CWH43 Is a Novel Tumor Suppressor Gene with Negative Regulation of TTK in Colorectal Cancer
Abstract
:1. Introduction
2. Results
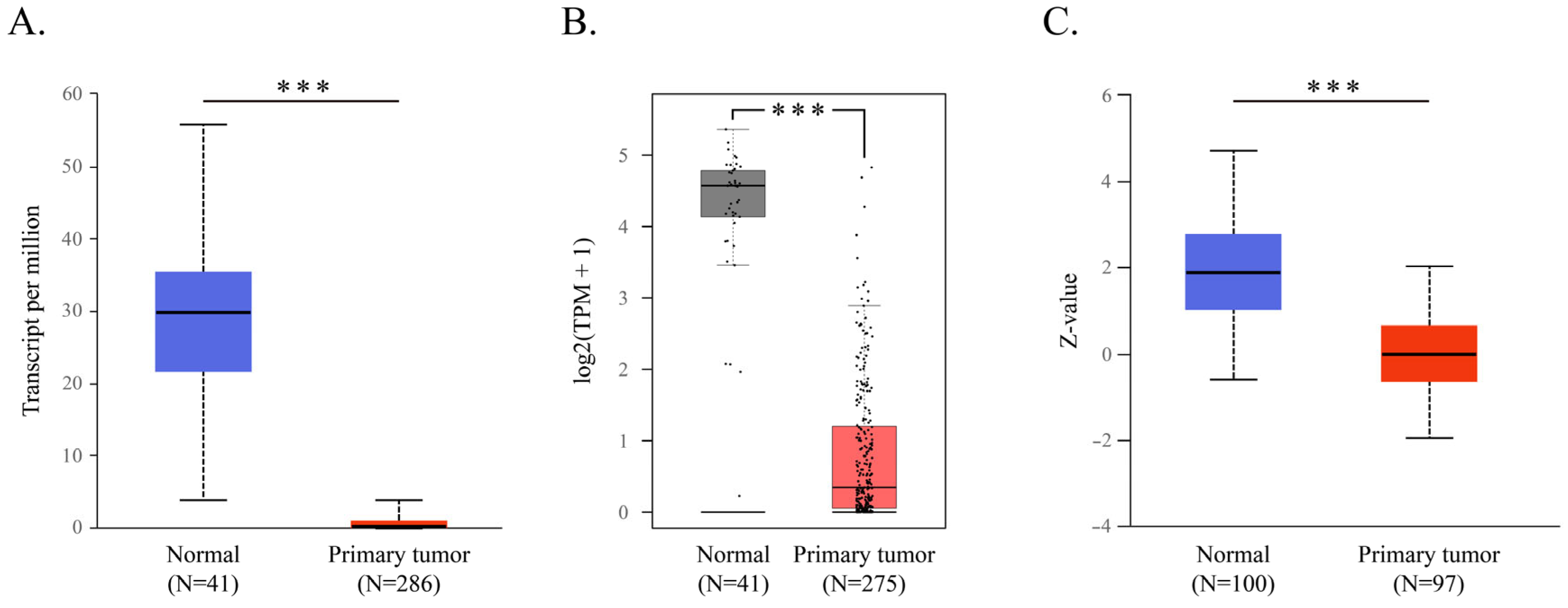
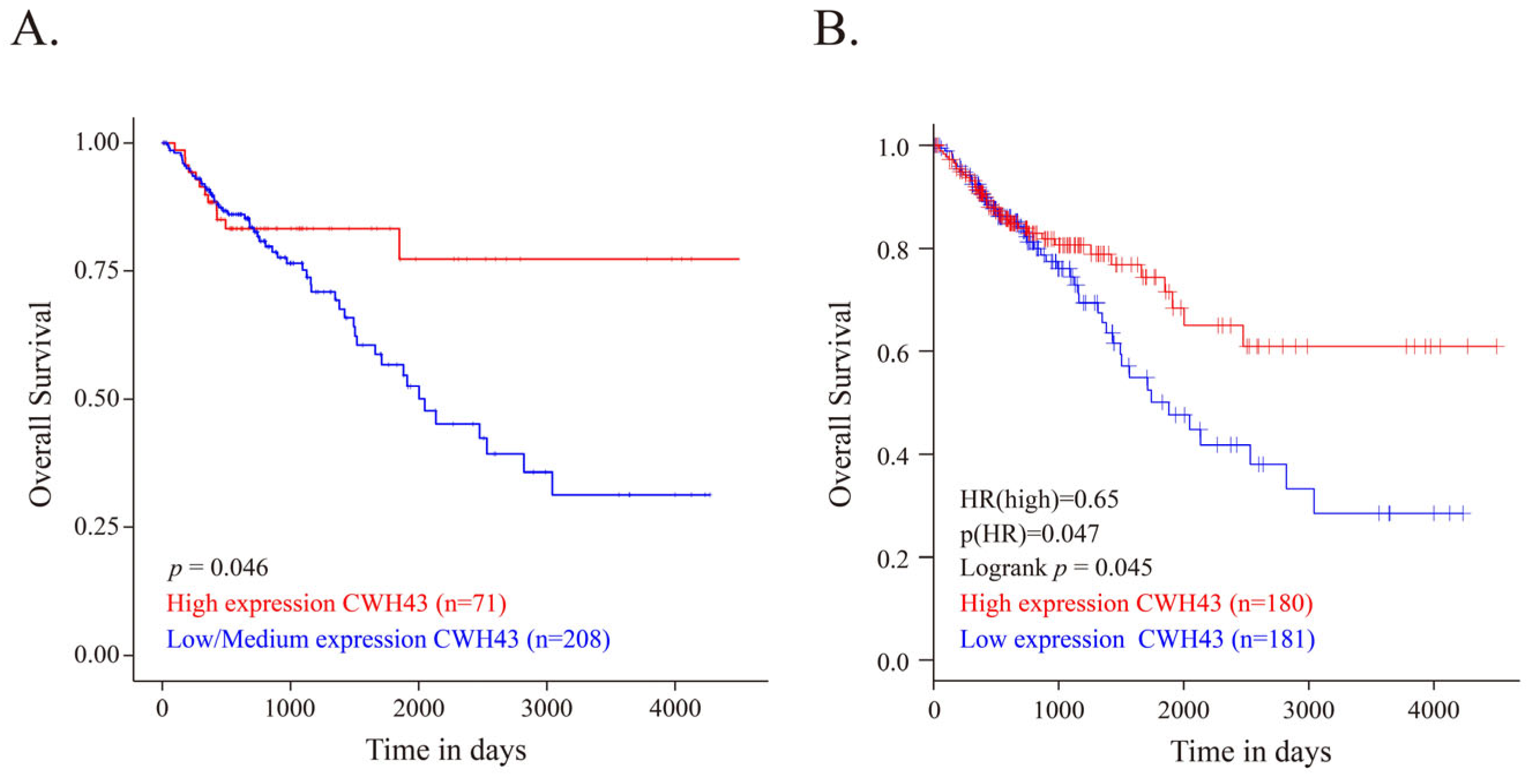
2.1. CWH43 in CRC and Its Implications for Patient Outcomes
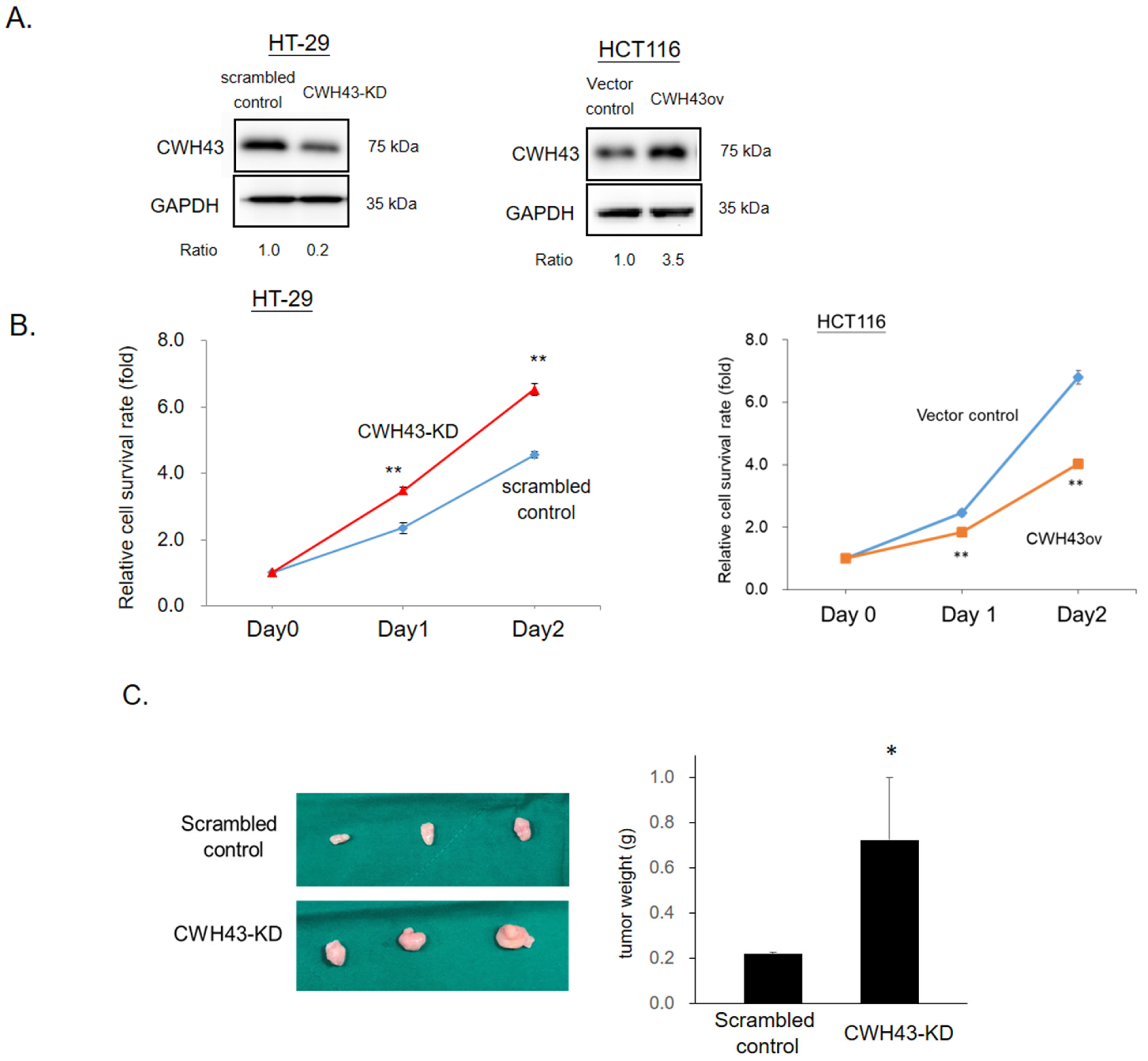
2.2. CWH43’s Influence on CRC Tumorigenesis and Cell Growth
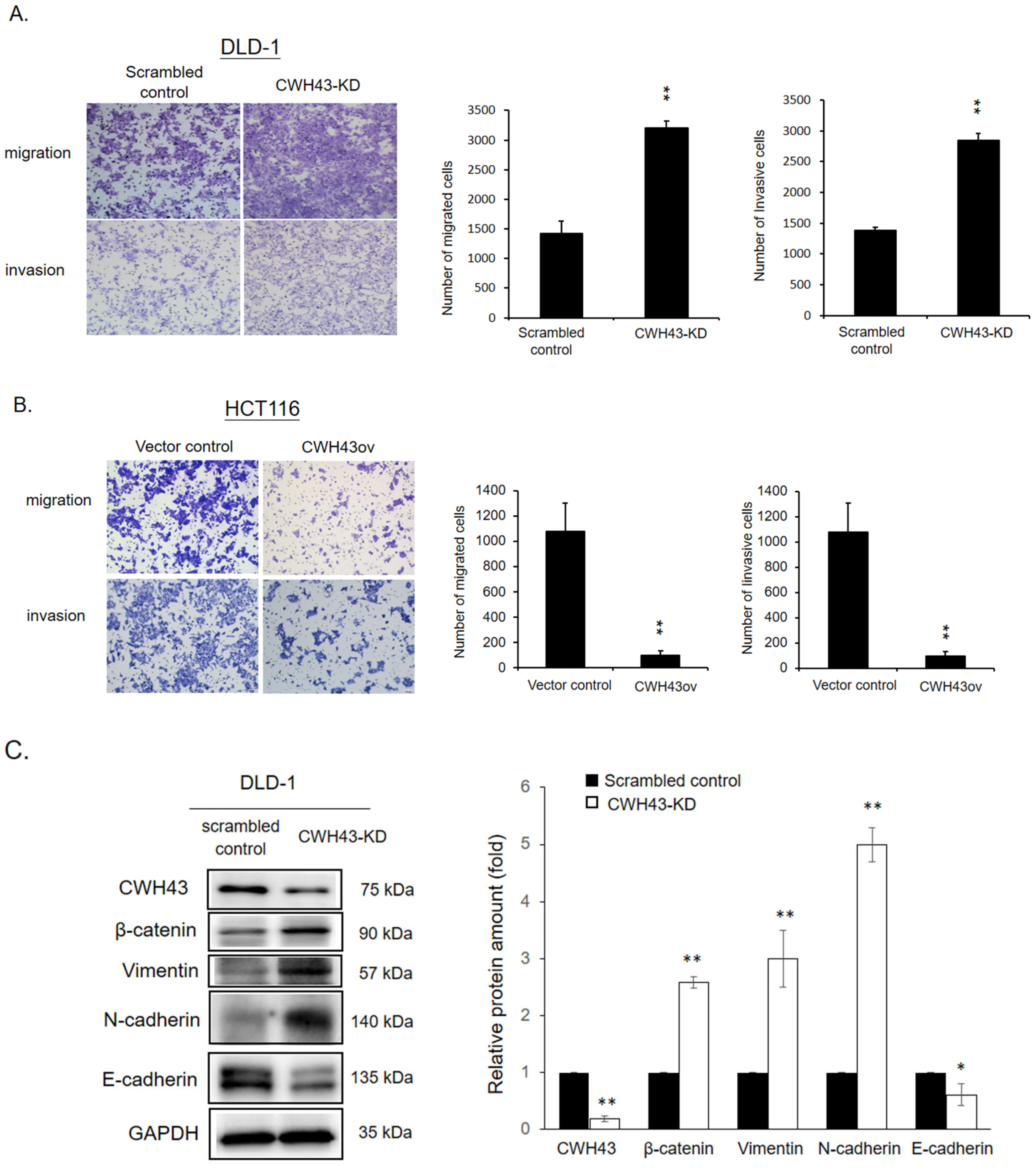
2.3. Involvement of CWH43 in CRC Migration, Invasion, and EMT Regulation
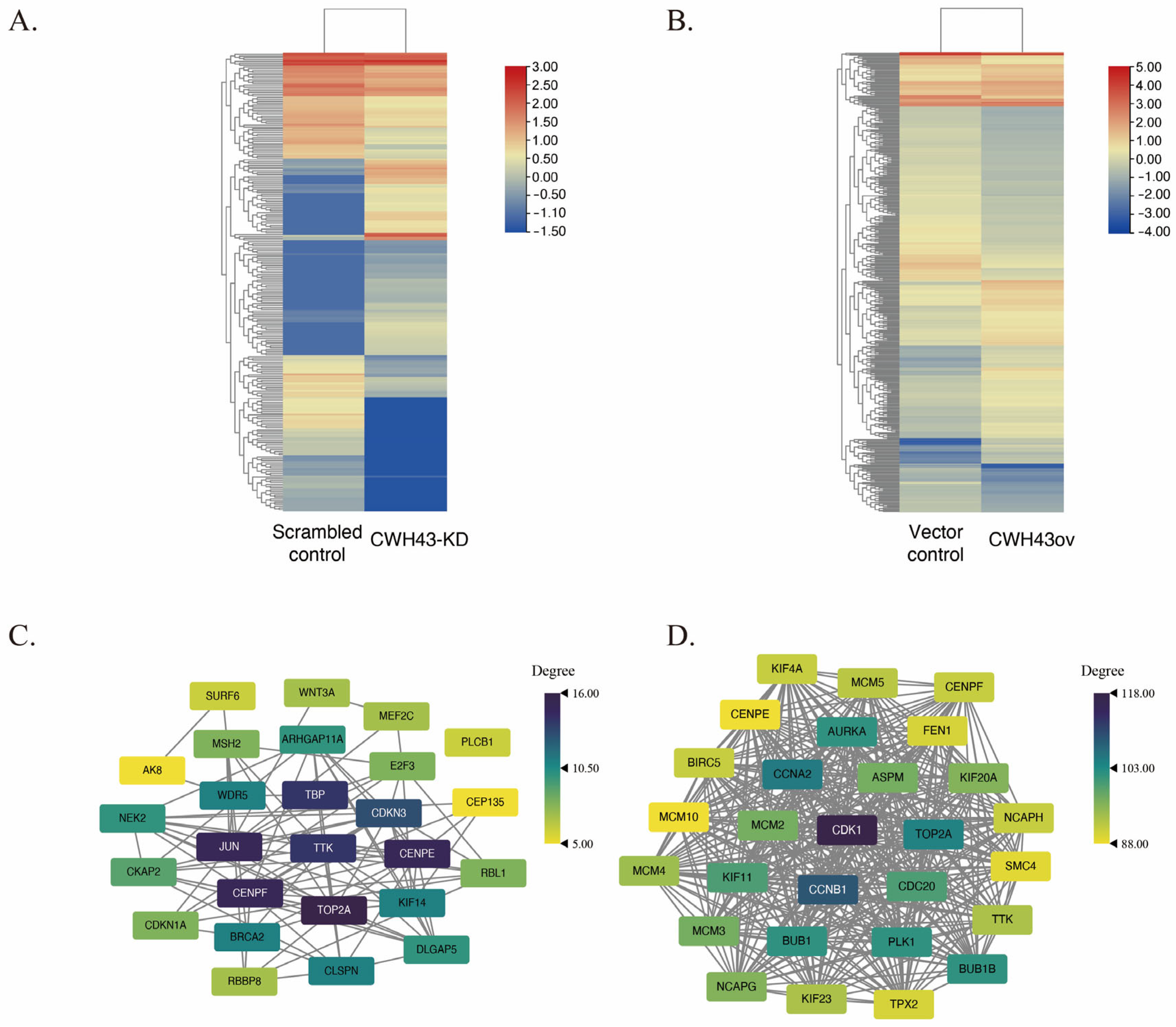
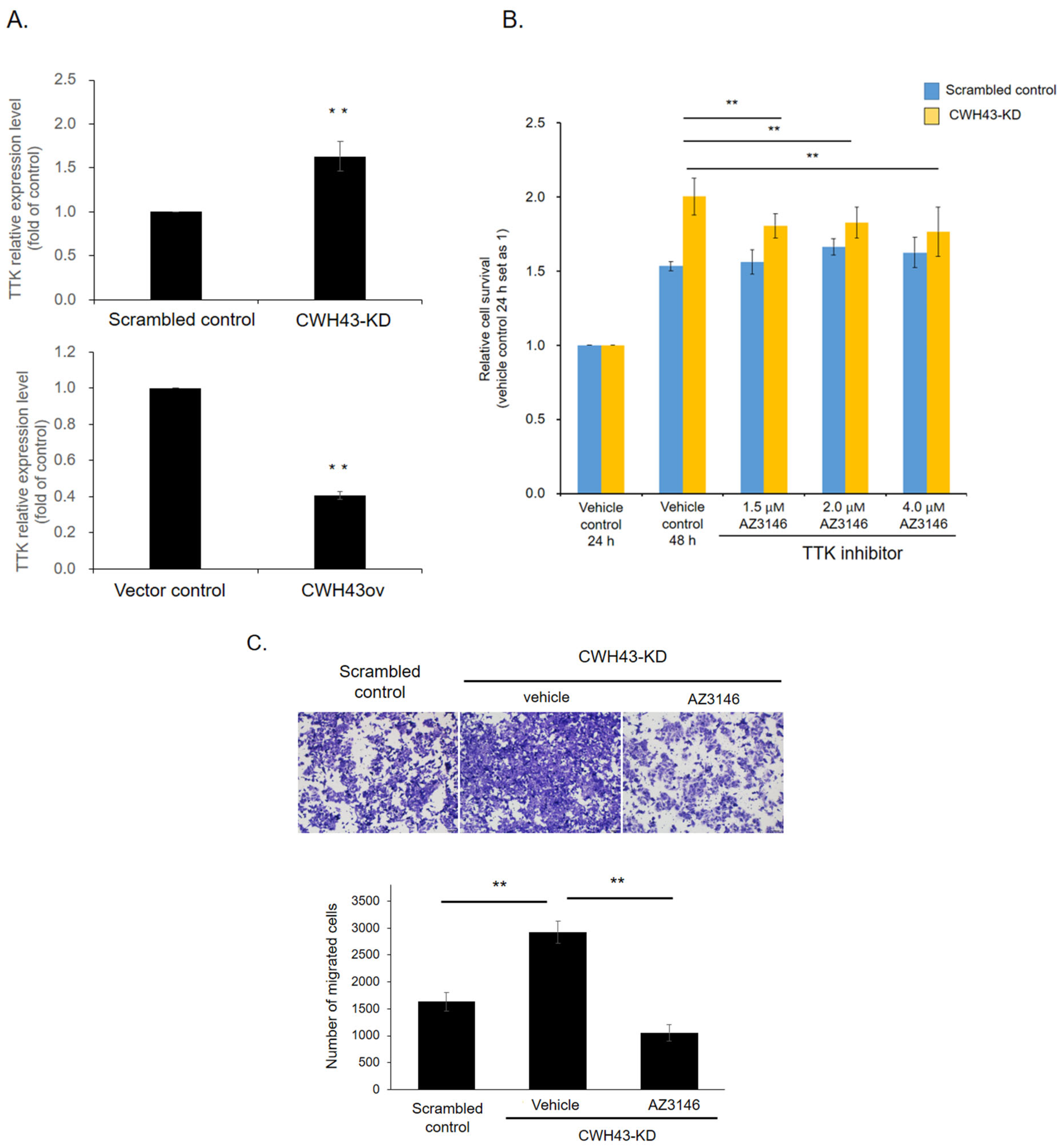
2.4. CWH43’s Regulatory Impact on Threonine Tyrosine Kinase (TTK) in CRC
2.5. KEGG Pathway Analysis and Its Regulatory Influence on TTK Expression in the Cell Cycle
2.6. TTK Inhibitor Reversed Tumor-Promoting Effect in CWH43-KD Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Gene Expression Level, Protein Expression Level, and Patient Survival Related to CWH43 in Colorectal Cancer
4.2. Functional Enrichment Analysis
4.3. Chemicals, Reagents, and Cell Culture
4.4. Transfection and Generation of Stable Colonies
4.5. Examination of Cell Viability
4.6. Transwell Migration and Invasion Assay
4.7. Animal Model
4.8. RNA Extraction, cDNA Synthesis, and Quantitative Polymerase Chain Reaction (qPCR) Analysis
4.9. Protein Extraction and Western Blot Assay
4.10. RNA Sequencing
4.11. PPI Network Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Wang, J.; Li, S.; Liu, Y.; Zhang, C.; Li, H.; Lai, B. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer Med. 2020, 9, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Morris, V.K.; Kennedy, E.B.; Baxter, N.N.; Benson, A.B., 3rd; Cercek, A.; Cho, M.; Ciombor, K.K.; Cremolini, C.; Davis, A.; Deming, D.A.; et al. Treatment of Metastatic Colorectal Cancer: ASCO Guideline. J. Clin. Oncol. 2023, 41, 678–700. [Google Scholar] [CrossRef] [PubMed]
- Paulick, M.G.; Bertozzi, C.R. The glycosylphosphatidylinositol anchor: A complex membrane-anchoring structure for proteins. Biochemistry 2008, 47, 6991–7000. [Google Scholar] [CrossRef]
- Kinoshita, T.; Fujita, M. Biosynthesis of GPI-anchored proteins: Special emphasis on GPI lipid remodeling. J. Lipid Res. 2015, 57, 6–24. [Google Scholar] [CrossRef]
- Umemura, M.; Fujita, M.; Yoko, O.T.; Fukamizu, A.; Jigami, Y. Saccharomyces cerevisiae CWH43 is involved in the remodeling of the lipid moiety of GPI anchors to ceramides. Mol. Biol. Cell 2007, 18, 4304–4316. [Google Scholar] [CrossRef]
- Fujita, M.; Kinoshita, T. GPI-anchor remodeling: Potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2012, 1821, 1050–1058. [Google Scholar] [CrossRef]
- Bosson, R.; Jaquenoud, M.; Conzelmann, A. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell 2006, 17, 2636–2645. [Google Scholar] [CrossRef]
- Novarino, G.; Fenstermaker, A.G.; Zaki, M.S.; Hofree, M.; Silhavy, J.L.; Heiberg, A.D.; Abdellateef, M.; Rosti, B.; Scott, E.; Mansour, L. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 2014, 343, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, A.; Lin, J.; Wu, L.; Zhang, H.; Yang, X.; Wan, X.; Miao, R.; Sang, X.; Zhao, H. Mps1/TTK: A novel target and biomarker for cancer. J. Drug Target. 2017, 25, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.M.; Wu, K.L.; Chang, Y.Y.; Hung, J.Y.; Chang, W.A.; Chang, C.Y.; Jian, S.F.; Tsai, P.H.; Huang, Y.C.; Chong, I.W.; et al. Upregulation of Thr/Tyr kinase Increases the Cancer Progression by Neurotensin and Dihydropyrimidinase-Like 3 in Lung Cancer. Int. J. Mol. Sci. 2020, 21, 1640. [Google Scholar] [CrossRef]
- Gao, Y.H.; Qu, S.S.; Cao, L.Q.; Yao, M. TTK predicts triple positive breast cancer prognosis and regulates tumor proliferation and invasion. Neoplasma 2022, 69, 274–282. [Google Scholar] [CrossRef]
- Liu, X.; Liao, W.; Yuan, Q.; Ou, Y.; Huang, J. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget 2015, 6, 34309–34320. [Google Scholar] [CrossRef]
- Liu, X.D.; Yao, D.W.; Xin, F. TTK contributes to tumor growth and metastasis of clear cell renal cell carcinoma by inducing cell proliferation and invasion. Neoplasma 2019, 66, 946–953. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, B.; Zhu, N.; Tao, M.; Jun, Y.; Chen, X.; Wang, Q.; Luo, C. Mitotic checkpoint kinase Mps1/TTK predicts prognosis of colon cancer patients and regulates tumor proliferation and differentiation via PKCalpha/ERK1/2 and PI3K/Akt pathway. Med. Oncol. 2020, 37, 5. [Google Scholar] [CrossRef]
- Huang, H.; Yang, Y.; Zhang, W.; Liu, X.; Yang, G. TTK regulates proliferation and apoptosis of gastric cancer cells through the Akt-mTOR pathway. FEBS Open Biol. 2020, 10, 1542–1549. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, M.; Liang, D.; Sun, W.; Zhang, C.; Jiang, M.; Liu, J.; Li, J.; Li, C.; Yang, X.; et al. Molecular design and anticancer activities of small-molecule monopolar spindle 1 inhibitors: A Medicinal chemistry perspective. Eur. J. Med. Chem. 2019, 175, 247–268. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-M.; Yao, C.-T.; Chang, Y.-T.; Chou, H.-L.; Chou, Y.-C.; Chen, K.-H.; Terng, H.-J.; Huang, C.-S.; Lee, C.-C.; Su, S.-L. Gene expression profiling of colorectal tumors and normal mucosa by microarrays meta-analysis using prediction analysis of microarray, artificial neural network, classification, and regression trees. Dis. Mark. 2014, 2014, 634123. [Google Scholar] [CrossRef] [PubMed]
- Lacalamita, A.; Piccinno, E.; Scalavino, V.; Bellotti, R.; Giannelli, G.; Serino, G. A Gene-Based Machine Learning Classifier Associated to the Colorectal Adenoma—Carcinoma Sequence. Biomedicines 2021, 9, 1937. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.W.; Lee, S.; Yang, D.; Dai, H.; Zhang, Y.; Han, L.; Zhao, S.; Zhang, S.; Ma, Y.; Johnson, M.F.; et al. Deletions in CWH43 cause idiopathic normal pressure hydrocephalus. EMBO Mol. Med. 2021, 13, e13249. [Google Scholar] [CrossRef]
- Tipton, P.W.; Atik, M.; Soto-Beasley, A.I.; Day, G.S.; Grewal, S.S.; Chaichana, K.; Fermo, O.P.; Ball, C.T.; Heckman, M.G.; White, L.J.; et al. CWH43 Variants Are Associated with Disease Risk and Clinical Phenotypic Measures in Patients with Normal Pressure Hydrocephalus. Neurol. Genet. 2023, 9, e200086. [Google Scholar] [CrossRef]
- Yeung, K.T.; Yang, J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef]
- Jonckheere, S.; Adams, J.; De Groote, D.; Campbell, K.; Berx, G.; Goossens, S. Epithelial-Mesenchymal Transition (EMT) as a Therapeutic Target. Cells Tissues Org. 2022, 211, 157–182. [Google Scholar] [CrossRef]
- Nantajit, D.; Lin, D.; Li, J.J. The network of epithelial-mesenchymal transition: Potential new targets for tumor resistance. J. Cancer Res. Clin. Oncol. 2015, 141, 1697–1713. [Google Scholar] [CrossRef]
- Yoko-o, T.; Umemura, M.; Komatsuzaki, A.; Ikeda, K.; Ichikawa, D.; Takase, K.; Kanzawa, N.; Saito, K.; Kinoshita, T.; Taguchi, R. Lipid moiety of glycosylphosphatidylinositol-anchored proteins contributes to the determination of their final destination in yeast. Genes Cells 2018, 23, 880–892. [Google Scholar] [CrossRef]
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2007, 1771, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, H.; Sun, Y.; Zeng, X.; Zhang, W. Identification of the hub genes in gastric cancer through weighted gene co-expression network analysis. PeerJ 2021, 9, e10682. [Google Scholar] [CrossRef]
- London, N.; Biggins, S. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 2014, 15, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Pachis, S.T.; Kops, G.J. Leader of the SAC: Molecular mechanisms of Mps1/TTK regulation in mitosis. Open Biol. 2018, 8, 180109. [Google Scholar]
- Thu, K.; Silvester, J.; Elliott, M.; Ba-Alawi, W.; Duncan, M.; Elia, A.; Mer, A.; Smirnov, P.; Safikhani, Z.; Haibe-Kains, B. Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E1570–E1577. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, Y.; Ni, C.; Meng, G.; Sheng, X. HLF/miR-132/TTK axis regulates cell proliferation, metastasis and radiosensitivity of glioma cells. Biomed. Pharmacother. 2016, 83, 898–904. [Google Scholar] [CrossRef]
- Lu, N.; Ren, L. TTK (threonine tyrosine kinase) regulates the malignant behaviors of cancer cells and is regulated by microRNA-582-5p in ovarian cancer. Bioengineered 2021, 12, 5759–5768. [Google Scholar] [CrossRef]
- Ling, Y.; Zhang, X.; Bai, Y.; Li, P.; Wei, C.; Song, T.; Zheng, Z.; Guan, K.; Zhang, Y.; Zhang, B. Overexpression of Mps1 in colon cancer cells attenuates the spindle assembly checkpoint and increases aneuploidy. Biochem. Biophys. Res. Commun. 2014, 450, 1690–1695. [Google Scholar] [CrossRef]
- Zhang, X.; Ling, Y.; Guo, Y.; Bai, Y.; Shi, X.; Gong, F.; Tan, P.; Zhang, Y.; Wei, C.; He, X. Mps1 kinase regulates tumor cell viability via its novel role in mitochondria. Cell Death Dis. 2016, 7, e2292. [Google Scholar] [CrossRef]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef]
- Maire, V.; Baldeyron, C.; Richardson, M.; Tesson, B.; Vincent-Salomon, A.; Gravier, E.; Marty-Prouvost, B.; De Koning, L.; Rigaill, G.; Dumont, A.; et al. TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS ONE 2013, 8, e63712. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, K.; Guan, X.; Xie, S.; Wang, Y.; Tong, Y.; Guo, L.; Zheng, H.; Lu, R. TTK is a potential therapeutic target for cisplatin-resistant ovarian cancer. J. Ovarian Res. 2021, 14, 128. [Google Scholar] [CrossRef]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef]
- Uitdehaag, J.C.M.; de Man, J.; Willemsen-Seegers, N.; Prinsen, M.B.W.; Libouban, M.A.A.; Sterrenburg, J.G.; de Wit, J.J.P.; de Vetter, J.R.F.; de Roos, J.; Buijsman, R.C.; et al. Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity. J. Mol. Biol. 2017, 429, 2211–2230. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.F.; Lee, C.Y.; Lai, L.C.; Tsai, M.H.; Lu, T.P.; Chuang, E.Y. CellExpress: A comprehensive microarray-based cancer cell line and clinical sample gene expression analysis online system. Database 2018, 2018, bax101. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Primer Sequences (5′ → 3′) |
|---|---|
| CWH43 | Forward: 5′-CCTCCTTCCAGGCTCCAAAT-3′ |
| Reverse: 5′-GACACCCCAAGCGCAAGA-3′ | |
| TTK | Forward: 5′-GCTTGTCAGTTGTCAACACCTTATG-3′ |
| Reverse: 5′-GGCAAGTATTTGATGCTGTTGCT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.-C.; Tsai, K.-Y.; Lee, A.-W.; Wei, P.-L.; Huang, C.-Y.; Batzorig, U.; Chang, Y.-J. CWH43 Is a Novel Tumor Suppressor Gene with Negative Regulation of TTK in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 15262. https://doi.org/10.3390/ijms242015262
Lee C-C, Tsai K-Y, Lee A-W, Wei P-L, Huang C-Y, Batzorig U, Chang Y-J. CWH43 Is a Novel Tumor Suppressor Gene with Negative Regulation of TTK in Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(20):15262. https://doi.org/10.3390/ijms242015262
Chicago/Turabian StyleLee, Cheng-Chin, Kuei-Yen Tsai, Ai-Wei Lee, Po-Li Wei, Chien-Yu Huang, Uyanga Batzorig, and Yu-Jia Chang. 2023. "CWH43 Is a Novel Tumor Suppressor Gene with Negative Regulation of TTK in Colorectal Cancer" International Journal of Molecular Sciences 24, no. 20: 15262. https://doi.org/10.3390/ijms242015262
APA StyleLee, C.-C., Tsai, K.-Y., Lee, A.-W., Wei, P.-L., Huang, C.-Y., Batzorig, U., & Chang, Y.-J. (2023). CWH43 Is a Novel Tumor Suppressor Gene with Negative Regulation of TTK in Colorectal Cancer. International Journal of Molecular Sciences, 24(20), 15262. https://doi.org/10.3390/ijms242015262






