Telomere Checkpoint in Development and Aging
Abstract
:1. Introduction
2. A Telomere Is a Dynamic DNA-RNA-Protein Complex
3. The Signaling Role of Telomeric RNA
4. Telomeres in Aging: Shortening or Dysfunction?
5. Nuclear Topology and Epigenetics of Telomeres
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Olovnikov, A.M. Principle of marginotomy in template synthesis of polynucleotides. Dokl Akad Nauk. SSSR 1971, 201, 1496–1499. [Google Scholar] [PubMed]
- Olovnikov, A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43 Pt 1, 405–413. [Google Scholar] [CrossRef]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of life-span by introduction of telomerase into normal human cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef]
- de Jesus, B.B.; Vera, E.; Schneeberger, K.; Tejera, A.M.; Ayuso, E.; Bosch, F.; Blasco, M.A. Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol. Med. 2012, 4, 691–704. [Google Scholar] [CrossRef]
- Lim, C.J.; Cech, T.R. Shaping human telomeres: From shelterin and CST complexes to telomeric chromatin organization. Nat. Rev. Mol. Cell Biol. 2021, 22, 283–298. [Google Scholar] [CrossRef]
- Shepelev, N.; Dontsova, O.; Rubtsova, M. Post-Transcriptional and Post-Translational Modifications in Telomerase Biogenesis and Recruitment to Telomeres. Int. J. Mol. Sci. 2023, 24, 5027. [Google Scholar] [CrossRef]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef]
- Garavis, M.; Gonzalez, C.; Villasante, A. On the origin of the eukaryotic chromosome: The role of noncanonical DNA structures in telomere evolution. Genome Biol. Evol. 2013, 5, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E.A.; Arkhipova, I.R. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc. Natl. Acad Sci. USA 2007, 104, 9352–9357. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, M.; Olovnikov, I.; Kalmykova, A. Transposon control mechanisms in telomere biology. Curr. Opin Genet. Dev. 2018, 49, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Pardue, M.L.; DeBaryshe, P.G. Drosophila telomeres: A variation on the telomerase theme. Fly 2008, 2, 101–110. [Google Scholar] [CrossRef]
- Casacuberta, E. Drosophila: Retrotransposons Making up Telomeres. Viruses 2017, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, H.; Osanai, M.; Matsumoto, T.; Kojima, K.K. Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm, Bombyx mori. Chromosome Res. 2005, 13, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Lukhtanov, V.A.; Pazhenkova, E.A. Diversity and evolution of telomeric motifs and telomere DNA organization in insects. Biol. J. Linn. Soc. 2023, blad068. [Google Scholar] [CrossRef]
- Morrish, T.A.; Garcia-Perez, J.L.; Stamato, T.D.; Taccioli, G.E.; Sekiguchi, J.; Moran, J.V. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 2007, 446, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Kopera, H.C.; Moldovan, J.B.; Morrish, T.A.; Garcia-Perez, J.L.; Moran, J.V. Similarities between long interspersed element-1 (LINE-1) reverse transcriptase and telomerase. Proc. Natl. Acad Sci. USA 2011, 108, 20345–20350. [Google Scholar] [CrossRef]
- Biessmann, H.; Donath, J.; Walter, M.F. Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol. Biol. 1996, 5, 11–20. [Google Scholar] [CrossRef]
- Compton, A.; Liang, J.; Chen, C.; Lukyanchikova, V.; Qi, Y.; Potters, M.; Settlage, R.; Miller, D.; Deschamps, S.; Mao, C.; et al. The Beginning of the End: A Chromosomal Assembly of the New World Malaria Mosquito Ends with a Novel Telomere. G3 (Bethesda) 2020, 10, 3811–3819. [Google Scholar] [CrossRef] [PubMed]
- Morrish, T.A.; Bekbolysnov, D.; Velliquette, D.; Morgan, M.; Ross, B.; Wang, Y.; Maine, I.P. Multiple Mechanisms Contribute To Telomere Maintenance. J. Cancer Biol. Res. 2013, 1, 3. [Google Scholar]
- Shay, J.W.; Reddel, R.R.; Wright, W.E. Cancer. Cancer and telomeres—An ALTernative to telomerase. Science 2012, 336, 1388–1390. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef]
- Cacchione, S.; Cenci, G.; Raffa, G.D. Silence at the End: How Drosophila Regulates Expression and Transposition of Telomeric Retroelements. J. Mol. Biol. 2020, 432, 4305–4321. [Google Scholar] [CrossRef]
- Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007, 8, 299–309. [Google Scholar] [CrossRef]
- D’Adda di Fagagna, F.; Teo, S.H.; Jackson, S.P. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004, 18, 1781–1799. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Bi, X.; Chen, J.; Srikanta, D.; Rong, Y.S. Mre11-Rad50-Nbs complex is required to cap telomeres during Drosophila embryogenesis. Proc. Natl. Acad Sci. USA 2009, 106, 10728–10733. [Google Scholar] [CrossRef]
- Oikemus, S.R.; McGinnis, N.; Queiroz-Machado, J.; Tukachinsky, H.; Takada, S.; Sunkel, C.E.; Brodsky, M.H. Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes Dev. 2004, 18, 1850–1861. [Google Scholar] [CrossRef]
- Bi, X.; Srikanta, D.; Fanti, L.; Pimpinelli, S.; Badugu, R.; Kellum, R.; Rong, Y.S. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc. Natl. Acad Sci. USA 2005, 102, 15167–15172. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Lingner, J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015, 25, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Rippe, K.; Luke, B. TERRA and the state of the telomere. Nat. Struct Mol. Biol. 2015, 22, 853–858. [Google Scholar] [CrossRef]
- Fernandes, R.V.; Feretzaki, M.; Lingner, J. The makings of TERRA R-loops at chromosome ends. Cell Cycle 2021, 20, 1745–1759. [Google Scholar] [CrossRef] [PubMed]
- Scheibe, M.; Arnoult, N.; Kappei, D.; Buchholz, F.; Decottignies, A.; Butter, F.; Mann, M. Quantitative interaction screen of telomeric repeat-containing RNA reveals novel TERRA regulators. Genome Res. 2013, 23, 2149–2157. [Google Scholar] [CrossRef]
- Ivanyi-Nagy, R.; Ahmed, S.M.; Peter, S.; Ramani, P.D.; Ong, P.F.; Dreesen, O.; Dröge, P. The RNA interactome of human telomerase RNA reveals a coding-independent role for a histone mRNA in telomere homeostasis. eLife 2018, 7, e40037. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.G.; Nager, A.C.; Lunardi, T.; Vancevska, A.; Lossaint, G.; Lingner, J. The human telomeric proteome during telomere replication. Nucleic Acids Res. 2021, 49, 12119–12135. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Liu, D.; Songyang, Z. The telosome/shelterin complex and its functions. Genome Biol. 2008, 9, 232. [Google Scholar] [CrossRef]
- Blasco, M.A. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007, 3, 640–649. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Ye, Q.; Apsley, A.T.; Etzel, L.; Hastings, W.J.; Kozlosky, J.T.; Walker, C.; Wolf, S.E.; Shalev, I. Telomere length and chronological age across the human lifespan: A systematic review and meta-analysis of 414 study samples including 743,019 individuals. Ageing Res. Rev. 2023, 90, 102031. [Google Scholar] [CrossRef]
- Remot, F.; Ronget, V.; Froy, H.; Rey, B.; Gaillard, J.M.; Nussey, D.H.; Lemaitre, J.F. Decline in telomere length with increasing age across nonhuman vertebrates: A meta-analysis. Mol. Ecol. 2022, 31, 5917–5932. [Google Scholar] [CrossRef]
- Rossiello, F.; Jurk, D.; Passos, J.F.; d’Adda di Fagagna, F. Telomere dysfunction in ageing and age-related diseases. Nat. Cell Biol. 2022, 24, 135–147. [Google Scholar] [CrossRef]
- Aguado, J.; d’Adda di Fagagna, F.; Wolvetang, E. Telomere transcription in ageing. Ageing Res Rev. 2020, 62, 101115. [Google Scholar] [CrossRef]
- Herbig, U.; Ferreira, M.; Condel, L.; Carey, D.; Sedivy, J.M. Cellular senescence in aging primates. Science 2006, 311, 1257. [Google Scholar] [CrossRef] [PubMed]
- Martinez, P.; Blasco, M.A. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer 2011, 11, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Segal-Bendirdjian, E.; Geli, V. Non-canonical Roles of Telomerase: Unraveling the Imbroglio. Front Cell Dev Biol. 2019, 7, 332. [Google Scholar] [CrossRef]
- Ye, J.; Renault, V.M.; Jamet, K.; Gilson, E. Transcriptional outcome of telomere signalling. Nat. Rev. Genet. 2014, 15, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Platt, J.M.; Ryvkin, P.; Wanat, J.J.; Donahue, G.; Ricketts, M.D.; Barrett, S.P.; Waters, H.J.; Song, S.; Chavez, A.; Abdallah, K.O.; et al. Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence. Genes. Dev. 2013, 27, 1406–1420. [Google Scholar] [CrossRef] [PubMed]
- Martínez, P.; Gómez-López, G.; García, F.; Mercken, E.; Mitchell, S.; Flores, J.M.; de Cabo, R.; Blasco, M.A. RAP1 protects from obesity through its extratelomeric role regulating gene expression. Cell Rep. 2013, 3, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Teo, H.; Ghosh, S.; Luesch, H.; Ghosh, A.; Wong, E.T.; Malik, N.; Tergaonkar, V. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat. Cell Biol. 2010, 12, 758–767. [Google Scholar] [CrossRef]
- Stock, A.J.; McDevitt, R.A.; Puligilla, C.; Wang, Y.; Zhang, Y.; Wang, K.; Sun, C.; Becker, K.G.; Lehrmann, E.; Wood, W.H.; et al. Aberrant expression and localization of the RAP1 shelterin protein contribute to age-related phenotypes. PLoS Genet. 2022, 18, e1010506. [Google Scholar] [CrossRef]
- Park, J.-I.; Venteicher, A.S.; Hong, J.Y.; Choi, J.; Jun, S.; Shkreli, M.; Chang, W.; Meng, Z.; Cheung, P.; Ji, H.; et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009, 460, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Southworth, L.K.; Sarin, K.Y.; Venteicher, A.S.; Ma, W.; Chang, W.; Artandi, S.E. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008, 4, e10. [Google Scholar] [CrossRef]
- Baruch-Eliyahu, N.; Rud, V.; Braiman, A.; Priel, E. Telomerase increasing compound protects hippocampal neurons from amyloid beta toxicity by enhancing the expression of neurotrophins and plasticity related genes. Sci. Rep. 2019, 9, 18118. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 2009, 35, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Porro, A.; Feuerhahn, S.; Reichenbach, P.; Lingner, J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell Biol. 2010, 30, 4808–4817. [Google Scholar] [CrossRef]
- Arora, R.; Azzalin, C.M. Telomere elongation chooses TERRA ALTernatives. RNA Biol. 2015, 12, 938–941. [Google Scholar] [CrossRef]
- Lopez de Silanes, I.; Stagno d’Alcontres, M.; Blasco, M.A. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 2010, 1, 33. [Google Scholar] [CrossRef]
- Petti, E.; Buemi, V.; Zappone, A.; Schillaci, O.; Broccia, P.V.; Dinami, R.; Matteoni, S.; Benetti, R.; Schoeftner, S. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat. Commun. 2019, 10, 1001. [Google Scholar] [CrossRef]
- Chu, H.-P.; Cifuentes-Rojas, C.; Kesner, B.; Aeby, E.; Lee, H.-G.; Wei, C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 2017, 170, 86–101.e16. [Google Scholar] [CrossRef]
- Wang, Z.; Deng, Z.; Dahmane, N.; Tsai, K.; Wang, P.; Williams, D.R.; Kossenkov, A.V.; Showe, L.C.; Zhang, R.; Huang, Q.; et al. Telomeric repeat-containing RNA (TERRA) constitutes a nucleoprotein component of extracellular inflammatory exosomes. Proc. Natl. Acad Sci. USA 2015, 112, E6293–E6300. [Google Scholar] [CrossRef]
- Wang, Z.; Lieberman, P.M. The crosstalk of telomere dysfunction and inflammation through cell-free TERRA containing exosomes. RNA Biol. 2016, 13, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Nassour, J.; Aguiar, L.G.; Correia, A.; Schmidt, T.T.; Mainz, L.; Przetocka, S.; Haggblom, C.; Tadepalle, N.; Williams, A.; Shokhirev, M.N.; et al. Telomere-to-mitochondria signalling by ZBP1 mediates replicative crisis. Nature 2023, 614, 767–773. [Google Scholar] [CrossRef]
- Morgunova, V.; Kordyukova, M.; Mikhaleva, E.A.; Butenko, I.; Pobeguts, O.V.; Kalmykova, A. Loss of telomere silencing is accompanied by dysfunction of Polo kinase and centrosomes during Drosophila oogenesis and early development. PLoS ONE 2021, 16, e0258156. [Google Scholar] [CrossRef]
- Morgunova, V.; Akulenko, N.; Radion, E.; Olovnikov, I.; Abramov, Y.; Olenina, L.V.; Shpiz, S.; Kopytova, D.V.; Georgieva, S.G.; Kalmykova, A. Telomeric repeat silencing in germ cells is essential for early development in Drosophila. Nucleic Acids Res. 2015, 43, 8762–8773. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, M.; Sokolova, O.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Glukhov, S.; Kalmykova, A. Nuclear Ccr4-Not mediates the degradation of telomeric and transposon transcripts at chromatin in the Drosophila germline. Nucleic Acids Res. 2019, 48, 141–156. [Google Scholar] [CrossRef]
- Aschacher, T.; Wolf, B.; Aschacher, O.; Enzmann, F.; Laszlo, V.; Messner, B.; Türkcan, A.; Weis, S.; Spiegl-Kreinecker, S.; Holzmann, K.; et al. Long interspersed element-1 ribonucleoprotein particles protect telomeric ends in alternative lengthening of telomeres dependent cells. Neoplasia 2020, 22, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Zhu, Y.; Xu, L.; Yu, H.; Yu, J.; Liu, X.; Fu, C.; Wang, X.; Ke, Y.; Huang, H.; et al. The 68-kDa telomeric repeat binding factor 1 (TRF1)-associated protein (TAP68) interacts with and recruits TRF1 to the spindle pole during mitosis. J. Biol. Chem. 2014, 289, 14145–14156. [Google Scholar] [CrossRef]
- Ohishi, T.; Hirota, T.; Tsuruo, T.; Seimiya, H. TRF1 mediates mitotic abnormalities induced by Aurora-A overexpression. Cancer Res. 2010, 70, 2041–2052. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Wright, W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, A.M. Chronographic theory of development, aging, and origin of cancer: Role of chronomeres and printomeres. Curr. Aging Sci. 2015, 8, 76–88. [Google Scholar] [CrossRef] [PubMed]
- D’Adda di Fagagna, F.; Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; Von Zglinicki, T.; Jackson, S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003, 426, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Takai, H.; Smogorzewska, A.; de Lange, T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003, 13, 1549–1556. [Google Scholar] [CrossRef]
- Rossiello, F.; Herbig, U.; Longhese, M.P.; Fumagalli, M.; d’Adda di Fagagna, F. Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing. Curr. Opin Genet. Dev. 2014, 26, 89–95. [Google Scholar] [CrossRef]
- Gorbunova, V.; Bozzella, M.J.; Seluanov, A. Rodents for comparative aging studies: From mice to beavers. Age 2008, 30, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Revy, P.; Kannengiesser, C.; Bertuch, A.A. Genetics of human telomere biology disorders. Nat. Rev. Genet. 2023, 24, 86–108. [Google Scholar] [CrossRef]
- DeBoy, E.A.; Tassia, M.G.; Schratz, K.E.; Yan, S.M.; Cosner, Z.L.; McNally, E.J.; Gable, D.L.; Xiang, Z.; Lombard, D.B.; Antonarakis, E.S.; et al. Familial Clonal Hematopoiesis in a Long Telomere Syndrome. N. Engl. J. Med. 2023, 388, 2422–2433. [Google Scholar] [CrossRef]
- Anderson, R.; Lagnado, A.; Maggiorani, D.; Walaszczyk, A.; Dookun, E.; Chapman, J.; Birch, J.; Salmonowicz, H.; Ogrodnik, M.; Jurk, D.; et al. Length-independent telomere damage drives post-mitotic cardiomyocyte senescence. EMBO J. 2019, 38, e100492. [Google Scholar] [CrossRef]
- Fumagalli, M.; Rossiello, F.; Clerici, M.; Barozzi, S.; Cittaro, D.; Kaplunov, J.M.; d’Adda di Fagagna, F. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 2012, 14, 355–365. [Google Scholar] [CrossRef]
- Hewitt, G.; Jurk, D.; Marques, F.D.; Correia-Melo, C.; Hardy, T.; Gackowska, A.; Anderson, R.; Taschuk, M.; Mann, J.; Passos, J.F. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 2012, 3, 708. [Google Scholar] [CrossRef]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef]
- Victorelli, S.; Lagnado, A.; Halim, J.; Moore, W.; Talbot, D.; Barrett, K.; Chapman, J.; Birch, J.; Ogrodnik, M.; Meves, A.; et al. Senescent human melanocytes drive skin ageing via paracrine telomere dysfunction. EMBO J. 2019, 38, e101982. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.L.; Lister, J.A.; Zeng, Z.; Ishizaki, H.; Anderson, C.; Kelsh, R.N.; Jackson, I.J.; Patton, E.E. Differentiated melanocyte cell division occurs in vivo and is promoted by mutations in Mitf. Development 2011, 138, 3579–3589. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Song, W.; Li, J.; Jing, Y.; Liang, C.; Zhang, L.; Liu, G.H. The landscape of aging. Sci. China Life Sci. 2022, 65, 2354–2454. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, D.; LaBella, K.A.; DePinho, R.A. Telomeres: History, health, and hallmarks of aging. Cell 2021, 184, 306–322. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Pereira-Smith, O.M. Expression of human telomerase (hTERT) does not prevent stress-induced senescence in normal human fibroblasts but protects the cells from stress-induced apoptosis and necrosis. J. Biol. Chem. 2002, 277, 38540–38549. [Google Scholar] [CrossRef] [PubMed]
- Correia-Melo, C.; Birch, J.; Fielder, E.; Rahmatika, D.; Taylor, J.; Chapman, J.; Passos, J.F. Rapamycin improves healthspan but not inflammaging in nfκb1(-/-) mice. Aging Cell 2019, 18, e12882. [Google Scholar] [CrossRef]
- Stout, M.B.; Steyn, F.J.; Jurczak, M.J.; Camporez, J.-P.G.; Zhu, Y.; Hawse, J.R.; Jurk, D.; Palmer, A.K.; Xu, M.; Pirtskhalava, T.; et al. 17α-Estradiol Alleviates Age-related Metabolic and Inflammatory Dysfunction in Male Mice Without Inducing Feminization. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2017, 72, 3–15. [Google Scholar] [CrossRef]
- Roos, C.M.; Zhang, B.; Palmer, A.K.; Ogrodnik, M.B.; Pirtskhalava, T.; Thalji, N.M.; Hagler, M.; Jurk, D.; Smith, L.A.; Casaclang-Verzosa, G.; et al. Chronic senolytic treatment alleviates established vasomotor dysfunction in aged or atherosclerotic mice. Aging Cell 2016, 15, 973–977. [Google Scholar] [CrossRef]
- Lagnado, A.; Leslie, J.; Ruchaud-Sparagano, M.; Victorelli, S.; Hirsova, P.; Ogrodnik, M.; Collins, A.L.; Vizioli, M.G.; Habiballa, L.; Saretzki, G.; et al. Neutrophils induce paracrine telomere dysfunction and senescence in ROS-dependent manner. EMBO J. 2021, 40, e106048. [Google Scholar] [CrossRef]
- Barnes, R.P.; de Rosa, M.; Thosar, S.A.; Detwiler, A.C.; Roginskaya, V.; Van Houten, B.; Bruchez, M.P.; Stewart-Ornstein, J.; Opresko, P.L. Telomeric 8-oxo-guanine drives rapid premature senescence in the absence of telomere shortening. Nat. Struct Mol. Biol. 2022, 29, 639–652. [Google Scholar] [CrossRef]
- Feretzaki, M.; Pospisilova, M.; Valador Fernandes, R.; Lunardi, T.; Krejci, L.; Lingner, J. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 2020, 587, 303–308. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Belotserkovskii, B.P.; Tornaletti, S.; D’Souza, A.D.; Hanawalt, P.C. R-loop generation during transcription: Formation, processing and cellular outcomes. DNA Repair 2018, 71, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, R.; Lynskey, M.L.; O’Sullivan, R.J. New twists to the ALTernative endings at telomeres. DNA Repair 2022, 115, 103342. [Google Scholar] [CrossRef]
- Gong, Y.; Liu, Y. R-Loops at Chromosome Ends: From Formation, Regulation, and Cellular Consequence. Cancers 2023, 15, 2178. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Voon, H.P.J.; Xella, B.; Scott, C.; Clynes, D.; Babbs, C.; Ayyub, H.; Kerry, J.; Sharpe, J.A.; Sloane-Stanley, J.A.; et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017, 18, 914–928. [Google Scholar] [CrossRef]
- Doksani, Y.; de Lange, T. Telomere-Internal Double-Strand Breaks Are Repaired by Homologous Recombination and PARP1/Lig3-Dependent End-Joining. Cell Rep. 2016, 17, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Balk, B.; Maicher, A.; Dees, M.; Klermund, J.; Luke-Glaser, S.; Bender, K.; Luke, B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct Mol. Biol. 2013, 20, 1199–1205. [Google Scholar] [CrossRef]
- Wang, S.S.; Zakian, V.A. Telomere-telomere recombination provides an express pathway for telomere acquisition. Nature 1990, 345, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; Hockemeyer, D.; Kibe, T.; de Lange, T. Functional dissection of human and mouse POT1 proteins. Mol. Cell Biol. 2009, 29, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, A.; Kabir, S.; van Overbeek, M.; Celli, G.B.; de Lange, T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 2010, 327, 1657–1661. [Google Scholar] [CrossRef]
- Liu, L.; Bailey, S.M.; Okuka, M.; Muñoz, P.; Li, C.; Zhou, L.; Wu, C.; Czerwiec, E.; Sandler, L.; Seyfang, A.; et al. Telomere lengthening early in development. Nat. Cell Biol. 2007, 9, 1436–1441. [Google Scholar] [CrossRef]
- Neumann, A.A.; Watson, C.M.; Noble, J.R.; Pickett, H.A.; Tam, P.P.; Reddel, R.R. Alternative lengthening of telomeres in normal mammalian somatic cells. Genes Dev. 2013, 27, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Burla, R.; La Torre, M.; Saggio, I. Mammalian telomeres and their partnership with lamins. Nucleus 2016, 7, 187–202. [Google Scholar] [CrossRef]
- Gonzalez-Suarez, I.; Redwood, A.B.; Perkins, S.M.; Vermolen, B.; Lichtensztejin, D.; Grotsky, D.A.; Morgado-Palacin, L.; Gapud, E.J.; Sleckman, B.P.; Sullivan, T.; et al. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009, 28, 2414–2427. [Google Scholar] [CrossRef]
- Pennarun, G.; Picotto, J.; Bertrand, P. Close Ties between the Nuclear Envelope and Mammalian Telomeres: Give Me Shelter. Genes 2023, 14, 775. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Doronin, S.A.; Khrameeva, E.E.; Kos, P.I.; Luzhin, A.V.; Starikov, S.S.; Galitsyna, A.A.; Nenasheva, V.V.; Ilyin, A.A.; Flyamer, I.M.; et al. Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 2019, 10, 1176. [Google Scholar] [CrossRef]
- Arnoult, N.; Schluth-Bolard, C.; Letessier, A.; Drascovic, I.; Bouarich-Bourimi, R.; Campisi, J.; Kim, S.-H.; Boussouar, A.; Ottaviani, A.; Magdinier, F.; et al. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 2010, 6, e1000920. [Google Scholar] [CrossRef]
- Turner, K.J.; Watson, E.M.; Skinner, B.M.; Griffin, D.K. Telomere Distribution in Human Sperm Heads and Its Relation to Sperm Nuclear Morphology: A New Marker for Male Factor Infertility? Int. J. Mol. Sci. 2021, 22, 7599. [Google Scholar] [CrossRef]
- Jurikova, K.; De Wulf, P.; Cusanelli, E. Nuclear Periphery and Telomere Maintenance: TERRA Joins the Stage. Trends Genet. 2021, 37, 608–611. [Google Scholar] [CrossRef]
- Radion, E.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Lavrov, S.; Abramov, Y.; Komarov, P.A.; Glukhov, S.I.; Olovnikov, I.; Kalmykova, A. Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline. Epigenetics Chromatin 2018, 11, 40. [Google Scholar] [CrossRef]
- Merigliano, C.; Chiolo, I. Multi-scale dynamics of heterochromatin repair. Curr. Opin Genet. Dev. 2021, 71, 206–215. [Google Scholar] [CrossRef]
- Tsouroula, K.; Furst, A.; Rogier, M.; Heyer, V.; Maglott-Roth, A.; Ferrand, A.; Reina-San-Martin, B.; Soutoglou, E. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol. Cell 2016, 63, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Amaral, N.; Ryu, T.; Li, X.; Chiolo, I. Nuclear Dynamics of Heterochromatin Repair. Trends Genet. 2017, 33, 86–100. [Google Scholar] [CrossRef]
- Ryu, T.; Spatola, B.; Delabaere, L.; Bowlin, K.; Hopp, H.; Kunitake, R.; Karpen, G.H.; Chiolo, I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 2015, 17, 1401–1411. [Google Scholar] [CrossRef] [PubMed]
- Chiolo, I.; Minoda, A.; Colmenares, S.U.; Polyzos, A.; Costes, S.V.; Karpen, G.H. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 2011, 144, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Lemaître, C.; Grabarz, A.; Tsouroula, K.; Andronov, L.; Furst, A.; Pankotai, T.; Heyer, V.; Rogier, M.; Attwood, K.M.; Kessler, P.; et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014, 28, 2450–2463. [Google Scholar] [CrossRef]
- Maestroni, L.; Reyes, C.; Vaurs, M.; Gachet, Y.; Tournier, S.; Géli, V.; Coulon, S. Nuclear envelope attachment of telomeres limits TERRA and telomeric rearrangements in quiescent fission yeast cells. Nucleic Acids Res. 2020, 48, 3029–3041. [Google Scholar] [CrossRef]
- Schober, H.; Ferreira, H.; Kalck, V.; Gehlen, L.R.; Gasser, S.M. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009, 23, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.W. The functional importance of lamins, actin, myosin, spectrin and the LINC complex in DNA repair. Exp. Biol. Med. 2019, 244, 1382–1406. [Google Scholar] [CrossRef]
- Wood, A.M.; Danielsen, J.M.R.; Lucas, C.A.; Rice, E.L.; Scalzo, D.; Shimi, T.; Goldman, R.D.; Smith, E.D.; Le Beau, M.M.; Kosak, S.T. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun. 2014, 5, 5467. [Google Scholar] [CrossRef]
- Marcelot, A.; Worman, H.J.; Zinn-Justin, S. Protein structural and mechanistic basis of progeroid laminopathies. FEBS J. 2021, 288, 2757–2772. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.P.; Donahue, G.; Otte, G.L.; Capell, B.C.; Nelson, D.M.; Cao, K.; Aggarwala, V.; Cruickshanks, H.A.; Rai, T.S.; McBryan, T.; et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013, 27, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, A.; Madsen-Osterbye, J.; Galigniana, N.M.; Collas, P. Restructuring of Lamina-Associated Domains in Senescence and Cancer. Cells 2022, 11, 1846. [Google Scholar] [CrossRef]
- Bi, S.; Liu, Z.; Wu, Z.; Wang, Z.; Liu, X.; Wang, S.; Ren, J.; Yao, Y.; Zhang, W.; Song, M.; et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 2020, 11, 483–504. [Google Scholar] [CrossRef]
- Hu, H.; Ji, Q.; Song, M.; Ren, J.; Liu, Z.; Wang, Z.; Liu, X.; Yan, K.; Hu, J.; Jing, Y.; et al. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 2020, 48, 6001–6018. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Liu, Z.; Song, M.; Li, W.; Wu, Z.; Wang, Z.; Wang, Q.; Wang, S.; Yan, K.; Sun, L.; et al. Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration. Cell Res. 2021, 31, 187–205. [Google Scholar] [CrossRef]
- Ghosh, S.; Zhou, Z. Genetics of aging, progeria and lamin disorders. Curr. Opin Genet Dev. 2014, 26, 41–46. [Google Scholar] [CrossRef]
- Huang, S.; Risques, R.A.; Martin, G.M.; Rabinovitch, P.S.; Oshima, J. Accelerated telomere shortening and replicative senescence in human fibroblasts overexpressing mutant and wild-type lamin A. Exp. Cell Res. 2008, 314, 82–91. [Google Scholar] [CrossRef]
- Sadaie, M.; Salama, R.; Carroll, T.; Tomimatsu, K.; Chandra, T.; Young, A.R.; Narita, M.; Pérez-Mancera, P.A.; Bennett, D.C.; Chong, H.; et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 2013, 27, 1800–1808. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Tyler, J.K. Epigenetics and aging. Sci. Adv. 2016, 2, e1600584. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Mather, K.A.; Jorm, A.F.; Parslow, R.A.; Christensen, H. Is telomere length a biomarker of aging? A review. J. Gerontol A Biol. Sci. Med. Sci. 2011, 66, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Benson, E.K.; Lee, S.W.; Aaronson, S.A. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J. Cell Sci. 2010, 123 Pt 15, 2605–2612. [Google Scholar] [CrossRef]
- Sengupta, D.; Sengupta, K. Lamin A and telomere maintenance in aging: Two to Tango. Mutat Res. 2022, 825, 111788. [Google Scholar] [CrossRef]
- Aguado, J.; Sola-Carvajal, A.; Cancila, V.; Revêchon, G.; Ong, P.F.; Jones-Weinert, C.W.; Arzt, E.W.; Lattanzi, G.; Dreesen, O.; Tripodo, C.; et al. Inhibition of DNA damage response at telomeres improves the detrimental phenotypes of Hutchinson-Gilford Progeria Syndrome. Nat. Commun. 2019, 10, 4990. [Google Scholar] [CrossRef]
- Morgunova, V.V.; Sokolova, O.A.; Sizova, T.V.; Malaev, L.G.; Babaev, D.S.; Kwon, D.A.; Kalmykova, A.I. Dysfunction of Lamin B and Physiological Aging Cause Telomere Instability in Drosophila Germline. Biochemistry 2022, 87, 1600–1610. [Google Scholar] [CrossRef]
- Wu, X.; Malkova, A. Break-induced replication mechanisms in yeast and mammals. Curr. Opin Genet Dev. 2021, 71, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Cevenini, E.; Monti, D.; Franceschi, C. Inflamm-ageing. Curr. Opin Clin. Nutr. Metab Care 2013, 16, 14–20. [Google Scholar] [CrossRef] [PubMed]
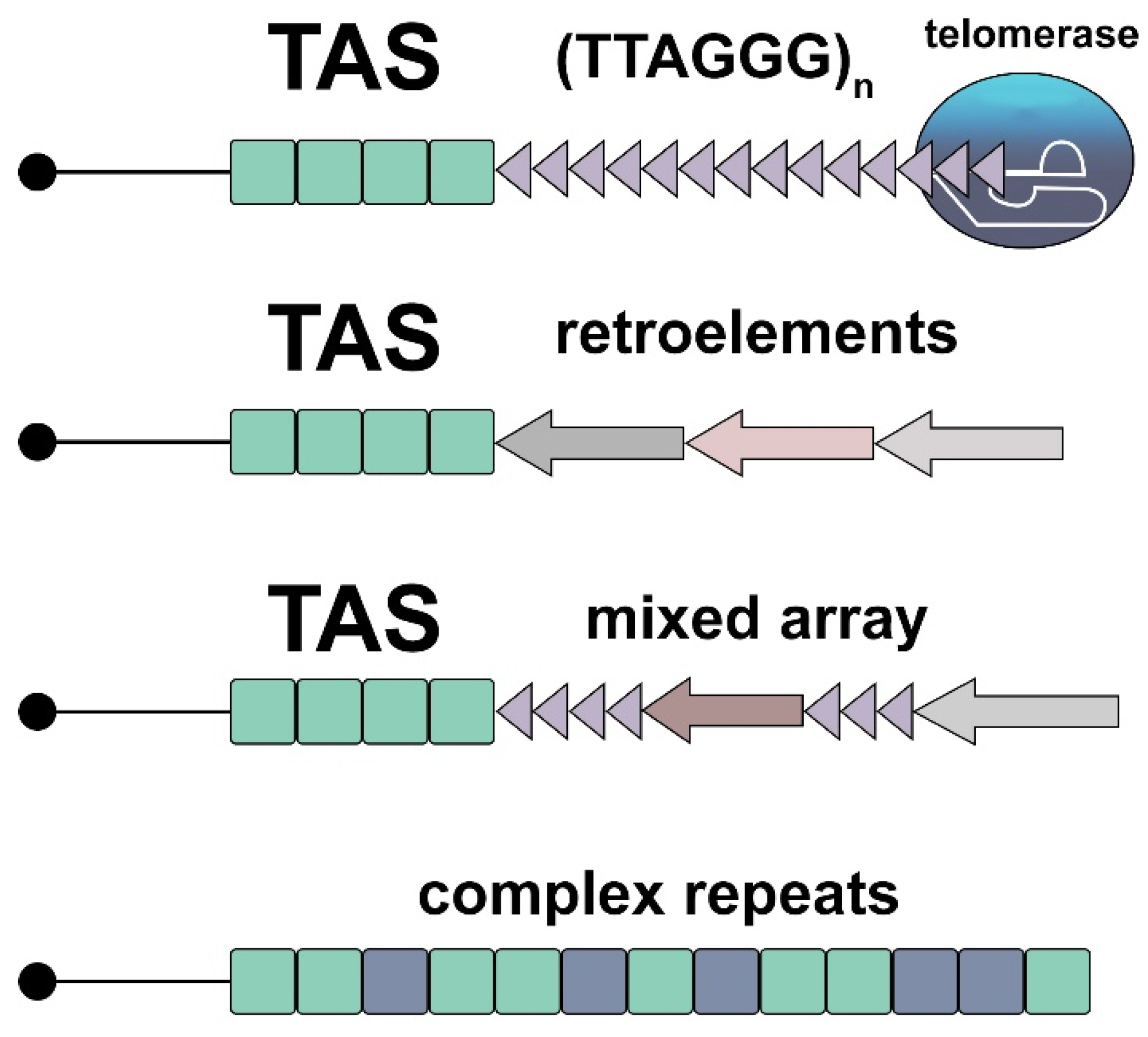

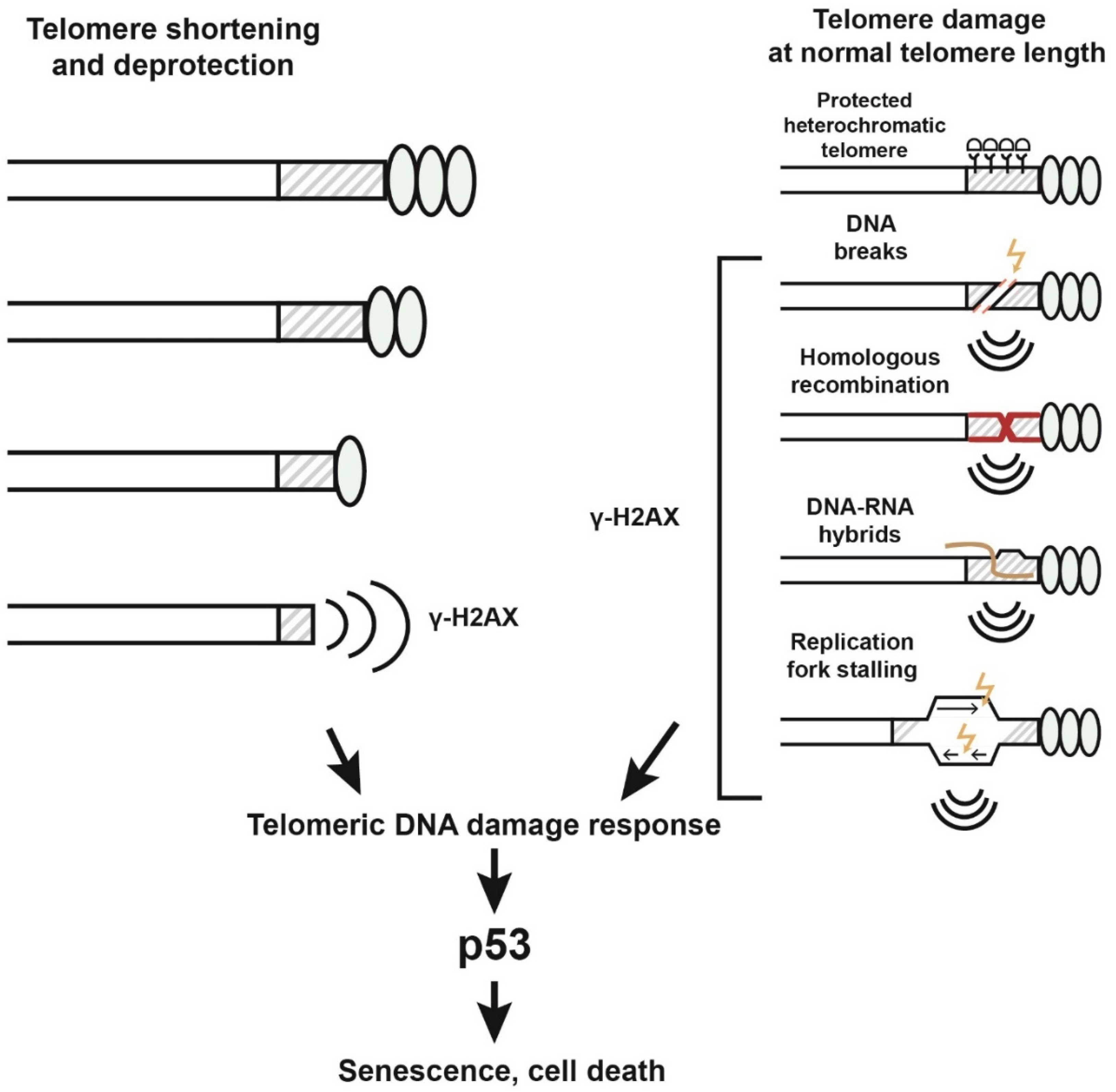
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalmykova, A. Telomere Checkpoint in Development and Aging. Int. J. Mol. Sci. 2023, 24, 15979. https://doi.org/10.3390/ijms242115979
Kalmykova A. Telomere Checkpoint in Development and Aging. International Journal of Molecular Sciences. 2023; 24(21):15979. https://doi.org/10.3390/ijms242115979
Chicago/Turabian StyleKalmykova, Alla. 2023. "Telomere Checkpoint in Development and Aging" International Journal of Molecular Sciences 24, no. 21: 15979. https://doi.org/10.3390/ijms242115979





