Altered Clock Gene Expression in Female APP/PS1 Mice and Aquaporin-Dependent Amyloid Accumulation in the Retina
Abstract
1. Introduction
2. Results
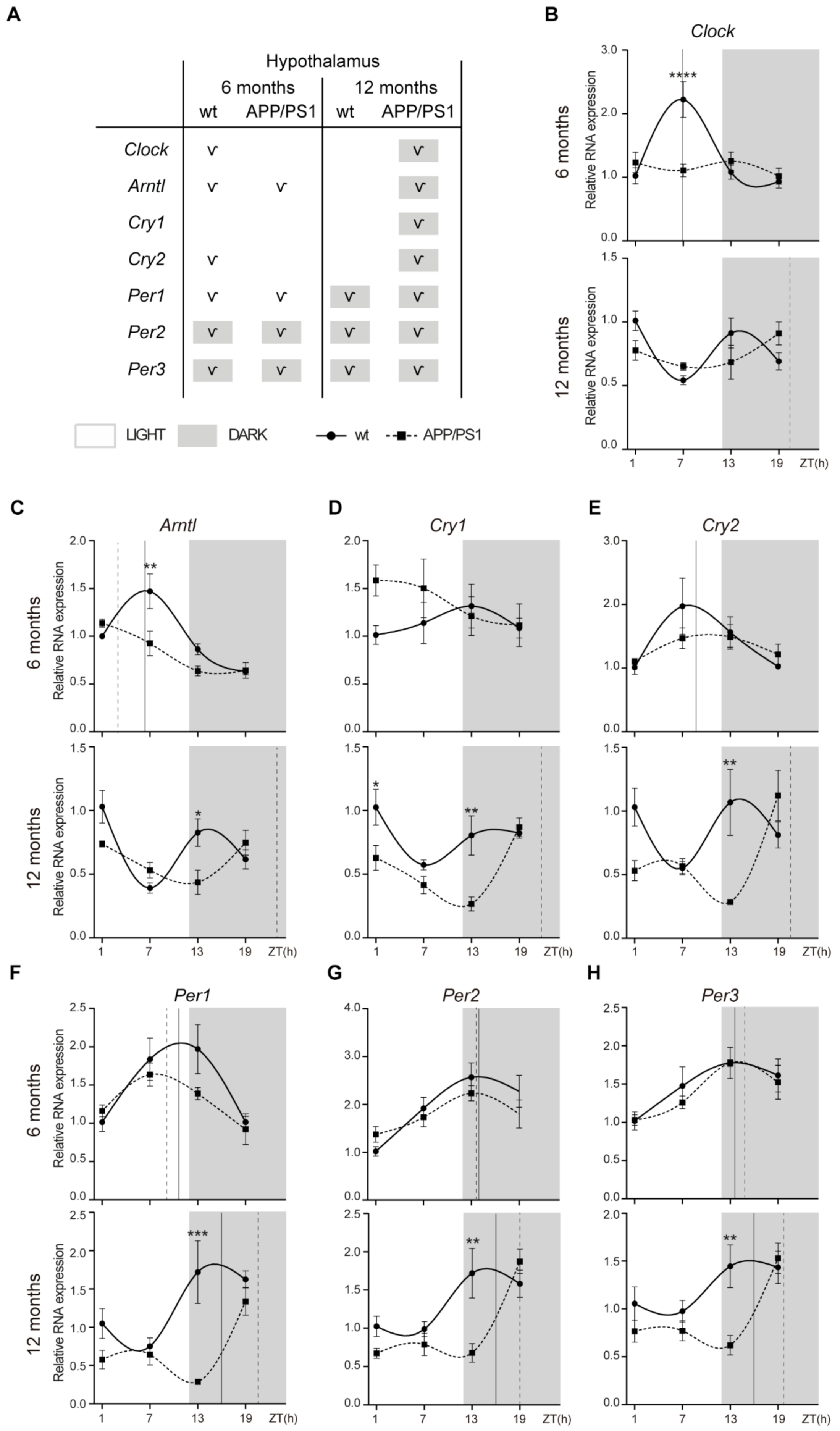
2.1. Alterations to the Molecular Circadian Clock in the Hypothalamus of Female APP/PS1 Mice
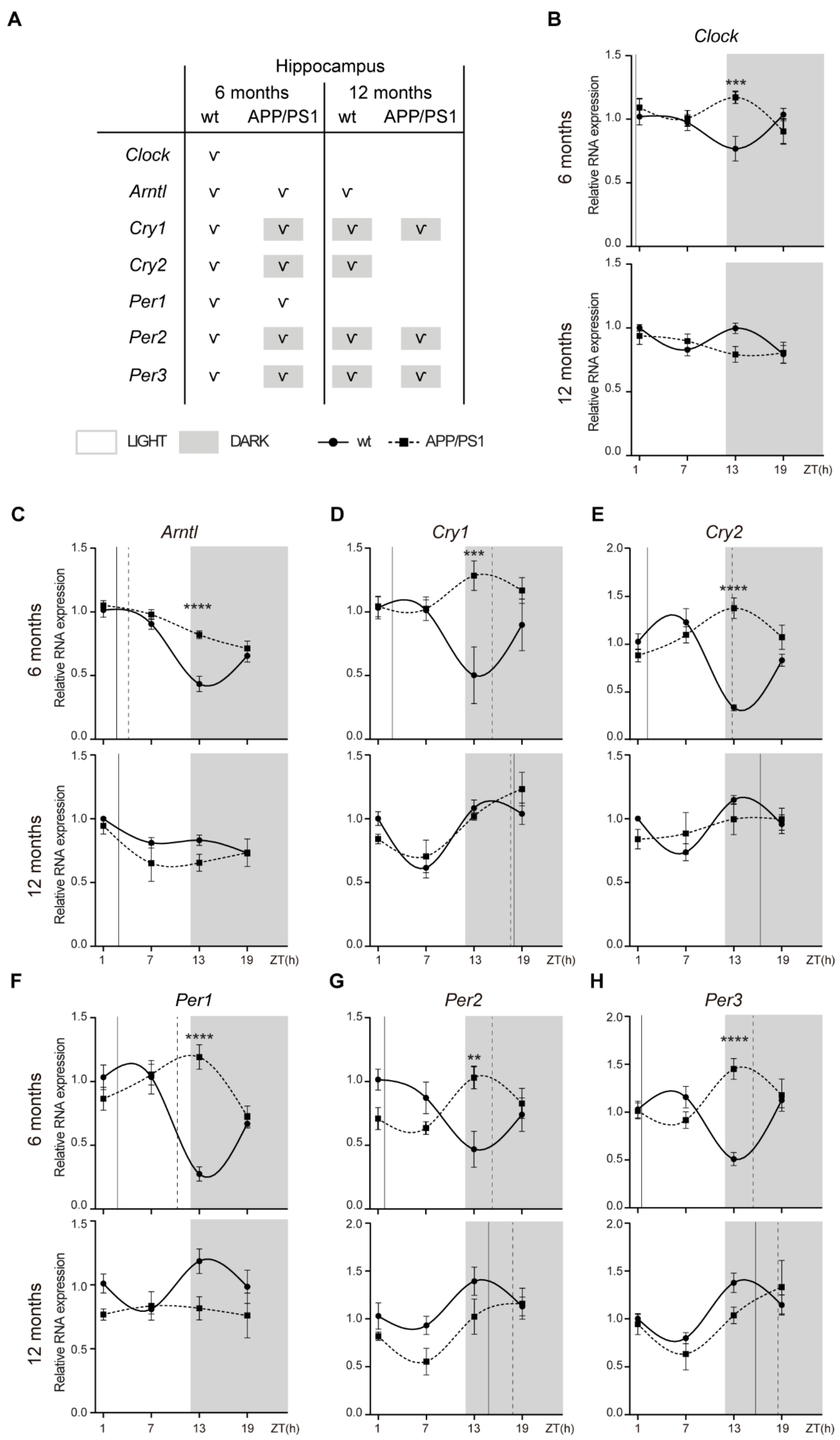
2.2. Alterations to the Molecular Circadian Clock in the Hippocampus of Female APP/PS1 Mice
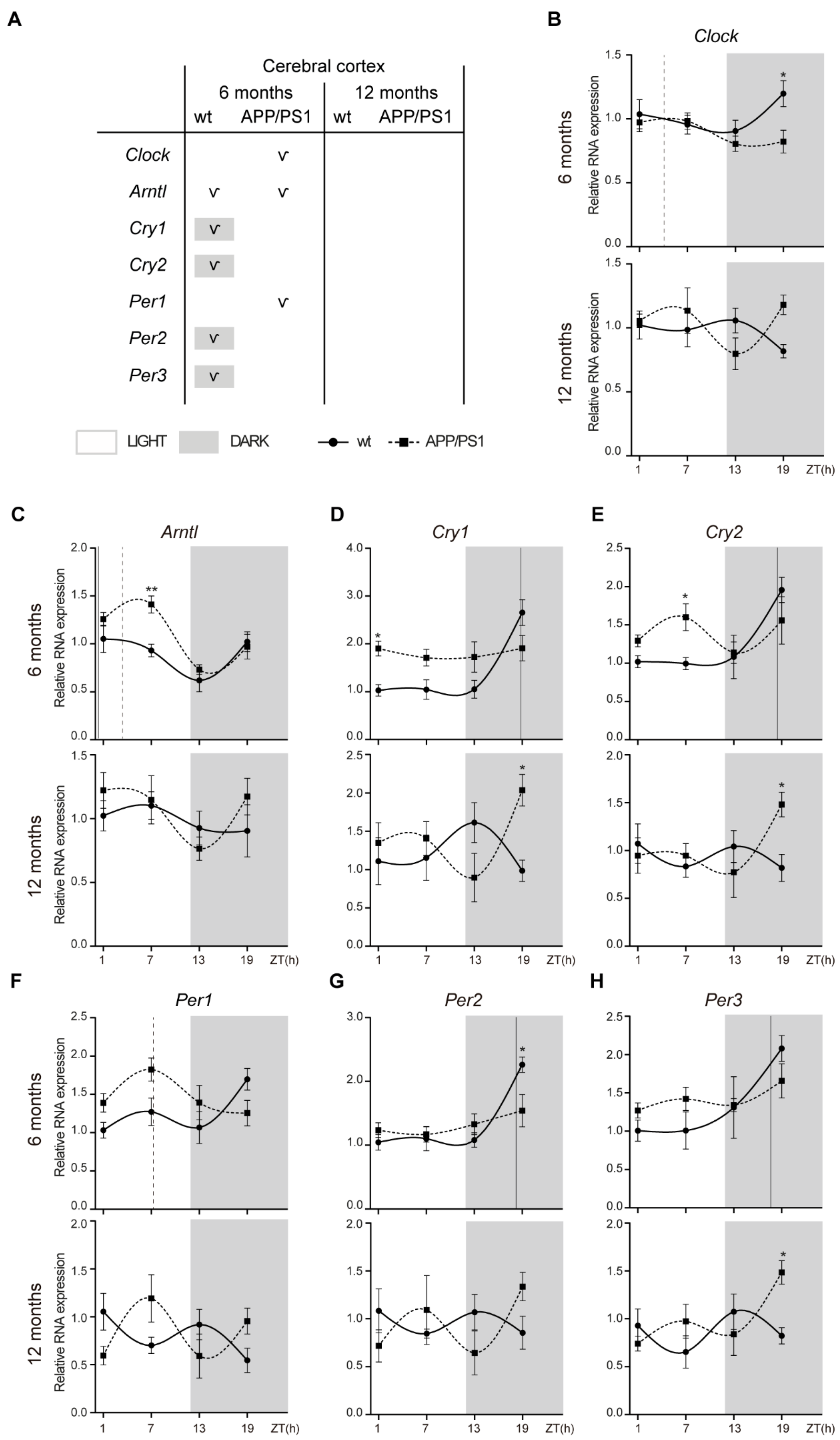
2.3. Alterations to the Molecular Circadian Clock in the Cerebral Cortex of Female APP/PS1 Mice
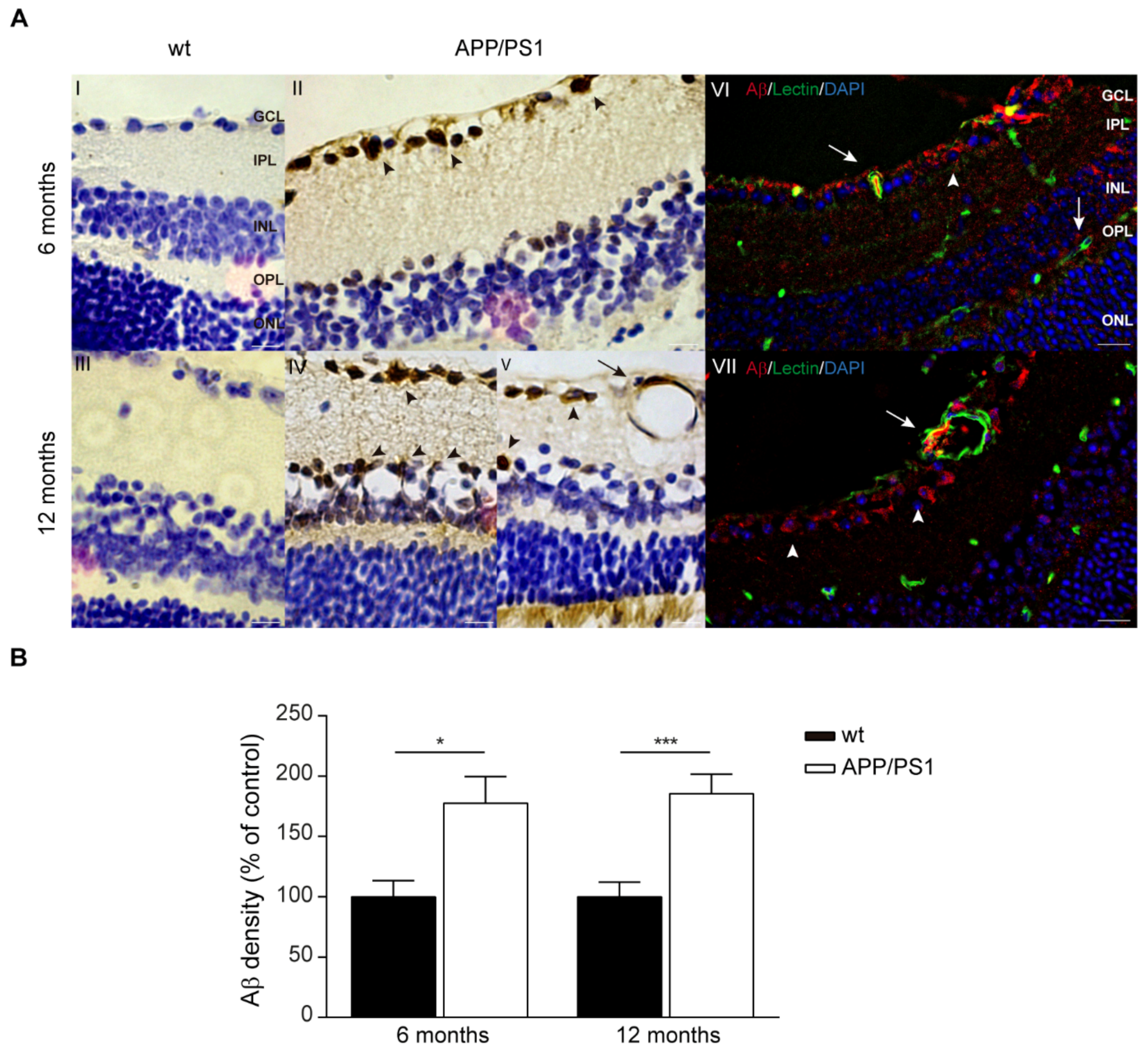
2.4. Aβ Accumulations in the Retinas of Female APP/PS1 Mice
2.5. AQPs Expression in the Retinas of APP/PS1 Mice
3. Discussion
4. Material and Methods
4.1. Animals
4.2. RNA Extraction and Quantification
4.3. Peroxidase-Based Immunostaining of Aβ
4.4. Immunofluorescent Staining of Retinal Cross-Sections
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Barnes, D.E.; Yaffe, K. The projected effect of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol. 2011, 10, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Masters, C.L.; Bateman, R.; Blennow, K.; Rowe, C.C.; Sperling, R.A.; Cummings, J.L. Alzheimer’s disease. Nat. Rev. Dis. Prim. 2015, 1, 15056. [Google Scholar] [CrossRef] [PubMed]
- Scheltens, P.; Blennow, K.; Breteler, M.M.; de Strooper, B.; Frisoni, G.B.; Salloway, S.; Van der Flier, W.M. Alzheimer’s disease. Lancet 2016, 388, 505–517. [Google Scholar] [CrossRef]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. Jama 1997, 278, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Riedel, B.C.; Thompson, P.M.; Brinton, R.D. Age, APOE and sex: Triad of risk of Alzheimer’s disease. J. Steroid Biochem. Mol. Biol. 2016, 160, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H.; Swaab, D.F. Disturbance and strategies for reactivation of the circadian rhythm system in aging and Alzheimer’s disease. Sleep. Med. 2007, 8, 623–636. [Google Scholar] [CrossRef]
- Musiek, E.S.; Bhimasani, M.; Zangrilli, M.A.; Morris, J.C.; Holtzman, D.M.; Ju, Y.S. Circadian Rest-Activity Pattern Changes in Aging and Preclinical Alzheimer Disease. JAMA Neurol. 2018, 75, 582–590. [Google Scholar] [CrossRef]
- Van Erum, J.; Van Dam, D.; De Deyn, P.P. Sleep and Alzheimer’s disease: A pivotal role for the suprachiasmatic nucleus. Sleep Med. Rev. 2018, 40, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.S.; Klein, H.U.; Yu, L.; Chibnik, L.B.; Ali, S.; Xu, J.; Bennett, D.A.; De Jager, P.L. Diurnal and seasonal molecular rhythms in human neocortex and their relation to Alzheimer’s disease. Nat. Commun. 2017, 8, 14931. [Google Scholar] [CrossRef] [PubMed]
- Fusilier, A.R.; Davis, J.A.; Paul, J.R.; Yates, S.D.; McMeekin, L.J.; Goode, L.K.; Mokashi, M.V.; Remiszewski, N.; van Groen, T.; Cowell, R.M.; et al. Dysregulated clock gene expression and abnormal diurnal regulation of hippocampal inhibitory transmission and spatial memory in amyloid precursor protein transgenic mice. Neurobiol. Dis. 2021, 158, 105454. [Google Scholar] [CrossRef] [PubMed]
- Spitschan, M.; Santhi, N.; Ahluwalia, A.; Fischer, D.; Hunt, L.; Karp, N.A.; Lévi, F.; Pineda-Torra, I.; Vidafar, P.; White, R. Sex differences and sex bias in human circadian and sleep physiology research. Elife 2022, 11, e65419. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.S.; Myers, A.J.; Yu, L.; Buchman, A.S.; Duffy, J.F.; De Jager, P.L.; Bennett, D.A. Sex difference in daily rhythms of clock gene expression in the aged human cerebral cortex. J. Biol. Rhythm. 2013, 28, 117–129. [Google Scholar] [CrossRef]
- Bailey, M.; Silver, R. Sex differences in circadian timing systems: Implications for disease. Front. Neuroendocrinol. 2014, 35, 111–139. [Google Scholar] [CrossRef]
- Oyegbami, O.; Collins, H.M.; Pardon, M.C.; Ebling, F.J.P.; Heery, D.M.; Moran, P.M. Abnormal Clock Gene Expression and Locomotor Activity Rhythms in Two Month-Old Female APPSwe/PS1dE9 Mice. Curr. Alzheimer Res. 2017, 14, 850–860. [Google Scholar] [CrossRef]
- Kim, E.; Nohara, K.; Wirianto, M.; Escobedo, G., Jr.; Lim, J.Y.; Morales, R.; Yoo, S.H.; Chen, Z. Effects of the Clock Modulator Nobiletin on Circadian Rhythms and Pathophysiology in Female Mice of an Alzheimer’s Disease Model. Biomolecules 2021, 11, 1004. [Google Scholar] [CrossRef]
- Gallagher, J.J.; Minogue, A.M.; Lynch, M.A. Impaired performance of female APP/PS1 mice in the Morris water maze is coupled with increased Aβ accumulation and microglial activation. Neurodegener. Dis. 2013, 11, 33–41. [Google Scholar] [CrossRef]
- Navakkode, S.; Gaunt, J.R.; Pavon, M.V.; Bansal, V.A.; Abraham, R.P.; Chong, Y.S.; Ch’ng, T.H.; Sajikumar, S. Sex-specific accelerated decay in time/activity-dependent plasticity and associative memory in an animal model of Alzheimer’s disease. Aging Cell 2021, 20, e13502. [Google Scholar] [CrossRef]
- Ordoñez-Gutierrez, L.; Fernandez-Perez, I.; Herrera, J.L.; Anton, M.; Benito-Cuesta, I.; Wandosell, F. AβPP/PS1 Transgenic Mice Show Sex Differences in the Cerebellum Associated with Aging. J. Alzheimers Dis. 2016, 54, 645–656. [Google Scholar] [CrossRef]
- Jiao, S.S.; Bu, X.L.; Liu, Y.H.; Zhu, C.; Wang, Q.H.; Shen, L.L.; Liu, C.H.; Wang, Y.R.; Yao, X.Q.; Wang, Y.J. Sex Dimorphism Profile of Alzheimer’s Disease-Type Pathologies in an APP/PS1 Mouse Model. Neurotox. Res. 2016, 29, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.E.; He, B.; Muhammad, N.; Oh, K.J.; Fahnestock, M.; Ikonomovic, M.D.; Mufson, E.J. Cholinotrophic basal forebrain system alterations in 3xTg-AD transgenic mice. Neurobiol. Dis. 2011, 41, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Creighton, S.D.; Mendell, A.L.; Palmer, D.; Kalisch, B.E.; MacLusky, N.J.; Prado, V.F.; Prado, M.A.M.; Winters, B.D. Dissociable cognitive impairments in two strains of transgenic Alzheimer’s disease mice revealed by a battery of object-based tests. Sci. Rep. 2019, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Stimmell, A.C.; Baglietto-Vargas, D.; Moseley, S.C.; Lapointe, V.; Thompson, L.M.; LaFerla, F.M.; McNaughton, B.L.; Wilber, A.A. Impaired Spatial Reorientation in the 3xTg-AD Mouse Model of Alzheimer’s Disease. Sci. Rep. 2019, 9, 1311. [Google Scholar] [CrossRef] [PubMed]
- Koronyo-Hamaoui, M.; Koronyo, Y.; Ljubimov, A.V.; Miller, C.A.; Ko, M.K.; Black, K.L.; Schwartz, M.; Farkas, D.L. Identification of amyloid plaques in retinas from Alzheimer’s patients and noninvasive in vivo optical imaging of retinal plaques in a mouse model. Neuroimage 2011, 54 (Suppl. 1), S204–S217. [Google Scholar] [CrossRef]
- La Morgia, C.; Ross-Cisneros, F.N.; Koronyo, Y.; Hannibal, J.; Gallassi, R.; Cantalupo, G.; Sambati, L.; Pan, B.X.; Tozer, K.R.; Barboni, P.; et al. Melanopsin retinal ganglion cell loss in Alzheimer disease. Ann. Neurol. 2016, 79, 90–109. [Google Scholar] [CrossRef]
- Koronyo, Y.; Biggs, D.; Barron, E.; Boyer, D.S.; Pearlman, J.A.; Au, W.J.; Kile, S.J.; Blanco, A.; Fuchs, D.T.; Ashfaq, A.; et al. Retinal amyloid pathology and proof-of-concept imaging trial in Alzheimer’s disease. JCI Insight 2017, 2, e93621. [Google Scholar] [CrossRef]
- den Haan, J.; Morrema, T.H.J.; Verbraak, F.D.; de Boer, J.F.; Scheltens, P.; Rozemuller, A.J.; Bergen, A.A.B.; Bouwman, F.H.; Hoozemans, J.J. Amyloid-beta and phosphorylated tau in post-mortem Alzheimer’s disease retinas. Acta Neuropathol. Commun. 2018, 6, 147. [Google Scholar] [CrossRef]
- Shi, H.; Koronyo, Y.; Rentsendorj, A.; Fuchs, D.T.; Sheyn, J.; Black, K.L.; Mirzaei, N.; Koronyo-Hamaoui, M. Retinal Vasculopathy in Alzheimer’s Disease. Front. Neurosci. 2021, 15, 731614. [Google Scholar] [CrossRef]
- Wang, X.; Lou, N.; Eberhardt, A.; Yang, Y.; Kusk, P.; Xu, Q.; Förstera, B.; Peng, S.; Shi, M.; Ladrón-de-Guevara, A.; et al. An ocular glymphatic clearance system removes β-amyloid from the rodent eye. Sci. Transl. Med. 2020, 12, eaaw3210. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Wang, M.X.; Ismail, O.; Braun, M.; Schindler, A.G.; Reemmer, J.; Wang, Z.; Haveliwala, M.A.; O’Boyle, R.P.; Han, W.Y.; et al. Loss of perivascular aquaporin-4 localization impairs glymphatic exchange and promotes amyloid β plaque formation in mice. Alzheimers Res. Ther. 2022, 14, 59. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, K.; Zhu, J. Glymphatic system: An emerging therapeutic approach for neurological disorders. Front. Mol. Neurosci. 2023, 16, 1138769. [Google Scholar] [CrossRef] [PubMed]
- Maroli, N. Aquaporin-4 Mediated Aggregation of Alzheimer’s Amyloid β-Peptide. ACS Chem. Neurosci. 2023, 14, 2683–2698. [Google Scholar] [CrossRef] [PubMed]
- Municio, C.; Carrero, L.; Antequera, D.; Carro, E. Choroid Plexus Aquaporins in CSF Homeostasis and the Glymphatic System: Their Relevance for Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 878. [Google Scholar] [CrossRef] [PubMed]
- Carrero, L.; Antequera, D.; Alcalde, I.; Megías, D.; Figueiro-Silva, J.; Merayo-Lloves, J.; Municio, C.; Carro, E. Disturbed circadian rhythm and retinal degeneration in a mouse model of Alzheimer’s disease. Acta Neuropathol. Commun. 2023, 11, 55. [Google Scholar] [CrossRef]
- Yang, Y.; Shiao, C.; Hemingway, J.F.; Jorstad, N.L.; Shalloway, B.R.; Chang, R.; Keene, C.D. Suppressed retinal degeneration in aged wild type and APPswe/PS1ΔE9 mice by bone marrow transplantation. PLoS ONE 2013, 8, e64246. [Google Scholar] [CrossRef]
- Grimaldi, A.; Brighi, C.; Peruzzi, G.; Ragozzino, D.; Bonanni, V.; Limatola, C.; Ruocco, G.; Di Angelantonio, S. Inflammation, neurodegeneration and protein aggregation in the retina as ocular biomarkers for Alzheimer’s disease in the 3xTg-AD mouse model. Cell Death Dis. 2018, 9, 685. [Google Scholar] [CrossRef]
- Chidlow, G.; Wood, J.P.; Manavis, J.; Finnie, J.; Casson, R.J. Investigations into Retinal Pathology in the Early Stages of a Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2017, 56, 655–675. [Google Scholar] [CrossRef]
- Iandiev, I.; Pannicke, T.; Biedermann, B.; Wiedemann, P.; Reichenbach, A.; Bringmann, A. Ischemia-reperfusion alters the immunolocalization of glial aquaporins in rat retina. Neurosci. Lett. 2006, 408, 108–112. [Google Scholar] [CrossRef]
- Buhr, E.D.; Takahashi, J.S. Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol. 2013, 217, 3–27. [Google Scholar] [CrossRef]
- DeBruyne, J.P.; Weaver, D.R.; Reppert, S.M. Peripheral circadian oscillators require CLOCK. Curr. Biol. 2007, 17, R538–R539. [Google Scholar] [CrossRef][Green Version]
- Byerly, M.S.; Blackshaw, S. Vertebrate retina and hypothalamus development. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 380–389. [Google Scholar] [CrossRef]
- Trost, A.; Lange, S.; Schroedl, F.; Bruckner, D.; Motloch, K.A.; Bogner, B.; Kaser-Eichberger, A.; Strohmaier, C.; Runge, C.; Aigner, L.; et al. Brain and Retinal Pericytes: Origin, Function and Role. Front. Cell Neurosci. 2016, 10, 20. [Google Scholar] [CrossRef]
- Johnson, R.F.; Moore, R.Y.; Morin, L.P. Loss of entrainment and anatomical plasticity after lesions of the hamster retinohypothalamic tract. Brain Res. 1988, 460, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.E.; Lumayag, S.; Kovacs, B.; Mufson, E.J.; Xu, S. Beta-amyloid deposition and functional impairment in the retina of the APPswe/PS1DeltaE9 transgenic mouse model of Alzheimer’s disease. Investig. Ophthalmol. Vis. Sci. 2009, 50, 793–800. [Google Scholar] [CrossRef]
- Ning, A.; Cui, J.; To, E.; Ashe, K.H.; Matsubara, J. Amyloid-beta deposits lead to retinal degeneration in a mouse model of Alzheimer disease. Investig. Ophthalmol. Vis. Sci. 2008, 49, 5136–5143. [Google Scholar] [CrossRef] [PubMed]
- Habiba, U.; Descallar, J.; Kreilaus, F.; Adhikari, U.K.; Kumar, S.; Morley, J.W.; Bui, B.V.; Koronyo-Hamaoui, M.; Tayebi, M. Detection of retinal and blood Aβ oligomers with nanobodies. Alzheimers Dement. 2021, 13, e12193. [Google Scholar] [CrossRef]
- Morin, P.J.; Abraham, C.R.; Amaratunga, A.; Johnson, R.J.; Huber, G.; Sandell, J.H.; Fine, R.E. Amyloid precursor protein is synthesized by retinal ganglion cells, rapidly transported to the optic nerve plasma membrane and nerve terminals, and metabolized. J. Neurochem. 1993, 61, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Koronyo, Y.; Rentsendorj, A.; Regis, G.C.; Sheyn, J.; Fuchs, D.T.; Kramerov, A.A.; Ljubimov, A.V.; Dumitrascu, O.M.; Rodriguez, A.R.; et al. Identification of early pericyte loss and vascular amyloidosis in Alzheimer’s disease retina. Acta Neuropathol. 2020, 139, 813–836. [Google Scholar] [CrossRef] [PubMed]
- Denniston, A.K.; Keane, P.A. Paravascular Pathways in the Eye: Is There an ‘Ocular Glymphatic System’? Investig. Ophthalmol. Vis. Sci. 2015, 56, 3955–3956. [Google Scholar] [CrossRef] [PubMed]
- Wostyn, P.; Van Dam, D.; Audenaert, K.; Killer, H.E.; De Deyn, P.P.; De Groot, V. A new glaucoma hypothesis: A role of glymphatic system dysfunction. Fluids Barriers CNS 2015, 12, 16. [Google Scholar] [CrossRef]
- Iliff, J.J.; Wang, M.; Liao, Y.; Plogg, B.A.; Peng, W.; Gundersen, G.A.; Benveniste, H.; Vates, G.E.; Deane, R.; Goldman, S.A.; et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 2012, 4, 147ra111. [Google Scholar] [CrossRef]
- Nakada, T.; Kwee, I.L.; Igarashi, H.; Suzuki, Y. Aquaporin-4 Functionality and Virchow-Robin Space Water Dynamics: Physiological Model for Neurovascular Coupling and Glymphatic Flow. Int. J. Mol. Sci. 2017, 18, 1798. [Google Scholar] [CrossRef]
- Municio, C.; Carro, E. Aquaporin 5 in Alzheimer’s disease: A link between oral and brain pathology? Neural Regen. Res. 2023, 18, 1491–1492. [Google Scholar] [CrossRef]
- Mestre, H.; Hablitz, L.M.; Xavier, A.L.; Feng, W.; Zou, W.; Pu, T.; Monai, H.; Murlidharan, G.; Castellanos Rivera, R.M.; Simon, M.J.; et al. Aquaporin-4-dependent glymphatic solute transport in the rodent brain. Elife 2018, 7, e40070. [Google Scholar] [CrossRef]
- Mader, S.; Brimberg, L. Aquaporin-4 Water Channel in the Brain and Its Implication for Health and Disease. Cells 2019, 8, 90. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Kanekiyo, T. Blood-Brain Barrier Dysfunction and the Pathogenesis of Alzheimer’s Disease. Int. J. Mol. Sci. 2017, 18, 1965. [Google Scholar] [CrossRef]
- Kim, I.B.; Oh, S.J.; Nielsen, S.; Chun, M.H. Immunocytochemical localization of aquaporin 1 in the rat retina. Neurosci. Lett. 1998, 244, 52–54. [Google Scholar] [CrossRef]
- Iandiev, I.; Pannicke, T.; Reichel, M.B.; Wiedemann, P.; Reichenbach, A.; Bringmann, A. Expression of aquaporin-1 immunoreactivity by photoreceptor cells in the mouse retina. Neurosci. Lett. 2005, 388, 96–99. [Google Scholar] [CrossRef]
- Kang, T.H.; Choi, Y.K.; Kim, I.B.; Oh, S.J.; Chun, M.H. Identification and characterization of an aquaporin 1 immunoreactive amacrine-type cell of the mouse retina. J. Comp. Neurol. 2005, 488, 352–367. [Google Scholar] [CrossRef]
- Qin, Y.; Xu, G.; Fan, J.; Witt, R.E.; Da, C. High-salt loading exacerbates increased retinal content of aquaporins AQP1 and AQP4 in rats with diabetic retinopathy. Exp. Eye Res. 2009, 89, 741–747. [Google Scholar] [CrossRef]
- Iandiev, I.; Pannicke, T.; Reichenbach, A.; Wiedemann, P.; Bringmann, A. Diabetes alters the localization of glial aquaporins in rat retina. Neurosci. Lett. 2007, 421, 132–136. [Google Scholar] [CrossRef]
- Trillo-Contreras, J.L.; Toledo-Aral, J.J.; Echevarría, M.; Villadiego, J. AQP1 and AQP4 Contribution to Cerebrospinal Fluid Homeostasis. Cells 2019, 8, 197. [Google Scholar] [CrossRef]
- Oshio, K.; Watanabe, H.; Song, Y.; Verkman, A.S.; Manley, G.T. Reduced cerebrospinal fluid production and intracranial pressure in mice lacking choroid plexus water channel Aquaporin-1. Faseb J. 2005, 19, 76–78. [Google Scholar] [CrossRef]
- Li, Q.; Aalling, N.N.; Förstera, B.; Ertürk, A.; Nedergaard, M.; Møllgård, K.; Xavier, A.L.R. Aquaporin 1 and the Na+/K+/2Cl− cotransporter 1 are present in the leptomeningeal vasculature of the adult rodent central nervous system. Fluids Barriers CNS 2020, 17, 15. [Google Scholar] [CrossRef]
- de Laurentis, C.; Cristaldi, P.; Arighi, A.; Cavandoli, C.; Trezza, A.; Sganzerla, E.P.; Giussani, C.G.; Di Cristofori, A. Role of aquaporins in hydrocephalus: What do we know and where do we stand? A systematic review. J. Neurol. 2021, 268, 4078–4094. [Google Scholar] [CrossRef]
- Damkier, H.H.; Brown, P.D.; Praetorius, J. Cerebrospinal fluid secretion by the choroid plexus. Physiol. Rev. 2013, 93, 1847–1892. [Google Scholar] [CrossRef]
- Boassa, D.; Stamer, W.D.; Yool, A.J. Ion channel function of aquaporin-1 natively expressed in choroid plexus. J. Neurosci. 2006, 26, 7811–7819. [Google Scholar] [CrossRef]
- González-Marrero, I.; Hernández-Abad, L.G.; González-Gómez, M.; Soto-Viera, M.; Carmona-Calero, E.M.; Castañeyra-Ruiz, L.; Castañeyra-Perdomo, A. Altered Expression of AQP1 and AQP4 in Brain Barriers and Cerebrospinal Fluid May Affect Cerebral Water Balance during Chronic Hypertension. Int. J. Mol. Sci. 2022, 23, 12277. [Google Scholar] [CrossRef]
- Wang, M.; Ma, W.; Zhao, L.; Fariss, R.N.; Wong, W.T. Adaptive Müller cell responses to microglial activation mediate neuroprotection and coordinate inflammation in the retina. J. Neuroinflamm. 2011, 8, 173. [Google Scholar] [CrossRef]
- Eberhardt, C.; Amann, B.; Feuchtinger, A.; Hauck, S.M.; Deeg, C.A. Differential expression of inwardly rectifying K+ channels and aquaporins 4 and 5 in autoimmune uveitis indicates misbalance in Müller glial cell-dependent ion and water homeostasis. Glia 2011, 59, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Hollborn, M.; Vogler, S.; Reichenbach, A.; Wiedemann, P.; Bringmann, A.; Kohen, L. Regulation of the hyperosmotic induction of aquaporin 5 and VEGF in retinal pigment epithelial cells: Involvement of NFAT5. Mol. Vis. 2015, 21, 360–377. [Google Scholar] [PubMed]
- Hollborn, M.; Rehak, M.; Iandiev, I.; Pannicke, T.; Ulbricht, E.; Reichenbach, A.; Wiedemann, P.; Bringmann, A.; Kohen, L. Transcriptional regulation of aquaporins in the ischemic rat retina: Upregulation of aquaporin-9. Curr. Eye Res. 2012, 37, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Bi, L.T.; Hou, S.P.; Zhao, X.L.; Song, Y.L.; Ma, T.H. Reduced lung water transport rate associated with downregulation of aquaporin-1 and aquaporin-5 in aged mice. Clin. Exp. Pharmacol. Physiol. 2009, 36, 734–738. [Google Scholar] [CrossRef]
- Kim, S.J.; Baek, K.W.; Jung, Y.K.; Kim, J.S.; Kim, B.G.; Yu, H.S.; Park, J.S.; Yoo, J.I. Changes in aquaporins expression due to acute water restriction in naturally aging mice. J. Physiol. Biochem. 2023, 79, 71–81. [Google Scholar] [CrossRef]
- Dinet, V.; Bruban, J.; Chalour, N.; Maoui, A.; An, N.; Jonet, L.; Buret, A.; Behar-Cohen, F.; Klein, C.; Tréton, J.; et al. Distinct effects of inflammation on gliosis, osmohomeostasis, and vascular integrity during amyloid beta-induced retinal degeneration. Aging Cell 2012, 11, 683–693. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, J.Y.; Jhee, K.H.; Yang, S.A. Amyloid-ß peptides inhibit the expression of AQP4 and glutamate transporter EAAC1 in insulin-treated C6 glioma cells. Toxicol. Rep. 2020, 7, 1083–1089. [Google Scholar] [CrossRef]
- Yao, D.; Li, R.; Hao, J.; Huang, H.; Wang, X.; Ran, L.; Fang, Y.; He, Y.; Wang, W.; Liu, X.; et al. Melatonin alleviates depression-like behaviors and cognitive dysfunction in mice by regulating the circadian rhythm of AQP4 polarization. Transl. Psychiatry 2023, 13, 310. [Google Scholar] [CrossRef]
- Murakami, A.; Tsuji, K.; Isoda, M.; Matsuo, M.; Abe, Y.; Yasui, M.; Okamura, H.; Tominaga, K. Prolonged Light Exposure Induces Circadian Impairment in Aquaporin-4-Knockout Mice. J. Biol. Rhythm. 2023, 38, 208–214. [Google Scholar] [CrossRef]
- Satou, R.; Sato, M.; Kimura, M.; Ishizuka, Y.; Tazaki, M.; Sugihara, N.; Shibukawa, Y. Temporal Expression Patterns of Clock Genes and Aquaporin 5/Anoctamin 1 in Rat Submandibular Gland Cells. Front. Physiol. 2017, 8, 320. [Google Scholar] [CrossRef]
- Uchida, H.; Nakamura, T.J.; Takasu, N.N.; Obana-Koshino, A.; Ono, H.; Todo, T.; Sakai, T.; Nakamura, W. The central clock controls the daily rhythm of Aqp5 expression in salivary glands. J. Physiol. Sci. 2018, 68, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Parsons, R.; Parsons, R.; Garner, N.; Oster, H.; Rawashdeh, O. CircaCompare: A method to estimate and statistically support differences in mesor, amplitude and phase, between circadian rhythms. Bioinformatics 2020, 36, 1208–1212. [Google Scholar] [CrossRef] [PubMed]


| Gene | Age | Genotype | Period (h) | Phase (h) | Amplitude | p-Value |
|---|---|---|---|---|---|---|
| Clock | 6 months | WT | 24.00 | 6.88 | 0.68 | 5.33 × 10−4 |
| APP/PS1 | 24.00 | 7.69 | 0.05 | 0.57 | ||
| 12 months | WT | 24.00 | 22.64 | 0.08 | 0.38 | |
| APP/PS1 | 24.00 | 20.38 | 0.14 | 0.04 | ||
| Arntl | 6 months | WT | 24.00 | 6.33 | 0.43 | 5.87 x 10−5 |
| APP/PS1 | 24.00 | 3.05 | 0.28 | 3.94 × 10−4 | ||
| 12 months | WT | 24.00 | 23.14 | 0.17 | 0.15 | |
| APP/PS1 | 24.00 | 22.66 | 0.19 | 3.90 × 10−4 | ||
| Cry1 | 6 months | WT | 24.00 | 12.34 | 0.16 | 0.23 |
| APP/PS1 | 24.00 | 4.07 | 0.27 | 0.13 | ||
| 12 months | WT | 24.00 | 21.84 | 0.16 | 0.09 | |
| APP/PS1 | 24.00 | 21.72 | 0.30 | 2.36 × 10−4 | ||
| Cry2 | 6 months | WT | 24.00 | 8.81 | 0.54 | 0.02 |
| APP/PS1 | 24.00 | 10.76 | 0.23 | 0.06 | ||
| 12 months | WT | 24.00 | 18.31 | 0.10 | 0.48 | |
| APP/PS1 | 24.00 | 20.59 | 0.30 | 0.02 | ||
| Per1 | 6 months | WT | 24.00 | 10.34 | 0.63 | 3.78 × 10−3 |
| APP/PS1 | 24.00 | 8.52 | 0.29 | 0.03 | ||
| 12 months | WT | 24.00 | 16.50 | 0.55 | 0.02 | |
| APP/PS1 | 24.00 | 20.41 | 0.40 | 0.01 | ||
| Per2 | 6 months | WT | 24.00 | 13.88 | 0.78 | 8.88 × 10−4 |
| APP/PS1 | 24.00 | 13.30 | 0.43 | 0.02 | ||
| 12 months | WT | 24.00 | 15.69 | 0.46 | 0.01 | |
| APP/PS1 | 24.00 | 18.98 | 0.54 | 1.75 × 10−4 | ||
| Per3 | 6 months | WT | 24.00 | 13.63 | 0.37 | 0.02 |
| APP/PS1 | 24.00 | 14.28 | 0.40 | 1.46 × 10−4 | ||
| 12 months | WT | 24.00 | 16.30 | 0.30 | 0.04 | |
| APP/PS1 | 24.00 | 19.73 | 0.39 | 4.11 × 10−4 |
| Gene | Age | Genotype | Period (h) | Phase (h) | Amplitude | p-Value |
|---|---|---|---|---|---|---|
| Clock | 6 months | WT | 24.00 | 0.35 | 0.12 | 0.02 |
| APP/PS1 | 24.00 | 9.69 | 0.04 | 0.42 | ||
| 12 months | WT | 24.00 | 8.74 | 0.02 | 0.68 | |
| APP/PS1 | 24.00 | 3.14 | 0.09 | 0.08 | ||
| Arntl | 6 months | WT | 24.00 | 2.58 | 0.32 | 2.35 × 1010 |
| APP/PS1 | 24.00 | 4.04 | 0.17 | 2.95 × 106 | ||
| 12 months | WT | 24.00 | 2.86 | 0.09 | 0.03 | |
| APP/PS1 | 24.00 | 23.99 | 0.16 | 0.05 | ||
| Cry1 | 6 months | WT | 24.00 | 2.08 | 0.26 | 0.02 |
| APP/PS1 | 24.00 | 15.26 | 0.14 | 0.047 | ||
| 12 months | WT | 24.00 | 18.06 | 0.22 | 2.64 × 103 | |
| APP/PS1 | 24.00 | 17.69 | 0.28 | 9.77 × 104 | ||
| Cry2 | 6 months | WT | 24.00 | 3.05 | 0.39 | 2.06 × 105 |
| APP/PS1 | 24.00 | 12.88 | 0.24 | 7.11 × 104 | ||
| 12 months | WT | 24.00 | 16.46 | 0.14 | 0.02 | |
| APP/PS1 | 24.00 | 15.27 | 0.10 | 0.24 | ||
| Per1 | 6 months | WT | 24.00 | 2.79 | 0.42 | 2.20 × 106 |
| APP/PS1 | 24.00 | 10.17 | 0.21 | 2.56 × 103 | ||
| 12 months | WT | 24.00 | 15.76 | 0.13 | 0.09 | |
| APP/PS1 | 24.00 | 9.25 | 0.04 | 0.56 | ||
| Per2 | 6 months | WT | 24.00 | 1.83 | 0.29 | 5.33 × 103 |
| APP/PS1 | 24.00 | 15.32 | 0.20 | 2.58 × 103 | ||
| 12 months | WT | 24.00 | 14.81 | 0.22 | 0.03 | |
| APP/PS1 | 24.00 | 17.82 | 0.33 | 4.28 × 103 | ||
| Per3 | 6 months | WT | 24.00 | 1.40 | 0.26 | 1.76 × 103 |
| APP/PS1 | 24.00 | 15.38 | 0.26 | 2.16 × 103 | ||
| 12 months | WT | 24.00 | 15.71 | 0.26 | 1.72 × 103 | |
| APP/PS1 | 24.00 | 18.51 | 0.35 | 0.01 |
| Gene | Age | Genotype | Period (h) | Phase (h) | Amplitude | p-Value |
|---|---|---|---|---|---|---|
| Clock | 6 months | WT | 24.00 | 20.70 | 0.13 | 0.07 |
| APP/PS1 | 24.00 | 3.99 | 0.12 | 0.03 | ||
| 12 months | WT | 24.00 | 7.64 | 0.10 | 0.18 | |
| APP/PS1 | 24.00 | 0.23 | 0.12 | 0.23 | ||
| Arntl | 6 months | WT | 24.00 | 0.26 | 0.21 | 0.02 |
| APP/PS1 | 24.00 | 3.37 | 0.31 | 3.07 × 104 | ||
| 12 months | WT | 24.00 | 5.17 | 0.11 | 0.37 | |
| APP/PS1 | 24.00 | 0.79 | 0.22 | 0.06 | ||
| Cry1 | 6 months | WT | 24.00 | 18.93 | 0.74 | 7.70 × 104 |
| APP/PS1 | 24.00 | 21.78 | 0.13 | 0.37 | ||
| 12 months | WT | 24.00 | 11.52 | 0.29 | 0.13 | |
| APP/PS1 | 24.00 | 21.09 | 0.37 | 0.08 | ||
| Cry2 | 6 months | WT | 24.00 | 18.43 | 0.45 | 1.46 × 103 |
| APP/PS1 | 24.00 | 4.67 | 0.07 | 0.61 | ||
| 12 months | WT | 24.00 | 5.65 | 0.04 | 0.75 | |
| APP/PS1 | 24.00 | 19.89 | 0.29 | 0.05 | ||
| Per1 | 6 months | WT | 24.00 | 18.30 | 0.18 | 0.22 |
| APP/PS1 | 24.00 | 7.22 | 0.30 | 0.01 | ||
| 12 months | WT | 24.00 | 5.52 | 0.14 | 0.31 | |
| APP/PS1 | 24.00 | 7.81 | 0.12 | 0.42 | ||
| Per2 | 6 months | WT | 24.00 | 18.23 | 0.54 | 1.01 × 103 |
| APP/PS1 | 24.00 | 17.89 | 0.18 | 0.10 | ||
| 12 months | WT | 24.00 | 5.98 | 0.03 | 0.85 | |
| APP/PS1 | 24.00 | 19.16 | 0.12 | 0.52 | ||
| Per3 | 6 months | WT | 24.00 | 17.67 | 0.53 | 9.38 × 106 |
| APP/PS1 | 24.00 | 14.60 | 0.09 | 0.45 | ||
| 12 months | WT | 24.00 | 15.17 | 0.09 | 0.47 | |
| APP/PS1 | 24.00 | 17.82 | 0.24 | 0.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrero, L.; Antequera, D.; Alcalde, I.; Megias, D.; Ordoñez-Gutierrez, L.; Gutierrez, C.; Merayo-Lloves, J.; Wandosell, F.; Municio, C.; Carro, E. Altered Clock Gene Expression in Female APP/PS1 Mice and Aquaporin-Dependent Amyloid Accumulation in the Retina. Int. J. Mol. Sci. 2023, 24, 15679. https://doi.org/10.3390/ijms242115679
Carrero L, Antequera D, Alcalde I, Megias D, Ordoñez-Gutierrez L, Gutierrez C, Merayo-Lloves J, Wandosell F, Municio C, Carro E. Altered Clock Gene Expression in Female APP/PS1 Mice and Aquaporin-Dependent Amyloid Accumulation in the Retina. International Journal of Molecular Sciences. 2023; 24(21):15679. https://doi.org/10.3390/ijms242115679
Chicago/Turabian StyleCarrero, Laura, Desireé Antequera, Ignacio Alcalde, Diego Megias, Lara Ordoñez-Gutierrez, Cristina Gutierrez, Jesús Merayo-Lloves, Francisco Wandosell, Cristina Municio, and Eva Carro. 2023. "Altered Clock Gene Expression in Female APP/PS1 Mice and Aquaporin-Dependent Amyloid Accumulation in the Retina" International Journal of Molecular Sciences 24, no. 21: 15679. https://doi.org/10.3390/ijms242115679
APA StyleCarrero, L., Antequera, D., Alcalde, I., Megias, D., Ordoñez-Gutierrez, L., Gutierrez, C., Merayo-Lloves, J., Wandosell, F., Municio, C., & Carro, E. (2023). Altered Clock Gene Expression in Female APP/PS1 Mice and Aquaporin-Dependent Amyloid Accumulation in the Retina. International Journal of Molecular Sciences, 24(21), 15679. https://doi.org/10.3390/ijms242115679







