birth defects by obesity, National Birth Defects Prevention Study, 1997–2011. Birth Defects Res. 2021, 113, 1084–1097. [Google Scholar] [CrossRef] [PubMed]
- Belmont, J.W. Considering the Genetic Architecture of Hypoplastic Left Heart Syndrome. J. Cardiovasc. Dev. Dis. 2022, 9, 315. [Google Scholar] [CrossRef]
- Mikryukov, A.A.; Mazine, A.; Wei, B.; Yang, D.; Miao, Y.; Gu, M.; Keller, G.M. BMP10 signaling promotes the development of endocardial cells from human pluripotent stem cell-derived cardiovascular progenitors. Cell Stem Cell 2021, 28, 96–111.e7. [Google Scholar] [CrossRef]
- Bao, X.; Bhute, V.J.; Han, T.; Qian, T.; Lian, X.; Palecek, S.P. Human pluripotent stem cell-derived epicardial progenitors can differentiate to endocardial-like endothelial cells. Bioeng. Transl. Med. 2017, 2, 191–201. [Google Scholar] [CrossRef]
- Neri, T.; Hiriart, E.; Van Vliet, P.P.; Faure, E.; Norris, R.A.; Farhat, B.; Jagla, B.; Lefrancois, J.; Sugi, Y.; Moore-Morris, T. Human pre-valvular endocardial cells derived from pluripotent stem cells recapitulate cardiac pathophysiological valvulogenesis. Nat. Commun. 2019, 10, 1929. [Google Scholar] [CrossRef]
- Zhu, L.; Liu, K.; Feng, Q.; Liao, Y. Cardiac Organoids: A 3D Technology for Modeling Heart Development and Disease. Stem Cell Rev. Rep. 2022, 18, 2593–2605. [Google Scholar] [CrossRef]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef] [PubMed]
- Musunuru, K. Genome editing: The recent history and perspective in cardiovascular diseases. J. Am. Coll. Cardiol. 2017, 70, 2808–2821. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Marti-Gutierrez, N.; Park, S.-W.; Wu, J.; Lee, Y.; Suzuki, K.; Koski, A.; Ji, D.; Hayama, T.; Ahmed, R. Correction of a pathogenic gene mutation in human embryos. Nature 2017, 548, 413–419. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Datta, S.; Cao, W.; Skillman, M.; Wu, M. Hypoplastic Left Heart Syndrome: Signaling & Molecular Perspectives, and the Road Ahead. Int. J. Mol. Sci. 2023, 24, 15249. https://doi.org/10.3390/ijms242015249
Datta S, Cao W, Skillman M, Wu M. Hypoplastic Left Heart Syndrome: Signaling & Molecular Perspectives, and the Road Ahead. International Journal of Molecular Sciences. 2023; 24(20):15249. https://doi.org/10.3390/ijms242015249
Chicago/Turabian StyleDatta, Sayantap, Wangjia Cao, Mikayla Skillman, and Mingfu Wu. 2023. "Hypoplastic Left Heart Syndrome: Signaling & Molecular Perspectives, and the Road Ahead" International Journal of Molecular Sciences 24, no. 20: 15249. https://doi.org/10.3390/ijms242015249
APA StyleDatta, S., Cao, W., Skillman, M., & Wu, M. (2023). Hypoplastic Left Heart Syndrome: Signaling & Molecular Perspectives, and the Road Ahead. International Journal of Molecular Sciences, 24(20), 15249. https://doi.org/10.3390/ijms242015249





