Differential Expression of Proteins Associated with Bipolar Disorder as Identified Using the PeptideShaker Software
Abstract
:1. Introduction
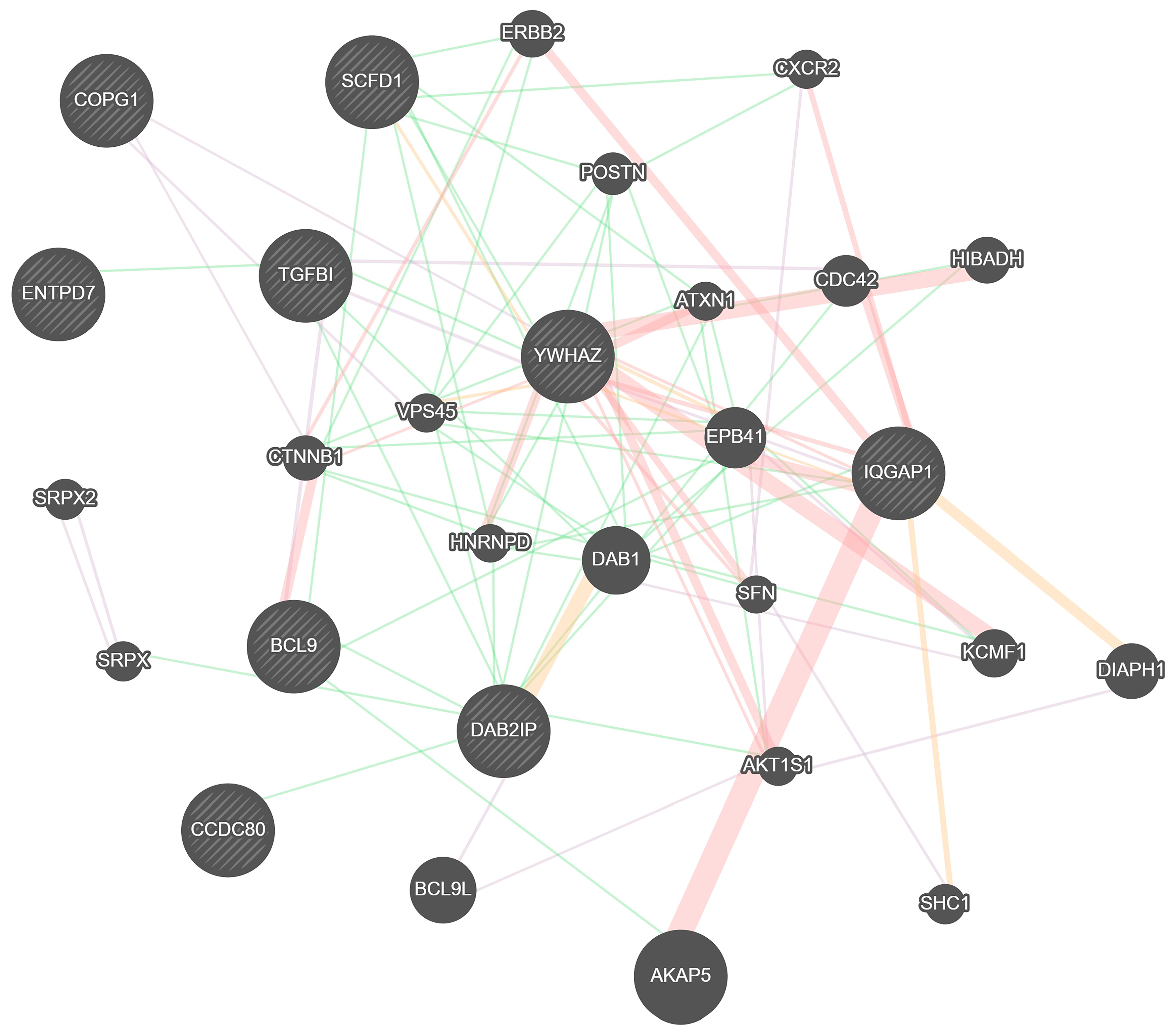
2. Results
3. Discussion
4. Materials and Methods
4.1. Patient Characteristics
4.2. Sample Preparation
4.3. Affinity Chromatography
4.4. One-Dimensional Laemmli PAG Electrophoresis
4.5. Trypsinolysis
4.6. Mass Spectrometry Analysis
4.7. Protein Identification and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferrari, A.J.; Stockings, E.; Khoo, J.P.; Erskine, H.E.; Degenhardt, L.; Vos, T.; Whiteford, H.A. The prevalence and burden of bipolar disorder: Findings from the Global Burden of Disease Study 2013. Bipolar Disord. 2016, 18, 440–450. [Google Scholar] [CrossRef]
- Carvalho, A.F.; Firth, J.; Vieta, E. Bipolar Disorder. N. Engl. J. Med. 2020, 383, 58–66. [Google Scholar] [CrossRef]
- Hirschfeld, R.M.; Calabrese, J.R.; Weissman, M.M.; Reed, M.; Davies, M.A.; Frye, M.A.; Keck, P.E., Jr.; Lewis, L.; McElroy, S.L.; McNulty, J.P.; et al. Screening for bipolar disorder in the community. J. Clin. Psychiatry 2003, 64, 53–59. [Google Scholar] [CrossRef]
- Cavazzoni, P.; Grof, P.; Duffy, A.; Grof, E.; Müller-Oerlinghausen, B.; Berghöfer, A.; Ahrens, B.; Zvolsky, P.; Robertson, C.; Davis, A.; et al. Heterogeneity of the risk of suicidal behavior in bipolar-spectrum disorders. Bipolar Disord. 2007, 9, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Kamali, M.; Reilly-Harrington, N.A.; Chang, W.C.; McInnis, M.; McElroy, S.L.; Ketter, T.A.; Shelton, R.C.; Deckersbach, T.; Tohen, M.; Kocsis, J.H.; et al. Bipolar depression and suicidal ideation: Moderators and mediators of a complex relationship. J. Affect. Disord. 2019, 1, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Angst, J.; Gamma, A. A new bipolar spectrum concept: a brief review. Bipolar Disord. 2002, 4, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Simutkin, G.G.; Bokhan, N.A.; Ivanova, S.A. Probabilistic Diagnosis of Bipolar Adjective Disorder: Modern Approaches, Possibly and Restrictions; Ovchinnikov, A.A., Aksenov, M.M., Eds.; Printing House Integrated Casework: Tomsk, Russia, 2023; pp. 13–21. ISBN 978-5-907509-34-4. (In Russian) [Google Scholar]
- Vieta, E.; Berk, M.; Schulze, T.G.; Carvalho, A.F.; Suppes, T.; Calabrese, J.R.; Gao, K.; Miskowiak, K.W.; Grande, I. Bipolar disorders. Nat. Rev. Dis. Primers 2018, 4, 18008. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, R.S.; Berk, M.; Brietzke, E.; Goldstein, B.I.; López-Jaramillo, C.; Kessing, L.V.; Malhi, G.S.; Nierenberg, A.A.; Rosenblat, J.D.; Majeed, A.; et al. Bipolar disorders. Lancet 2020, 396, 1841–1856. [Google Scholar] [CrossRef]
- Teixeira, A.L.; Colpo, G.D.; Fries, G.R.; Bauer, I.E.; Selvaraj, S. Biomarkers for bipolar disorder: Current status and challenges ahead. Expert. Rev. Neurother. 2019, 19, 67–81. [Google Scholar] [CrossRef]
- Weiner, M.; Warren, L.; Fiedorowicz, J.G. Cardiovascular morbidity and mortality in bipolar disorder. Ann. Clin. Psychiatry Off. J. Am. Acad. Clin. Psychiatr. 2011, 23, 40–47. [Google Scholar]
- Knežević, V.; Nedić, A. Influence of misdiagnosis on the course of bipolar disorder. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 1542–1545. [Google Scholar] [PubMed]
- Schaffer, A.; Isometsa, E.T.; Tondo, L.; Moreno, D.H.; Turecki, G.; Reis, C.; Cassidy, F.; Sinyor, M.; Azorin, J.M.; Kessing, L.V.; et al. International Society for Bipolar Disorders Task Force on Suicide: Meta-analyses and meta-regression of correlates of suicide attempts and suicide deaths in bipolar disorder. Bipolar Disord. 2015, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lish, J.D.; Dime-Meenan, S.; Whybrow, P.C.; Price, R.A.; Hirschfeld, R.M. The national Depressive and Manic-Depressive Association (DMDA) survey of bi-polai members. J. Affect. Disord. 1994, 31, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Geoffroy, P.A.; Leboyer, M.; Scott, J. Predicting bipolar disorder: what can we learn from prospective cohort studies? Encephale 2015, 41, 10–16. [Google Scholar] [CrossRef]
- Gore, F.M.; Bloem, P.J.; Patton, G.C.; Ferguson, J.; Joseph, V.; Coffey, C.; Sawyer, S.M.; Mathers, C.D. Global burden of disease in young people aged 10–24 years: A systematic analysis. Lancet 2011, 377, 2093–2102. [Google Scholar] [CrossRef]
- Bebbington, P.; Ramana, R. The epidemiology of bipolar affective disorder. Soc. Psychiatry Psychiatr. Epidemiol. 1995, 30, 279–292. [Google Scholar] [CrossRef]
- Hirschfeld, R.M.; Lewis, L.; Vornik, L.A. Perceptions and impact of bipolar disorder: How far have we really come? Results of the National Depressive and Manic-Depressive Association 2000 survey of individuals with bipolar disorder. J. Clin. Psychiatry 2003, 64, 161–174. [Google Scholar] [CrossRef]
- Geoffroy, P.A.; Scott, J. Prodrome or risk syndrome: What’s in a name? Int. J. Bipolar Disord. 2017, 5, 7. [Google Scholar] [CrossRef]
- McIntyre, R.S.; Cha, D.S.; Jerrell, J.M.; Swardfager, W.; Kim, R.D.; Costa, L.G.; Baskaran, A.; Soczynska, J.K.; Woldeyohannes, H.O.; Mansur, R.B.; et al. Advancing biomarker research: Utilizing ‘Big Data’ approaches for the characterization and prevention of bipolar disorder. Bipolar Disord. 2014, 16, 531–547. [Google Scholar] [CrossRef]
- de Jesus, J.R.; de Campos, B.K.; Galazzi, R.M.; Martinez, J.L.; Arruda, M.A. Bipolar disorder: Recent advances and future trends in bioanalytical developments for biomarker discovery. Anal. Bioanal. Chem. 2015, 407, 661–667. [Google Scholar] [CrossRef]
- Saia-Cereda, V.M.; Cassoli, J.S.; Martins-de-Souza, D.; Nascimento, J.M. Psychiatric disorders biochemical pathways unraveled by human brain proteomics. Eur. Arch. Psychiatry Clin. Neurosci. 2017, 267, 3–17. [Google Scholar] [CrossRef]
- Martins-de-Souza, D.; Maccarrone, G.; Wobrock, T.; Zerr, I.; Gormanns, P.; Reckow, S.; Falkai, P.; Schmitt, A.; Turck, C.W. Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. J. Psychiatr. Res. 2010, 44, 1176–1189. [Google Scholar] [CrossRef]
- Taurines, R.; Dudley, E.; Grassl, J.; Warnke, A.; Gerlach, M.; Coogan, A.N.; Thome, J. Proteomic research in psychiatry. J. Psychopharmacol. 2011, 25, 151–196. [Google Scholar] [CrossRef]
- Martins-de-Souza, D. Biomarkers for psychiatric disorders: where are we standing? Dis. Markers 2013, 35, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Seregin, A.; Boksha, I.; Dmitrieva, E.; Simutkin, G.; Kornetova, E.; Savushkina, O.; Letova, A.; Bokhan, N.; Ivanova, S.; et al. The difference in serum proteomes in schizophrenia and bipolar disorder. BMC Genom. 2019, 20, 535. [Google Scholar] [CrossRef]
- Dmitrieva, E.; Smirnova, L.; Seregin, A.; Zgoda, V.; Semke, A.; Ivanova, S. Proteomic profile of serum from patients with schizophrenia spectrum disorders. PeerJ 2022, 10, e13907. [Google Scholar] [CrossRef] [PubMed]
- Silva-Costa, L.C.; Carlson, P.T.C.; Guest, P.C.; de Almeida, V.; Martins-de-Souza, D. Proteomic Markers for Depression. Adv. Exp. Med. Biol. 2019, 1118, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Novikova, S.I.; He, F.; Cutrufello, N.J.; Lidow, M.S. Identification of protein biomarkers for schizophrenia and bipolar disorder in the postmortem prefrontal cortex using SELDI-TOF-MS ProteinChip profiling combined with MALDI-TOF-PSD-MS analysis. Neurobiol. Dis. 2006, 23, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Beasley, C.L.; Pennington, K.; Behan, A.; Wait, R.; Dunn, M.J.; Cotter, D. Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: Evidence for disease-associated changes. Proteomics 2006, 6, 3414–3425. [Google Scholar] [CrossRef]
- Behan, A.T.; Byrne, C.; Dunn, M.J.; Cagney, G.; Cotter, D.R. Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol. Psychiatry 2009, 14, 601–613. [Google Scholar] [CrossRef]
- Guest, P.C.; Chan, M.K.; Gottschalk, M.G.; Bahn, S. The use of proteomic biomarkers for improved diagnosis and stratification of schizophrenia patients. Biomark. Med. 2014, 8, 15–27. [Google Scholar] [CrossRef]
- Comes, A.L.; Papiol, S.; Mueller, T.; Geyer, P.E.; Mann, M.; Schulze, T.G. Proteomics for blood biomarker exploration of severe mental illness: Pitfalls of the past and potential for the future. Transl. Psychiat. 2018, 8, 160. [Google Scholar] [CrossRef]
- Ding, Y.H.; Guo, J.H.; Hu, Q.Y.; Jiang, W.; Wang, K.Z. Protein Biomarkers in Serum of Patients with Schizophrenia. Cell Biochem. Biophys. 2015, 72, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, C.; Song, Y.; Shi, H.; Wu, D.; Yang, Y.; Rao, C.; Liao, L.; Wu, Y.; Tang, J. Comparative proteomic analysis of plasma from bipolar depression and depressive disorder: identification of proteins associated with immune regulatory. Protein Cell 2015, 6, 908–911. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.; Guest, P.C. A Clinical Study Protocol to Identify Serum Biomarkers Predictive of Response to Antipsychotics in Schizophrenia Patients. Adv. Exp. Med. Biol. 2017, 974, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Seregin, A.A.; Smirnova, L.P.; Dmitrieva, E.M.; Vasil’eva, S.N.; Semke, A.V.; Ivanova, S.A. Glutamate Level’s in Blood Serum of Patients with Schisophrenic Spectrum and Bipolar Affective Disorder. Psikhiatriya 2020, 18, 22–31. [Google Scholar] [CrossRef]
- Ivanova, S.A.; Smirnova, L.P.; Shchigoreva, Y.G.; Semke, A.V.; Bokhan, N.A. Serum Glutathione in Patients with Schizophrenia in Dynamics of Antipsychotic Therapy. Bull. Exp. Biol. Med. 2015, 160, 283–285. [Google Scholar] [CrossRef]
- Sabherwal, S.; English, J.A.; Föcking, M.; Cagney, G.; Cotter, D.R. Blood biomarker discovery in drug-free schizophrenia: the contribution of proteomics and multiplex immunoassays. Expert. Rev. Proteom. 2016, 13, 1141–1155. [Google Scholar] [CrossRef]
- Seregin, A.A.; Smirnova, L.P.; Dmitrieva, E.M.; Boksha, I.S.; Savushkina, O.K.; Simutkin, G.G.; Ivanova, S.A. Correlations between clinical features of bipolar affective disorder and serum concentrations of ANKRD12 gene product, coagulation factor XIII, and cadherin 5. Zhurnal Nevrol. I Psikhiatrii Im. SS Korsakova 2022, 122, 137–142. (In Russian) [Google Scholar] [CrossRef]
- Rhee, S.J.; Shin, D.; Shin, D.; Song, Y.; Joo, E.J.; Jung, H.Y.; Roh, S.; Lee, S.H.; Kim, H.; Bang, M.; et al. Latent class analysis of psychotic-affective disorders with data-driven plasma proteomics. Transl. Psychiatry 2023, 13, 44. [Google Scholar] [CrossRef]
- Bantscheff, M.; Lemeer, S.; Savitski, M.M.; Kuster, B. Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012, 404, 939–965. [Google Scholar] [CrossRef] [PubMed]
- Latosinska, A.; Vougas, K.; Makridakis, M.; Klein, J.; Mullen, W.; Abbas, M.; Stravodimos, K.; Katafigiotis, I.; Merseburger, A.S.; Zoidakis, J.; et al. Comparative Analysis of Label-Free and 8-Plex iTRAQ Approach for Quantitative Tissue Proteomic Analysis. PLoS ONE 2015, 10, e0137048. [Google Scholar] [CrossRef]
- Martins-de-Souza, D.; Guest, P.C.; Vanattou-Saifoudine, N.; Harris, L.W.; Bahn, S. Proteomic technologies for biomarker studies in psychiatry: advances and needs. Int. Rev. Neurobiol. 2011, 101, 65–94. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef]
- Shteynberg, D.; Nesvizhskii, A.I.; Moritz, R.L.; Deutsch, E.W. Combining results of multiple search engines in proteomics. Mol. Cell Proteom. 2013, 12, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteom. 2010, 73, 2092–2123. [Google Scholar] [CrossRef]
- Krey, J.F.; Wilmarth, P.A.; Shin, J.B.; Klimek, J.; Sherman, N.E.; Jeffery, E.D.; Choi, D.; David, L.L.; Barr-Gillespie, P.G. Accurate label-free protein quantitation with high- and low-resolution mass spectrometers. J. Proteome Res. 2014, 13, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Välikangas, T.; Suomi, T.; Elo, L.L. A comprehensive evaluation of popular proteomics software workflows for label-free proteome quantification and imputation. Brief. Bioinform. 2018, 19, 1344–1355. [Google Scholar] [CrossRef]
- Kramps, T.; Peter, O.; Brunner, E.; Nellen, D.; Froesch, B.; Chatterjee, S.; Murone, M.; Züllig, S.; Basler, K. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 2002, 109, 47–60. [Google Scholar] [CrossRef]
- Wiese, K.E.; Nusse, R.; van Amerongen, R. Wnt signalling: conquering complexity. Development 2018, 145, dev165902. [Google Scholar] [CrossRef]
- Lie, D.C.; Colamarino, S.A.; Song, H.J.; Désiré, L.; Mira, H.; Consiglio, A.; Lein, E.S.; Jessberger, S.; Lansford, H.; Dearie, A.R.; et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature 2005, 437, 1370–1375. [Google Scholar] [CrossRef] [PubMed]
- Zandi, P.P.; Belmonte, P.L.; Willour, V.L.; Goes, F.S.; Badner, J.A.; Simpson, S.G.; Gershon, E.S.; McMahon, F.J.; DePaulo, J.R., Jr.; Potash, J.B.; et al. Association study of Wnt signaling pathway genes in bipolar disorder. Arch. Gen. Psychiatry 2008, 65, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Cuellar-Barboza, A.B.; Winham, S.J.; McElroy, S.L.; Geske, J.R.; Jenkins, G.D.; Colby, C.L.; Prieto, M.L.; Ryu, E.; Cunningham, J.M.; Frye, M.A.; et al. Accumulating evidence for a role of TCF7L2 variants in bipolar disorder with elevated body mass index. Bipolar Disord. 2016, 18, 124–135. [Google Scholar] [CrossRef]
- Guérit, S.; Fidan, E.; Macas, J.; Czupalla, C.J.; Figueiredo, R.; Vijikumar, A.; Yalcin, B.H.; Thom, S.; Winter, P.; Gerhardt, H.; et al. Astrocyte-derived Wnt growth factors are required for endothelial blood-brain barrier maintenance. Prog. Neurobiol. 2021, 199, 101937. [Google Scholar] [CrossRef] [PubMed]
- Gastfriend, B.D.; Nishihara, H.; Canfield, S.G.; Foreman, K.L.; Engelhardt, B.; Palecek, S.P.; Shusta, E.V. Wnt signaling mediates acquisition of blood–brain barrier properties in naïve endothelium derived from human pluripotent stem cells. eLife 2021, 10, e70992. [Google Scholar] [CrossRef] [PubMed]
- Baxter, H.C.; Fraser, J.R.; Liu, W.G.; Forster, J.L.; Clokie, S.; Steinacker, P.; Otto, M.; Bahn, E.; Wiltfang, J.; Aitken, A. Specific 14-3-3 isoform detection and immunolocalization in prion diseases. Biochem. Soc. Trans. 2002, 30, 387–391. [Google Scholar] [CrossRef]
- Jones, D.H.; Ley, S.; Aitken, A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995, 368, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Obsil, T.; Obsilova, V. Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 2011, 22, 663–672. [Google Scholar] [CrossRef]
- Brunet, A.; Kanai, F.; Stehn, J.; Xu, J.; Sarbassova, D.; Frangioni, J.V.; Dalal, S.N.; DeCaprio, J.A.; Greenberg, M.E.; Yaffe, M.B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002, 156, 817–828. [Google Scholar] [CrossRef]
- Sluchanko, N.N.; Gusev, N.B. Moonlighting chaperone-like activity of the universal regulatory 14-3-3 proteins. FEBS J. 2017, 284, 1279–1295. [Google Scholar] [CrossRef]
- Bridges, D.; Moorhead, G.B. 14-3-3 proteins: a number of functions for a numbered protein. Sci. STKE 2005, 2005, re10. [Google Scholar] [CrossRef]
- Jin, J.; Smith, F.D.; Stark, C.; Wells, C.D.; Fawcett, J.P.; Kulkarni, S.; Metalnikov, P.; O’Donnell, P.; Taylor, P.; Taylor, L.; et al. Proteomic functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004, 14, 1436–1450. [Google Scholar] [CrossRef]
- van Hemert, M.J.; Steensma, H.Y.; van Heusden, G.P. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 2001, 23, 936–946. [Google Scholar] [CrossRef]
- Freeman, A.K.; Morrison, D.K. 14-3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011, 22, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Gardino, A.K.; Yaffe, M.B. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin. Cell Dev. Biol. 2011, 22, 688–695. [Google Scholar] [CrossRef]
- Baxter, H.C.; Liu, W.G.; Forster, J.L.; Aitken, A.; Fraser, J.R. Immunolocalisation of 14-3-3 isoforms in normal and scrapie-infected murine brain. Neuroscience 2002, 109, 5–14. [Google Scholar] [CrossRef]
- Broadie, K.; Rushton, E.; Skoulakis, E.M.; Davis, R.L. Leonardo, a Drosophila 14-3-3 protein involved in learning, regulates presynaptic function. Neuron 1997, 19, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Schopperle, W.M.; Murrey, H.; Jaramillo, A.; Dagan, D.; Griffith, L.C.; Levitan, I.B. A dynamically regulated 14-3-3, Slob, and Slowpoke potassium channel complex in Drosophila presynaptic nerve terminals. Neuron 1999, 22, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, T.; Isobe, T.; Okuyama, T.; Yamauchi, T.; Fujisawa, H. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+, calmodulin-dependent protein kinase II. FEBS Lett. 1987, 219, 79–82. [Google Scholar] [CrossRef]
- Wang, J.; Lou, H.; Pedersen, C.J.; Smith, A.D.; Perez, R.G. 14-3-3zeta contributes to tyrosine hydroxylase activity in MN9D cells: localization of dopamine regulatory proteins to mitochondria. J. Biol. Chem. 2009, 284, 14011–14019. [Google Scholar] [CrossRef]
- Aitken, A. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 2006, 16, 162–172. [Google Scholar] [CrossRef]
- Toyo-oka, K.; Wachi, T.; Hunt, R.F.; Baraban, S.C.; Taya, S.; Ramshaw, H.; Kaibuchi, K.; Schwarz, Q.P.; Lopez, A.F.; Wynshaw-Boris, A. 14-3-3ε and ζ regulate neurogenesis and differentiation of neuronal progenitor cells in the developing brain. J. Neurosci. 2014, 34, 12168–12181. [Google Scholar] [CrossRef] [PubMed]
- Toyo-oka, K.; Shionoya, A.; Gambello, M.J.; Cardoso, C.; Leventer, R.; Ward, H.L.; Ayala, R.; Tsai, L.H.; Dobyns, W.; Ledbetter, D.; et al. 14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat. Genet. 2003, 34, 274–285. [Google Scholar] [CrossRef]
- Taya, S.; Shinoda, T.; Tsuboi, D.; Asaki, J.; Nagai, K.; Hikita, T.; Kuroda, S.; Kuroda, K.; Shimizu, M.; Hirotsune, S.; et al. DISC1 regulates the transport of the NUDEL/LIS1/14-3-3epsilon complex through kinesin-1. J. Neurosci. 2007, 27, 15–26. [Google Scholar] [CrossRef]
- Jaehne, E.J.; Ramshaw, H.; Xu, X.; Saleh, E.; Clark, S.R.; Schubert, K.O.; Lopez, A.; Schwarz, Q.; Baune, B.T. In-vivo administration of clozapine affects behaviour but does not reverse dendritic spine deficits in the 14-3-3ζ KO mouse model of schizophrenia-like disorders. Pharmacol. Biochem. Behav. 2015, 138, 1–8. [Google Scholar] [CrossRef]
- Xu, X.; Jaehne, E.J.; Greenberg, Z.; McCarthy, P.; Saleh, E.; Parish, C.L.; Camera, D.; Heng, J.; Haas, M.; Baune, B.T.; et al. 14-3-3ζ deficient mice in the BALB/c background display behavioural and anatomical defects associated with neurodevelopmental disorders. Sci. Rep. 2015, 24, 12434. [Google Scholar] [CrossRef]
- Wong, A.H.; Likhodi, O.; Trakalo, J.; Yusuf, M.; Sinha, A.; Pato, C.N.; Pato, M.T.; Van Tol, H.H.; Kennedy, J.L. Genetic and post-mortem mRNA analysis of the 14-3-3 genes that encode phosphoserine/threonine-binding regulatory proteins in schizophrenia and bipolar disorder. Schizophr. Res. 2005, 78, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yu, X.; Zhang, B.; Yuan, Y.; Xu, Q.; Shen, Y.; Shen, Y. An association study between polymorphisms in three genes of 14-3-3 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein) family and paranoid schizophrenia in northern Chinese population. Eur. Psychiatry 2004, 19, 377–379. [Google Scholar] [CrossRef] [PubMed]
- Cheah, P.S.; Ramshaw, H.S.; Thomas, P.Q.; Toyo-Oka, K.; Xu, X.; Martin, S.; Coyle, P.; Guthridge, M.A.; Stomski, F.; van den Buuse, M.; et al. Neurodevelopmental and neuropsychiatric behaviour defects arise from 14-3-3ζ deficiency. Mol. Psychiatry 2012, 17, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Fromer, M.; Pocklington, A.J.; Kavanagh, D.H.; Williams, H.J.; Dwyer, S.; Gormley, P.; Georgieva, L.; Rees, E.; Palta, P.; Ruderfer, D.M.; et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014, 506, 179–184. [Google Scholar] [CrossRef]
- Vawter, M.P.; Barrett, T.; Cheadle, C.; Sokolov, B.P.; Wood, W.H., 3rd; Donovan, D.M.; Webster, M.; Freed, W.J.; Becker, K.G. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res. Bull. 2001, 55, 641–650. [Google Scholar] [CrossRef]
- English, J.A.; Pennington, K.; Dunn, M.J.; Cotter, D.R. The neuroproteomics of schizophrenia. Biol. Psychiatry 2011, 69, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Potash, J.B.; Zandi, P.P.; Willour, V.L.; Lan, T.H.; Huo, Y.; Avramopoulos, D.; Shugart, Y.Y.; MacKinnon, D.F.; Simpson, S.G.; McMahon, F.J.; et al. Suggestive linkage to chromosomal regions 13q31 and 22q12 in families with psychotic bipolar disorder. Am. J. Psychiatry 2003, 160, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Fallin, M.D.; Lasseter, V.K.; Avramopoulos, D.; Nicodemus, K.K.; Wolyniec, P.S.; McGrath, J.A.; Steel, G.; Nestadt, G.; Liang, K.Y.; Huganir, R.L.; et al. Bipolar I disorder and schizophrenia: A 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am. J. Hum. Genet. 2005, 77, 918–936. [Google Scholar] [CrossRef] [PubMed]
- Grover, D.; Verma, R.; Goes, F.S.; Mahon, P.L.; Gershon, E.S.; McMahon, F.J.; Potash, J.B.; Gershon, E.S.; McMahon, F.J.; Potash, J.B. Family-based association of YWHAH in psychotic bipolar disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009, 150B, 977–983. [Google Scholar] [CrossRef]
- Pers, T.H.; Hansen, N.T.; Lage, K.; Koefoed, P.; Dworzynski, P.; Miller, M.L.; Flint, T.J.; Mellerup, E.; Dam, H.; Andreassen, O.A.; et al. Meta-analysis of heterogeneous data sources for genome-scale identification of risk genes in complex phenotypes. Genet. Epidemiol. 2011, 35, 318–332. [Google Scholar] [CrossRef]
- Elashoff, M.; Higgs, B.W.; Yolken, R.H.; Knable, M.B.; Weis, S.; Webster, M.J.; Barci, B.M.; Torrey, E.F. Meta-analysis of 12 genomic studies in bipolar disorder. J. Mol. Neurosci. 2007, 31, 221–243. [Google Scholar] [CrossRef]
- Thapa, N.; Lee, B.H.; Kim, I.S. TGFBIp/betaig-h3 protein: A versatile matrix molecule induced by TGF-beta. Int. J. Biochem. Cell Biol. 2007, 39, 2183–2194. [Google Scholar] [CrossRef]
- Billings, P.C.; Whitbeck, J.C.; Adams, C.S.; Abrams, W.R.; Cohen, A.J.; Engelsberg, B.N.; Howard, P.S.; Rosenbloom, J. The transforming growth factor-beta-inducible matrix protein (beta)ig-h3 interacts with fibronectin. J. Biol. Chem. 2002, 277, 28003–28009. [Google Scholar] [CrossRef]
- Kim, J.E.; Jeong, H.W.; Nam, J.O.; Lee, B.H.; Choi, J.Y.; Park, R.W.; Park, J.Y.; Kim, I.S. Identification of motifs in the fasciclin domains of the transforming growth factor-beta-induced matrix protein betaig-h3 that interact with the alphavbeta5 integrin. J. Biol. Chem. 2002, 277, 46159–46165. [Google Scholar] [CrossRef]
- Reinboth, B.; Thomas, J.; Hanssen, E.; Gibson, M.A. Beta ig-h3 interacts directly with biglycan and decorin, promotes collagen VI aggregation, and participates in ternary complexing with these macromolecules. J. Biol. Chem. 2006, 281, 7816–7824. [Google Scholar] [CrossRef]
- Lee, B.H.; Bae, J.S.; Park, R.W.; Kim, J.E.; Park, J.Y.; Kim, I.S. betaig-h3 triggers signaling pathways mediating adhesion and migration of vascular smooth muscle cells through alphavbeta5 integrin. Exp. Mol. Med. 2006, 38, 153–161. [Google Scholar] [CrossRef]
- Thapa, N.; Kang, K.B.; Kim, I.S. Beta ig-h3 mediates osteoblast adhesion and inhibits differentiation. Bone 2005, 36, 232–242. [Google Scholar] [CrossRef]
- Yun, S.J.; Kim, M.O.; Kim, S.O.; Park, J.; Kwon, Y.K.; Kim, I.S.; Lee, E.H. Induction of TGF-beta-inducible gene-h3 (betaig-h3) by TGF-beta1 in astrocytes: implications for astrocyte response to brain injury. Brain Res. Mol. Brain Res. 2002, 107, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.D.; Kukar, T.; Wang, C.Y.; Li, Q.Z.; Cruz, P.E.; Davoodi-Semiromi, A.; Yang, P.; Gu, Y.; Lian, W.; Wu, D.H.; et al. Molecular cloning and characterization of a novel mammalian endo-apyrase (LALP1). J. Biol. Chem. 2001, 276, 17474–17478. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Osorio, J.S.; Schmitt, E.; Corrêa, M.N.; Bertoni, G.; Trevisi, E.; Loor, J.J. Hepatic purinergic signaling gene network expression and its relationship with inflammation and oxidative stress biomarkers in blood from peripartal dairy cattle. J. Dairy Sci. 2014, 97, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Tordella, L.; Khan, S.; Hohmeyer, A.; Banito, A.; Klotz, S.; Raguz, S.; Martin, N.; Dhamarlingam, G.; Carroll, T.; González Meljem, J.M.; et al. SWI/SNF regulates a transcriptional program that induces senescence to prevent liver cancer. Genes. Dev. 2016, 30, 2187–2198. [Google Scholar] [CrossRef]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tseng, C.P.; Pong, R.C.; Chen, H.; McConnell, J.D.; Navone, N.; Hsieh, J.T. The mechanism of growth-inhibitory effect of DOC-2/DAB2 in prostate cancer. Characterization of a novel GTPase-activating protein associated with N-terminal domain of DOC-2/DAB2. J. Biol. Chem. 2002, 277, 12622–12631. [Google Scholar] [CrossRef]
- Zhang, H.; He, Y.; Dai, S.; Xu, Z.; Luo, Y.; Wan, T.; Luo, D.; Jones, D.; Tang, S.; Chen, H.; et al. AIP1 functions as an endogenous inhibitor of VEGFR2-mediated signaling and inflammatory angiogenesis in mice. J. Clin. Investig. 2008, 118, 3904–3916. [Google Scholar] [CrossRef]
- Zhou, H.J.; Chen, X.; Huang, Q.; Liu, R.; Zhang, H.; Wang, Y.; Jin, Y.; Liang, X.; Lu, L.; Xu, Z.; et al. AIP1 mediates vascular endothelial cell growth factor receptor-3-dependent angiogenic and lymphangiogenic responses. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 603–615. [Google Scholar] [CrossRef]
- Clarke, H.J.; Chambers, J.E.; Liniker, E.; Marciniak, S.J. Endoplasmic reticulum stress in malignancy. Cancer Cell. 2014, 25, 563–573. [Google Scholar] [CrossRef]
- Luo, D.; He, Y.; Zhang, H.; Yu, L.; Chen, H.; Xu, Z.; Tang, S.; Urano, F.; Min, W. AIP1 is critical in transducing IRE1-mediated endoplasmic reticulum stress response. J. Biol. Chem. 2008, 283, 11905–11912. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Lin, Y.; Li, J.; Pober, J.S.; Min, W. RIP1-mediated AIP1 phosphorylation at a 14-3-3-binding site is critical for tumor necrosis factor-induced ASK1-JNK/p38 activation. J. Biol. Chem. 2007, 282, 14788–14796. [Google Scholar] [CrossRef]
- Di Minin, G.; Bellazzo, A.; Dal Ferro, M.; Chiaruttini, G.; Nuzzo, S.; Bicciato, S.; Piazza, S.; Rami, D.; Bulla, R.; Sommaggio, R.; et al. Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol. Cell. 2014, 56, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; He, X.; Liu, W.; Lu, M.; Hsieh, J.T.; Min, W. AIP1 mediates TNF-alpha-induced ASK1 activation by facilitating dissociation of ASK1 from its inhibitor 14-3-3. J. Clin. Investig. 2003, 111, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Homayouni, R. Dab2IP Regulates Neuronal Positioning, Rap1 Activity and Integrin Signaling in the Developing Cortex. Dev. Neurosci. 2015, 37, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Kim, S.H.; Heck, D.; Goldowitz, D.; LeDoux, M.S.; Homayouni, R. Dab2IP GTPase activating protein regulates dendrite development and synapse number in cerebellum. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacol 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Shoichet, B.K.; Kobilka, B.K. Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol. Sci. 2012, 33, 268–272. [Google Scholar] [CrossRef]
- Denis, C.; Saulière, A.; Galandrin, S.; Sénard, J.M.; Galés, C. Probing heterotrimeric G protein activation: applications to biased ligands. Curr. Pharm. Des. 2012, 18, 128–144. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Sriram, K.; Insel, P.A. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol. Pharmacol. 2018, 93, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Purcell, R.H.; Hall, R.A. Adhesion G Protein-Coupled Receptors as Drug Targets. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 429–449. [Google Scholar] [CrossRef]
- Hamann, J.; Aust, G.; Araç, D.; Engel, F.B.; Formstone, C.; Fredriksson, R.; Hall, R.A.; Harty, B.L.; Kirchhoff, C.; Knapp, B.; et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015, 67, 338–367. [Google Scholar] [CrossRef]
- Ganesh, R.A.; Venkataraman, K.; Sirdeshmukh, R. GPR56: An adhesion GPCR involved in brain development, neurological disorders and cancer. Brain Res. 2020, 1747, 147055. [Google Scholar] [CrossRef]
- Langenhan, T.; Piao, X.; Monk, K.R. Adhesion G protein-coupled receptors in nervous system development and disease. Nat. Rev. Neurosci. 2016, 17, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Duman, J.G.; Tzeng, C.P.; Tu, Y.K.; Munjal, T.; Schwechter, B.; Ho, T.S.; Tolias, K.F. The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. J. Neurosci. 2013, 33, 6964–6978. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Li, C.; Swanson, A.M.; Villalba, R.M.; Guo, J.; Zhang, Z.; Matheny, S.; Murakami, T.; Stephenson, J.R.; Daniel, S.; et al. BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. J. Clin. Investig. 2015, 125, 1497–1508. [Google Scholar] [CrossRef]
- Stephenson, J.R.; Paavola, K.J.; Schaefer, S.A.; Kaur, B.; Van Meir, E.G.; Hall, R.A. Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J. Biol. Chem. 2013, 288, 22248–22256. [Google Scholar] [CrossRef]
- Tu, Y.K.; Duman, J.G.; Tolias, K.F. The Adhesion-GPCR BAI1 Promotes Excitatory Synaptogenesis by Coordinating Bidirectional Trans-synaptic Signaling. J. Neurosci. 2018, 38, 8388–8406. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.M.; Rizo, J. At the junction of SNARE and SM protein function. Curr. Opin. Cell Biol. 2010, 22, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Hou, N.; Yang, Y.; Scott, I.C.; Lou, X. The Sec domain protein Scfd1 facilitates trafficking of ECM components during chondrogenesis. Dev. Biol. 2017, 421, 8–15. [Google Scholar] [CrossRef]
- Nogueira, C.; Erlmann, P.; Villeneuve, J.; Santos, A.J.; Martínez-Alonso, E.; Martínez-Menárguez, J.Á.; Malhotra, V. SLY1 and Syntaxin 18 specify a distinct pathway for procollagen VII export from the endoplasmic reticulum. eLife 2014, 19, e02784. [Google Scholar] [CrossRef]
- Bando, Y.; Katayama, T.; Taniguchi, M.; Ishibashi, T.; Matsuo, N.; Ogawa, S.; Tohyama, M. RA410/Sly1 suppresses MPP+ and 6-hydroxydopamine-induced cell death in SH-SY5Y cells. Neurobiol. Dis. 2005, 18, 143–151. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Q.; Gu, X.; Wei, Q.; Cao, B.; Liu, H.; Hou, Y.; Shang, H. An association study between SCFD1 rs10139154 variant and amyotrophic lateral sclerosis in a Chinese cohort. Amyotroph. Lateral Scler. Front. Degener 2018, 19, 413–418. [Google Scholar] [CrossRef]
- Aoki, Y.; Manzano, R.; Lee, Y.; Dafinca, R.; Aoki, M.; Douglas, A.G.L.; Varela, M.A.; Sathyaprakash, C.; Scaber, J.; Barbagallo, P.; et al. C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. Brain 2017, 140, 887–897. [Google Scholar] [CrossRef]
- Jovičić, A.; Mertens, J.; Boeynaems, S.; Bogaert, E.; Chai, N.; Yamada, S.B.; Paul, J.W., 3rd; Sun, S.; Herdy, J.R.; Bieri, G.; et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015, 18, 1226–1229. [Google Scholar] [CrossRef] [PubMed]
- Theuns, J.; Verstraeten, A.; Sleegers, K.; Wauters, E.; Gijselinck, I.; Smolders, S.; Crosiers, D.; Corsmit, E.; Elinck, E.; Sharma, M.; et al. GEO-PD Consortium. Global investigation and meta-analysis of the C9orf72 (G4C2)n repeat in Parkinson disease. Neurology 2014, 83, 1906–1913. [Google Scholar] [CrossRef] [PubMed]
- Conlon, E.G.; Fagegaltier, D.; Agius, P.; Davis-Porada, J.; Gregory, J.; Hubbard, I.; Kang, K.; Kim, D.; Phatnani, H.; Shneider, N.A.; et al. Unexpected similarities between C9ORF72 and sporadic forms of ALS/FTD suggest a common disease mechanism. eLife 2018, 7, e37754. [Google Scholar] [CrossRef]
- Haeusler, A.R.; Donnelly, C.J.; Periz, G.; Simko, E.A.; Shaw, P.G.; Kim, M.S.; Maragakis, N.J.; Troncoso, J.C.; Pandey, A.; Sattler, R.; et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014, 507, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Muresan, V.; Ladescu Muresan, Z. Shared Molecular Mechanisms in Alzheimer’s Disease and Amyotrophic Lateral Sclerosis: Neurofilament-Dependent Transport of sAPP, FUS, TDP-43 and SOD1, with Endoplasmic Reticulum-like Tubules. Neurodegener. Dis. 2016, 16, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Dardiotis, E.; Karampinis, E.; Siokas, V.; Aloizou, A.M.; Rikos, D.; Ralli, S.; Papadimitriou, D.; Bogdanos, D.P.; Hadjigeorgiou, G.M. ERCC6L2 rs591486 polymorphism and risk for amyotrophic lateral sclerosis in Greek population. Neurol. Sci. 2019, 40, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Jouroukhin, Y.; Ostritsky, R.; Assaf, Y.; Pelled, G.; Giladi, E.; Gozes, I. NAP (davunetide) modifies disease progression in a mouse model of severe neurodegeneration: protection against impairments in axonal transport. Neurobiol. Dis. 2013, 56, 79–94. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Dawe, A.L.; Caldwell, K.A.; Harris, P.M.; Morris, N.R.; Caldwell, G.A. Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans. Dev. Genes. Evol. 2001, 211, 434–441. [Google Scholar] [CrossRef]
- Cockell, M.M.; Baumer, K.; Gönczy, P. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J. Cell Sci. 2004, 117, 4571–4582. [Google Scholar] [CrossRef]
- Locke, C.J.; Williams, S.N.; Schwarz, E.M.; Caldwell, G.A.; Caldwell, K.A. Genetic interactions among cortical malformation genes that influence susceptibility to convulsions in C. elegans. Brain Res. 2006, 1120, 23–34. [Google Scholar] [CrossRef]
- Williams, S.N.; Locke, C.J.; Braden, A.L.; Caldwell, K.A.; Caldwell, G.A. Epileptic-like convulsions associated with LIS-1 in the cytoskeletal control of neurotransmitter signaling in Caenorhabditis elegans. Hum. Mol. Genet. 2004, 13, 2043–2059. [Google Scholar] [CrossRef]
- Johnson, M.; Sharma, M.; Brocardo, M.G.; Henderson, B.R. IQGAP1 translocates to the nucleus in early S-phase and contributes to cell cycle progression after DNA replication arrest. Int. J. Biochem. Cell Biol. 2011, 43, 65–73. [Google Scholar] [CrossRef]
- Li, Z.; McNulty, D.E.; Marler, K.J.; Lim, L.; Hall, C.; Annan, R.S.; Sacks, D.B. IQGAP1 promotes neurite outgrowth in a phosphorylation-dependent manner. J. Biol. Chem. 2005, 280, 13871–13878. [Google Scholar] [CrossRef]
- Wang, Y.N.; Wang, H.; Yamaguchi, H.; Lee, H.J.; Lee, H.H.; Hung, M.C. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem. Biophys. Res. Commun. 2010, 399, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Lin, X.; Chen, K. Integrated analysis of differential gene expression profiles in hippocampi to identify candidate genes involved in Alzheimer’s disease. Mol. Med. Rep. 2015, 12, 6679–6687. [Google Scholar] [CrossRef]
- Bettayeb, K.; Hooli, B.V.; Parrado, A.R.; Randolph, L.; Varotsis, D.; Aryal, S.; Gresack, J.; Tanzi, R.E.; Greengard, P.; Flajolet, M. Relevance of the COPI complex for Alzheimer’s disease progression in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 5418–5423. [Google Scholar] [CrossRef] [PubMed]
- Jain Goyal, M.; Zhao, X.; Bozhinova, M.; Andrade-López, K.; de Heus, C.; Schulze-Dramac, S.; Müller-McNicoll, M.; Klumperman, J.; Béthune, J. A paralog-specific role of COPI vesicles in the neuronal differentiation of mouse pluripotent cells. Life Sci. Alliance 2020, 3, e202000714. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Rusanov, A.L.; Kozhin, P.M.; Tikhonova, O.V.; Zgoda, V.G.; Loginov, D.S.; Chlastáková, A.; Selinger, M.; Sterba, J.; Grubhoffer, L.; Luzgina, N.G. Proteome Profiling of PMJ2-R and Primary Peritoneal Macrophages. Int. J. Mol. Sci. 2021, 22, 6323. [Google Scholar] [CrossRef]
- Geer, L.Y.; Markey, S.P.; Kowalak, J.A.; Wagner, L.; Xu, M.; Maynard, D.M.; Yang, X.; Shi, W.; Bryant, S.H. Open mass spectrometry search algorithm. J. Proteome Res. 2004, 3, 958–964. [Google Scholar] [CrossRef]
- Craig, R.; Beavis, R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 2004, 20, 1466–1467. [Google Scholar] [CrossRef]
- Barsnes, H.; Vaudel, M. SearchGUI: A Highly Adaptable Common Interface for Proteomics Search and de Novo Engines. J. Proteome Res. 2018, 17, 2552–2555. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 2010, 604, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Vaudel, M.; Burkhart, J.M.; Zahedi, R.P.; Oveland, E.; Berven, F.S.; Sickmann, A.; Martens, L.; Barsnes, H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [Google Scholar] [CrossRef]
- Vaudel, M.; Breiter, D.; Beck, F.; Rahnenführer, J.; Martens, L.; Zahedi, R.P. D-score: a search engine independent MD-score. Proteomics 2013, 13, 1036–1041. [Google Scholar] [CrossRef]
- Taus, T.; Köcher, T.; Pichler, P.; Paschke, C.; Schmidt, A.; Henrich, C.; Mechtler, K. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 2011, 10, 5354–5362. [Google Scholar] [CrossRef] [PubMed]
- Barsnes, H.; Vaudel, M.; Colaert, N.; Helsens, K.; Sickmann, A.; Berven, F.S.; Martens, L. Compomics-utilities: an open-source Java library for computational proteomics. BMC Bioinform. 2011, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, A.C.; Parmely, T.J.; Tomomori-Sato, C.; Sato, S.; Zhu, D.; Conaway, R.C.; Conaway, J.W.; Florens, L.; Washburn, M.P. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. USA 2006, 103, 18928–18933. [Google Scholar] [CrossRef]
- McIlwain, S.; Mathews, M.; Bereman, M.S.; Rubel, E.W.; MacCoss, M.J.; Noble, W.S. Estimating relative abundances of proteins from shotgun proteomics data. BMC Bioinform. 2012, 13, 308. [Google Scholar] [CrossRef] [PubMed]
- Degroeve, S.; Staes, A.; De Bock, P.J.; Martens, L. The effect of peptide identification search algorithms on MS2-based label-free protein quantification. OMICS 2012, 16, 443–448. [Google Scholar] [CrossRef]
- Florens, L.; Carozza, M.J.; Swanson, S.K.; Fournier, M.; Coleman, M.K.; Workman, J.L.; Washburn, M.P. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 2006, 40, 303–311. [Google Scholar] [CrossRef]
- Geraghty, N.J.; Satapathy, S.; Kelly, M.; Cheng, F.; Lee, A.; Wilson, M.R. Expanding the family of extracellular chaperones: Identification of human plasma proteins with chaperone activity. Protein Sci. 2021, 30, 2272–2286. [Google Scholar] [CrossRef]
- Rossouw, S.C.; Bendou, H.; Blignaut, R.J.; Bell, L.; Rigby, J.; Christoffels, A. Evaluation of Protein Purification Techniques and Effects of Storage Duration on LC-MS/MS Analysis of Archived FFPE Human CRC Tissues. Pathol. Oncol. Res. 2021, 27, 622855. [Google Scholar] [CrossRef] [PubMed]
- Van der Pan, K.; Kassem, S.; Khatri, I.; de Ru Arnoud, H.; Janssen George, M.C.; Tjokrodirijo Rayman, T.N.; al Makindji, F.; Stavrakaki, E.; de Jager Anniek, L.; Naber Brigitta, A.E.; et al. Quantitative proteomics of small numbers of closely-related cells: Selection of the optimal method for a clinical setting. Front. Med. 2022, 9, 997305. [Google Scholar] [CrossRef] [PubMed]
| Uniprot Code | Protein Name | Gene | NSAF BD Mean | NSAF Control Mean | Student’s t-Test, p |
|---|---|---|---|---|---|
| Q15582 | Transforming growth factor-beta-induced protein ig-h3 | TGFBI | 0.005941 | 0.001980 | 0.0342 |
| Q5VWQ8 | Disabled homolog 2-interacting protein | DAB2IP | 0.000014 | 0.004689 | 0.01055 |
| Q76M96 | Coiled-coil domain-containing protein 80 | CCDC80 | 0.000015 | 0.001359 | 0.01064 |
| O00512 | B-cell CLL/lymphoma 9 protein | BCL9 | 0.018721 | 0.000029 | 0.01092 |
| Q9Y678 | Coatomer subunit gamma-1 | COPG1 | 0.000018 | 0.002829 | 0.01281 |
| P46940 | Ras GTPase-activating-like protein IQGAP1 | IQGAP1 | 0.0000019 | 0.000765 | 0.01446 |
| Q9NQZ7 | Ectonucleoside triphosphate diphosphohydrolase 7 | ENTPD7 | 0.002105 | 0.000012 | 0.01655 |
| O14514 | Adhesion G-protein-coupled receptor B1 | ADGRB1 | 0.000015 | 0.003524 | 0.02106 |
| Q8WVM8 | Sec1 family domain-containing protein | SCFD1 | 0.000017 | 0.001866 | 0.00486 |
| P63104 | 14-3-3 protein zeta/delta | YWHAZ | 0.009346 | 0.000026 | 0.005644 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seregin, A.A.; Smirnova, L.P.; Dmitrieva, E.M.; Zavialova, M.G.; Simutkin, G.G.; Ivanova, S.A. Differential Expression of Proteins Associated with Bipolar Disorder as Identified Using the PeptideShaker Software. Int. J. Mol. Sci. 2023, 24, 15250. https://doi.org/10.3390/ijms242015250
Seregin AA, Smirnova LP, Dmitrieva EM, Zavialova MG, Simutkin GG, Ivanova SA. Differential Expression of Proteins Associated with Bipolar Disorder as Identified Using the PeptideShaker Software. International Journal of Molecular Sciences. 2023; 24(20):15250. https://doi.org/10.3390/ijms242015250
Chicago/Turabian StyleSeregin, Alexander A., Liudmila P. Smirnova, Elena M. Dmitrieva, Maria G. Zavialova, German G. Simutkin, and Svetlana A. Ivanova. 2023. "Differential Expression of Proteins Associated with Bipolar Disorder as Identified Using the PeptideShaker Software" International Journal of Molecular Sciences 24, no. 20: 15250. https://doi.org/10.3390/ijms242015250





