RNA Profiles of Tear Fluid Extracellular Vesicles in Patients with Dry Eye-Related Symptoms
Abstract
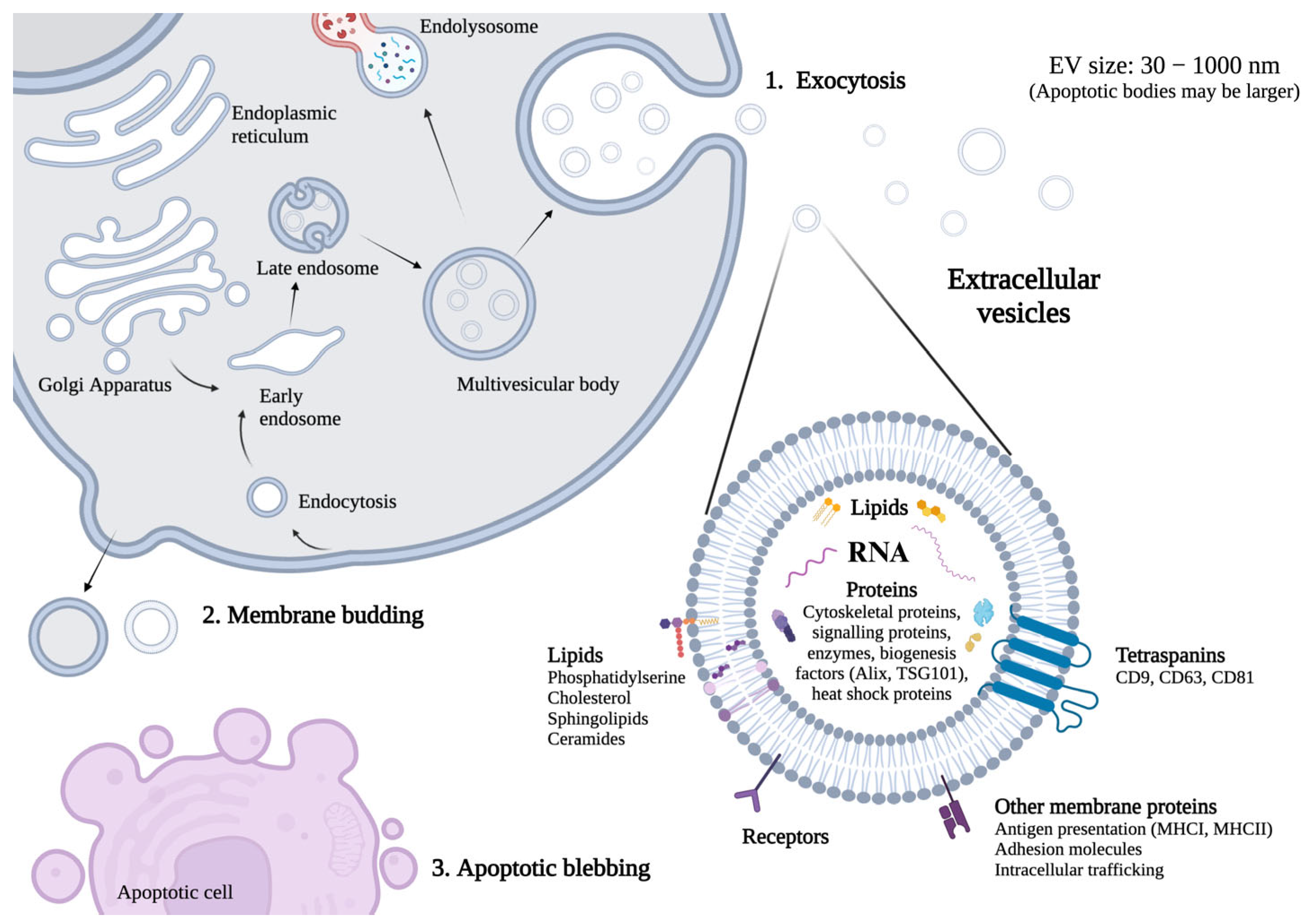
1. Introduction
2. Results
2.1. EV Characterization
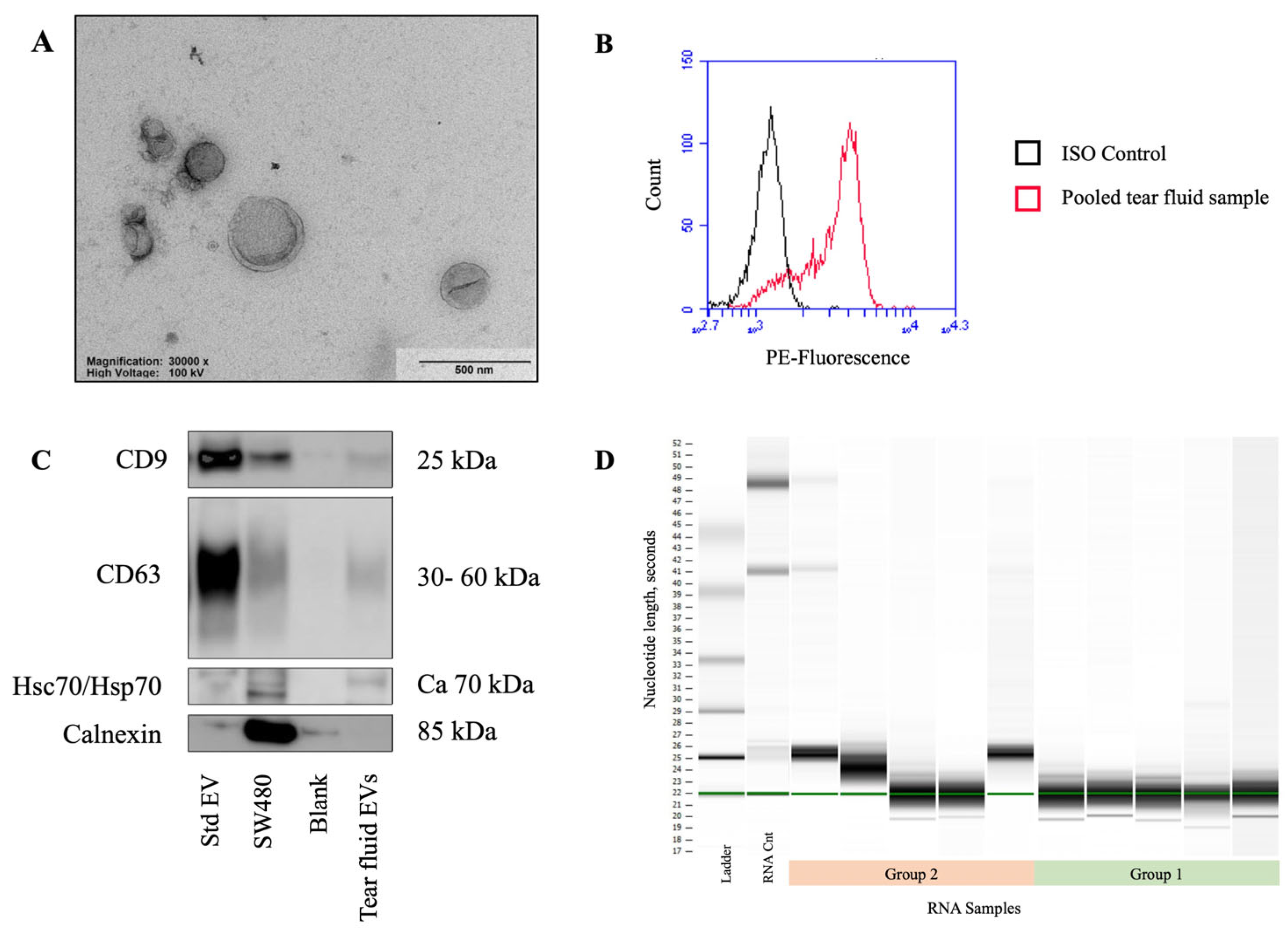
2.1.1. Nanoparticle Tracking Analysis
2.1.2. Transmission Electron Microscopy
2.1.3. Flow Cytometry
2.1.4. Western Blotting
2.1.5. EV-RNA Characterization
2.2. Tear Fluid EV-RNA Analysis Using Microarrays
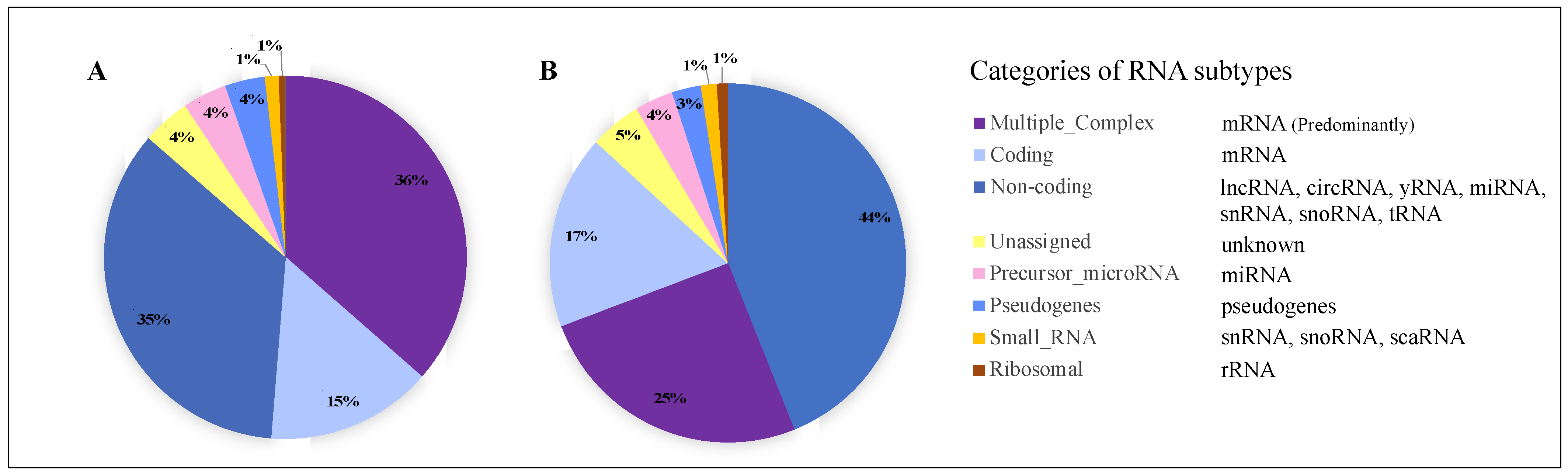
2.2.1. RNA Subtypes Identified
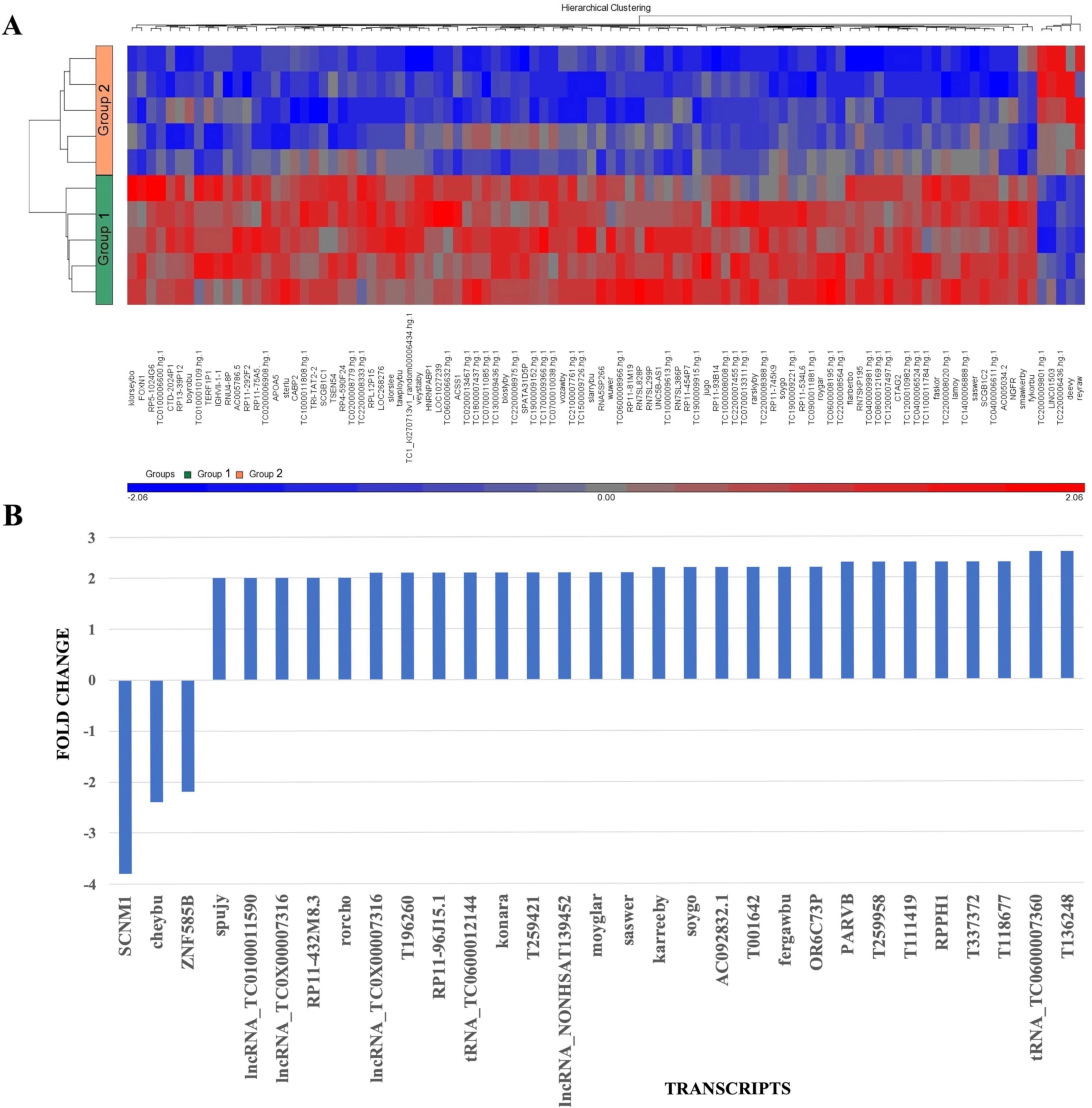
2.2.2. Comparing Patients with TBUT 2 s and TBUT 10 s
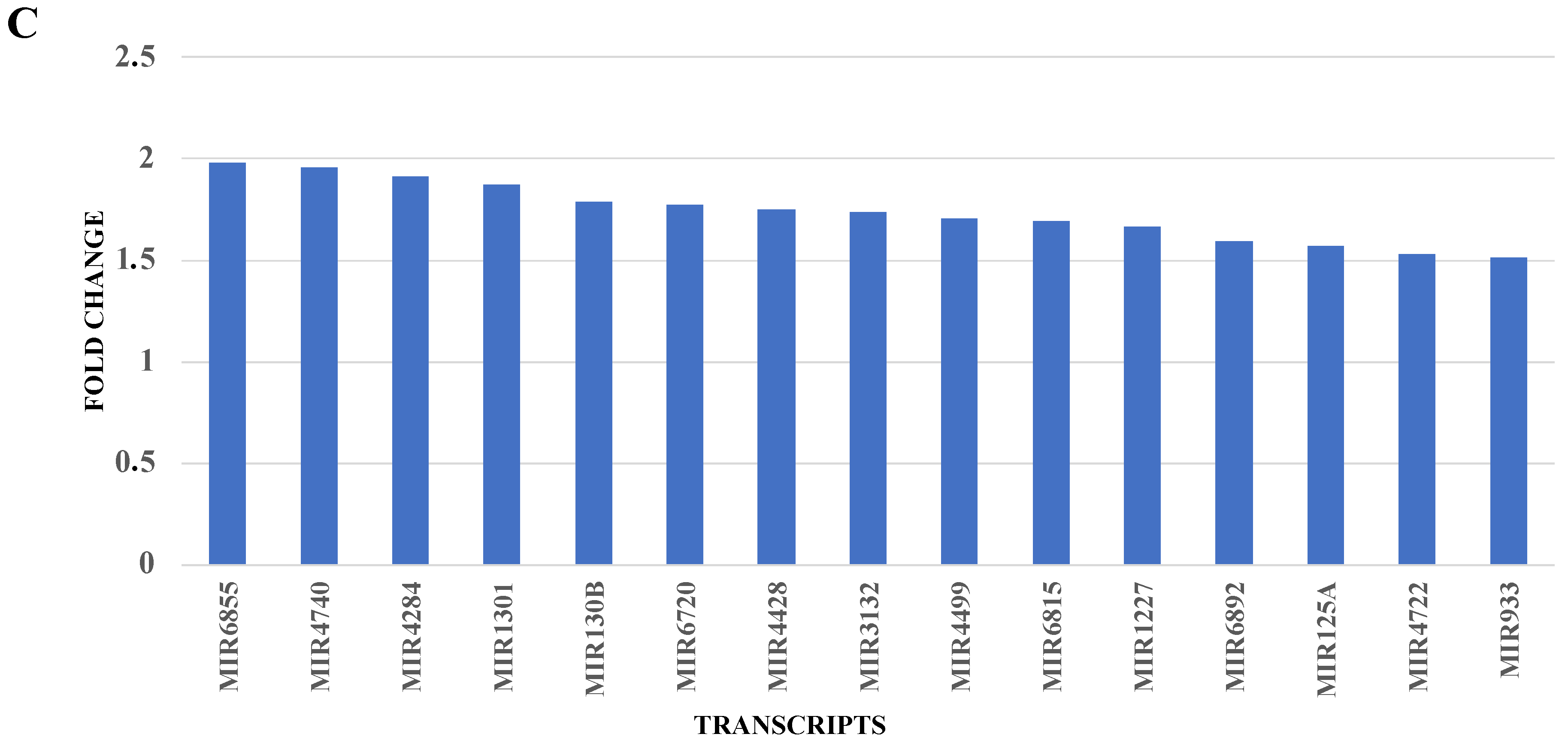
2.2.3. RNA Validation
3. Discussion
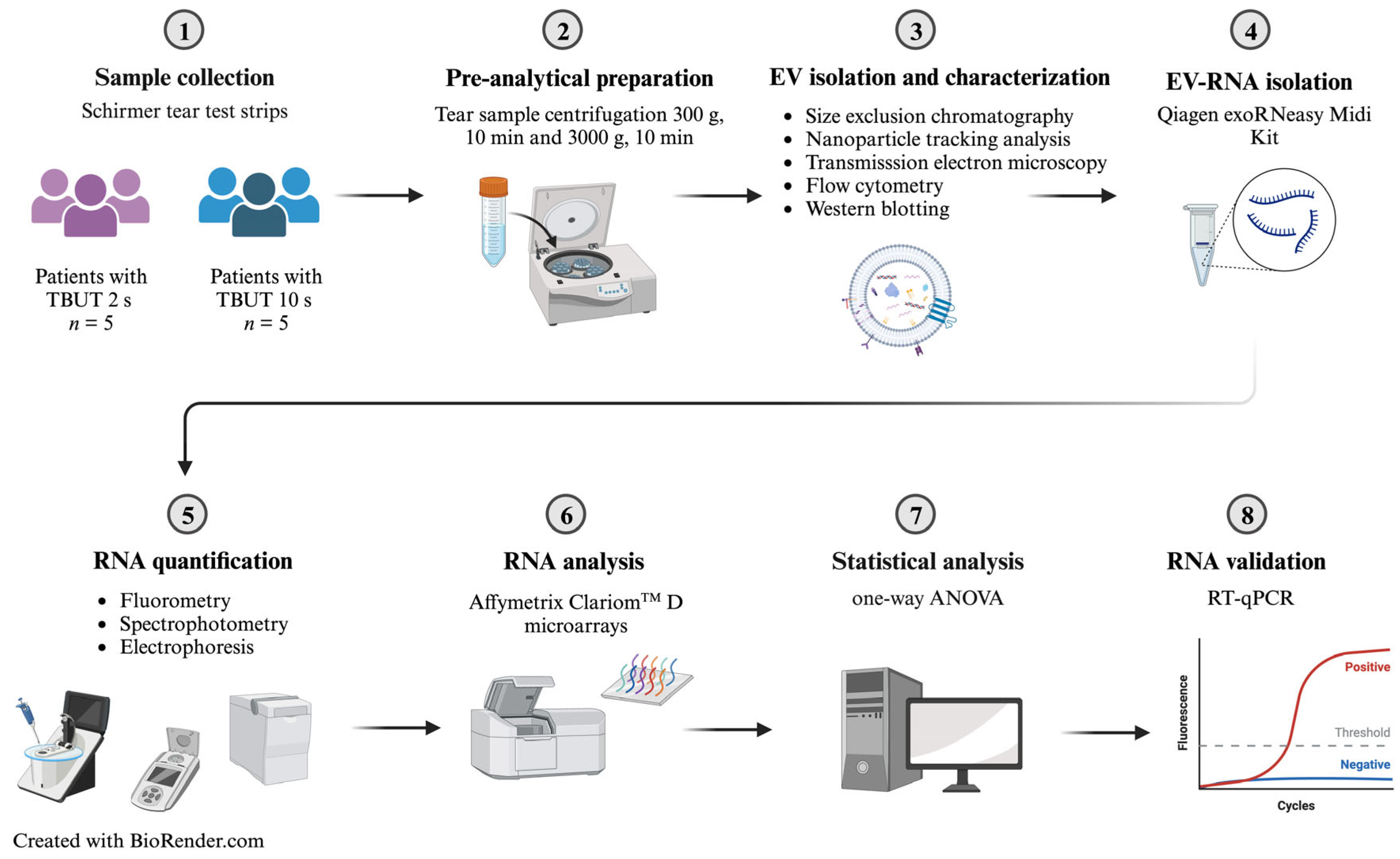
4. Materials and Methods
4.1. Study Participants
4.2. Tear Fluid Sample Collection
4.3. Tear Fluid Sample Preparation: Differential Centrifugation
4.4. EV Isolation and Characterization: Nanoparticle Tracking Analysis (NTA)
4.5. EV Characterization: Transmission Electron Microscopy
4.6. EV Characterization: Flow Cytometry Analysis
4.7. EV Isolation and Characterization: Western Blotting of EV-Proteins Isolated from Tear Fluid
4.8. Tear Fluid EV-RNA Isolation Using a Qiagen exoRNeasy Midi Kit
4.9. Sample RNA Quantification and Purity Assessment Using a Qubit® 2.0 Fluorometer and a Nanodrop™ One Spectrophotometer, Respectively
4.10. Sample RNA Characterization Using an Agilent 2100 Bioanalyzer
4.11. Extracellular Vesicle RNA Analysis Using Affymetrix ClariomTM D Microarrays
4.12. RNA Validation Using RT-qPCR
4.13. Bioinformatic Processing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Craig, J.P.; Nichols, K.K.; Akpek, E.K.; Caffery, B.; Dua, H.S.; Joo, C.K.; Liu, Z.; Nelson, J.D.; Nichols, J.J.; Tsubota, K.; et al. TFOS DEWS II Definition and Classification Report. Ocul. Surf. 2017, 15, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Guo, O.L.; Akpek, E. The negative effects of dry eye disease on quality of life and visual function. Turk. J. Med. Sci. 2020, 50, 1611–1615. [Google Scholar] [CrossRef]
- Stapleton, F.; Alves, M.; Bunya, V.Y.; Jalbert, I.; Lekhanont, K.; Malet, F.; Na, K.S.; Schaumberg, D.; Uchino, M.; Vehof, J.; et al. TFOS DEWS II Epidemiology Report. Ocul. Surf. 2017, 15, 334–365. [Google Scholar] [CrossRef] [PubMed]
- Farrand, K.F.; Fridman, M.; Stillman, I.O.; Schaumberg, D.A. Prevalence of Diagnosed Dry Eye Disease in the United States Among Adults Aged 18 Years and Older. Am. J. Ophthalmol. 2017, 182, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Hindman, H.B. Aging: A predisposition to dry eyes. J. Ophthalmol. 2014, 2014, 781683. [Google Scholar] [CrossRef]
- Ding, J.; Sullivan, D.A. Aging and dry eye disease. Exp. Gerontol. 2012, 47, 483–490. [Google Scholar] [CrossRef]
- Pflugfelder, S.C.; Stern, M.E. Biological functions of tear film. Exp. Eye Res. 2020, 197, 108115. [Google Scholar] [CrossRef]
- Pflugfelder, S.C.; de Paiva, C.S. The Pathophysiology of Dry Eye Disease: What We Know and Future Directions for Research. Ophthalmology 2017, 124, S4–S13. [Google Scholar] [CrossRef]
- Wolffsohn, J.S.; Arita, R.; Chalmers, R.; Djalilian, A.; Dogru, M.; Dumbleton, K.; Gupta, P.K.; Karpecki, P.; Lazreg, S.; Pult, H.; et al. TFOS DEWS II Diagnostic Methodology report. Ocul. Surf. 2017, 15, 539–574. [Google Scholar] [CrossRef]
- Barabino, S.; Aragona, P.; di Zazzo, A.; Rolando, M.; with the Contribution of Selected Ocular Surface Experts from the Societa Italiana di Dacriologia e Superficie Oculare. Updated definition and classification of dry eye disease: Renewed proposals using the nominal group and Delphi techniques. Eur. J. Ophthalmol. 2021, 31, 42–48. [Google Scholar] [CrossRef]
- Yamaguchi, T. Inflammatory Response in Dry Eye. Invest. Ophthalmol. Vis. Sci. 2018, 59, DES192–DES199. [Google Scholar] [CrossRef] [PubMed]
- Raeder, S.; Klyve, P.; Utheim, T.P. Dry eye disease—Diagnosis and treatment. Tidsskr. Nor. Laegeforen. 2019, 139. [Google Scholar] [CrossRef]
- Rhee, M.K.; Mah, F.S. Inflammation in Dry Eye Disease: How Do We Break the Cycle? Ophthalmology 2017, 124, S14–S19. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Bazzan, E.; Tine, M.; Casara, A.; Biondini, D.; Semenzato, U.; Cocconcelli, E.; Balestro, E.; Damin, M.; Radu, C.M.; Turato, G.; et al. Critical Review of the Evolution of Extracellular Vesicles’ Knowledge: From 1946 to Today. Int. J. Mol. Sci. 2021, 22, 6417. [Google Scholar] [CrossRef]
- McKay, T.B.; Yeung, V.; Hutcheon, A.E.K.; Guo, X.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles in the Cornea: Insights from Other Tissues. Anal. Cell Pathol. 2021, 2021, 9983900. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 2021, 23, 631–641. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front. Genet. 2013, 4, 142. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- van Balkom, B.W.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef] [PubMed]
- Lasser, C.; Eldh, M.; Lotvall, J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012, 59, e3037. [Google Scholar]
- Tang, Y.T.; Huang, Y.Y.; Zheng, L.; Qin, S.H.; Xu, X.P.; An, T.X.; Xu, Y.; Wu, Y.S.; Hu, X.M.; Ping, B.H.; et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med. 2017, 40, 834–844. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lasser, C.; Szabo, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzas, E.I.; Lotvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2, 20677. [Google Scholar] [CrossRef]
- Narang, P.; Shah, M.; Beljanski, V. Exosomal RNAs in diagnosis and therapies. Noncoding RNA Res. 2022, 7, 7–15. [Google Scholar] [CrossRef]
- Console, L.; Scalise, M.; Indiveri, C. Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta 2019, 488, 165–171. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Wachter, A. Gene regulation by structured mRNA elements. Trends Genet. 2014, 30, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Veziroglu, E.M.; Mias, G.I. Characterizing Extracellular Vesicles and Their Diverse RNA Contents. Front. Genet. 2020, 11, 700. [Google Scholar] [CrossRef] [PubMed]
- Bron, A.J.; Tomlinson, A.; Foulks, G.N.; Pepose, J.S.; Baudouin, C.; Geerling, G.; Nichols, K.K.; Lemp, M.A. Rethinking dry eye disease: A perspective on clinical implications. Ocul. Surf. 2014, 12, S1–S31. [Google Scholar] [CrossRef]
- Miller, K.L.; Walt, J.G.; Mink, D.R.; Satram-Hoang, S.; Wilson, S.E.; Perry, H.D.; Asbell, P.A.; Pflugfelder, S.C. Minimal clinically important difference for the ocular surface disease index. Arch. Ophthalmol. 2010, 128, 94–101. [Google Scholar] [CrossRef]
- Sullivan, B.D.; Crews, L.A.; Messmer, E.M.; Foulks, G.N.; Nichols, K.K.; Baenninger, P.; Geerling, G.; Figueiredo, F.; Lemp, M.A. Correlations between commonly used objective signs and symptoms for the diagnosis of dry eye disease: Clinical implications. Acta. Ophthalmol. 2014, 92, 161–166. [Google Scholar] [CrossRef]
- Methodologies to diagnose and monitor dry eye disease: Report of the Diagnostic Methodology Subcommittee of the International Dry Eye WorkShop (2007). Ocul. Surf. 2007, 5, 108–152. [CrossRef]
- Iturrate, A.; Rivera-Barahona, A.; Flores, C.L.; Otaify, G.A.; Elhossini, R.; Perez-Sanz, M.L.; Nevado, J.; Tenorio-Castano, J.; Trivino, J.C.; Garcia-Gonzalo, F.R.; et al. Mutations in SCNM1 cause orofaciodigital syndrome due to minor intron splicing defects affecting primary cilia. Am. J. Hum. Genet. 2022, 109, 1828–1849. [Google Scholar] [CrossRef]
- Howell, V.M.; Jones, J.M.; Bergren, S.K.; Li, L.; Billi, A.C.; Avenarius, M.R.; Meisler, M.H. Evidence for a direct role of the disease modifier SCNM1 in splicing. Hum. Mol. Genet. 2007, 16, 2506–2516. [Google Scholar] [CrossRef]
- Bai, R.; Wan, R.; Wang, L.; Xu, K.; Zhang, Q.; Lei, J.; Shi, Y. Structure of the activated human minor spliceosome. Science 2021, 371, eabg0879. [Google Scholar] [CrossRef]
- Pucker, A.D.; Ngo, W.; Postnikoff, C.K.; Fortinberry, H.; Nichols, J.J. Tear Film miRNAs and Their Association With Human Dry Eye Disease. Curr. Eye Res. 2022, 47, 1479–1487. [Google Scholar] [CrossRef]
- Zeng, Z.; Dai, Y.; Deng, S.; Zou, S.; Dou, T.; Wei, F. Synovial mesenchymal stem cell-derived extracellular vesicles alleviate chondrocyte damage during osteoarthritis through microRNA-130b-3p-mediated inhibition of the LRP12/AKT/beta-catenin axis. Immunopharmacol. Immunotoxicol. 2022, 44, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yan, J.; Song, T.; Zhong, C.; Kuang, J.; Mo, Y.; Tan, J.; Li, D.; Sui, Z.; Cai, K.; et al. microRNA-130b-3p Contained in MSC-Derived EVs Promotes Lung Cancer Progression by Regulating the FOXO3/NFE2L2/TXNRD1 Axis. Mol. Ther. Oncolytics 2021, 20, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, H.; Bang, S.; Jee, S.; Jang, K. MicroRNA-130b functions as an oncogene and is a predictive marker of poor prognosis in lung adenocarcinoma. Lab Investig. 2021, 101, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Grigor’eva, A.E.; Tamkovich, S.N.; Eremina, A.V.; Tupikin, A.E.; Kabilov, M.R.; Chernykh, V.V.; Vlassov, V.V.; Laktionov, P.P.; Ryabchikova, E.I. Characteristics of exosomes andmicroparticles discovered in human tears. Biomed. Khim. 2016, 62, 99–106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aqrawi, L.A.; Galtung, H.K.; Vestad, B.; Ovstebo, R.; Thiede, B.; Rusthen, S.; Young, A.; Guerreiro, E.M.; Utheim, T.P.; Chen, X.; et al. Identification of potential saliva and tear biomarkers in primary Sjogren’s syndrome, utilising the extraction of extracellular vesicles and proteomics analysis. Arthritis. Res. Ther. 2017, 19, 14. [Google Scholar] [CrossRef]
- Aqrawi, L.A.; Galtung, H.K.; Guerreiro, E.M.; Ovstebo, R.; Thiede, B.; Utheim, T.P.; Chen, X.; Utheim, O.A.; Palm, O.; Skarstein, K.; et al. Proteomic and histopathological characterisation of sicca subjects and primary Sjogren’s syndrome patients reveals promising tear, saliva and extracellular vesicle disease biomarkers. Arthritis. Res. Ther. 2019, 21, 181. [Google Scholar] [CrossRef]
- Pieragostino, D.; Lanuti, P.; Cicalini, I.; Cufaro, M.C.; Ciccocioppo, F.; Ronci, M.; Simeone, P.; Onofrj, M.; van der Pol, E.; Fontana, A.; et al. Proteomics characterization of extracellular vesicles sorted by flow cytometry reveals a disease-specific molecular cross-talk from cerebrospinal fluid and tears in multiple sclerosis. J. Proteom. 2019, 204, 103403. [Google Scholar] [CrossRef]
- Tamkovich, S.; Grigor’eva, A.; Eremina, A.; Tupikin, A.; Kabilov, M.; Chernykh, V.; Vlassov, V.; Ryabchikova, E. What information can be obtained from the tears of a patient with primary open angle glaucoma? Clin. Chim. Acta 2019, 495, 529–537. [Google Scholar] [CrossRef]
- Inubushi, S.; Kawaguchi, H.; Mizumoto, S.; Kunihisa, T.; Baba, M.; Kitayama, Y.; Takeuchi, T.; Hoffman, R.M.; Tanino, H.; Sasaki, R. Oncogenic miRNAs Identified in Tear Exosomes From Metastatic Breast Cancer Patients. Anticancer Res. 2020, 40, 3091–3096. [Google Scholar] [CrossRef]
- Takeuchi, T.; Mori, K.; Sunayama, H.; Takano, E.; Kitayama, Y.; Shimizu, T.; Hirose, Y.; Inubushi, S.; Sasaki, R.; Tanino, H. Antibody-Conjugated Signaling Nanocavities Fabricated by Dynamic Molding for Detecting Cancers Using Small Extracellular Vesicle Markers from Tears. J. Am. Chem. Soc. 2020, 142, 6617–6624. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, L.; Ma, H.; Ni, F.; Liu, F.; Chen, H. Detection of Tear Components Using Matrix-Assisted Laser Desorption Ionization/Time-of-Flight Mass Spectrometry for Rapid Dry Eye Diagnosis. J. Proteome Res. 2020, 19, 3644–3651. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Kim, S.E.; Jin, J.Q.; Park, N.R.; Lee, J.Y.; Kim, H.L.; Lee, S.B.; Yang, S.W.; Lim, D.J. Tear-Derived Exosome Proteins Are Increased in Patients with Thyroid Eye Disease. Int. J. Mol. Sci. 2021, 22, 1115. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yuan, W.; Pang, Q.; Xue, C.; Yan, X. Single-particle analysis of tear fluid reveals abundant presence of tissue factor-exposing extracellular vesicles with strong coagulation activity. Talanta 2022, 239, 123089. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, T.; Ma, H.; Pan, Y.; Wang, S.; Liu, X.; Dai, X.; Zheng, Y.; Lee, L.P.; Liu, F. Discovering the Secret of Diseases by Incorporated Tear Exosomes Analysis via Rapid-Isolation System: iTEARS. ACS Nano. 2022, 16, 11720–11732. [Google Scholar] [CrossRef]
- Ma, J.Y.W.; Sze, Y.H.; Bian, J.F.; Lam, T.C. Critical role of mass spectrometry proteomics in tear biomarker discovery for multifactorial ocular diseases (Review). Int. J. Mol. Med. 2021, 47, 1–15. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Cross, T.; Haug, K.B.F.; Brusletto, B.S.; Ommundsen, S.K.; Troseid, A.S.; Aspelin, T.; Olstad, O.K.; Aass, H.C.D.; Galtung, H.K.; Utheim, T.P.; et al. Non-Coding RNA in Salivary Extracellular Vesicles: A New Frontier in Sjogren’s Syndrome Diagnostics? Int. J. Mol. Sci. 2023, 24, 13409. [Google Scholar] [CrossRef]
| Group | Age | Sex | TBUT (s) | OSDI | Schirmer (mm) | OSS |
|---|---|---|---|---|---|---|
| 1 | 59 | F | 2 | 33.3 | 9 | 0 |
| 1 | 66 | F | 2 | 47.9 | 20 | 0 |
| 1 | 22 | M | 2 | 37.5 | 6 | 1 |
| 1 | 20 | M | 2 | 39.6 | 14 | 2 |
| 1 | 75 | F | 2 | 45.5 | 35 | 3 |
| 2 | 47 | M | 10 | 8.3 | 5 | 2 |
| 2 | 51 | F | 10 | 20.8 | 30 | 1 |
| 2 | 39 | M | 10 | 22.9 | 13 | 1 |
| 2 | 71 | F | 10 | 0 | 18 | 3 |
| 2 | 30 | M | 10 | 34.1 | 3 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cross, T.; Øvstebø, R.; Brusletto, B.S.; Trøseid, A.-M.S.; Olstad, O.K.; Aspelin, T.; Jackson, C.J.; Chen, X.; Utheim, T.P.; Haug, K.B.F. RNA Profiles of Tear Fluid Extracellular Vesicles in Patients with Dry Eye-Related Symptoms. Int. J. Mol. Sci. 2023, 24, 15390. https://doi.org/10.3390/ijms242015390
Cross T, Øvstebø R, Brusletto BS, Trøseid A-MS, Olstad OK, Aspelin T, Jackson CJ, Chen X, Utheim TP, Haug KBF. RNA Profiles of Tear Fluid Extracellular Vesicles in Patients with Dry Eye-Related Symptoms. International Journal of Molecular Sciences. 2023; 24(20):15390. https://doi.org/10.3390/ijms242015390
Chicago/Turabian StyleCross, Tanya, Reidun Øvstebø, Berit Sletbakk Brusletto, Anne-Marie Siebke Trøseid, Ole Kristoffer Olstad, Trude Aspelin, Catherine Joan Jackson, Xiangjun Chen, Tor Paaske Utheim, and Kari Bente Foss Haug. 2023. "RNA Profiles of Tear Fluid Extracellular Vesicles in Patients with Dry Eye-Related Symptoms" International Journal of Molecular Sciences 24, no. 20: 15390. https://doi.org/10.3390/ijms242015390
APA StyleCross, T., Øvstebø, R., Brusletto, B. S., Trøseid, A.-M. S., Olstad, O. K., Aspelin, T., Jackson, C. J., Chen, X., Utheim, T. P., & Haug, K. B. F. (2023). RNA Profiles of Tear Fluid Extracellular Vesicles in Patients with Dry Eye-Related Symptoms. International Journal of Molecular Sciences, 24(20), 15390. https://doi.org/10.3390/ijms242015390







