The Quest for Immunity: Exploring Human Herpesviruses as Vaccine Vectors
Abstract
1. Introduction
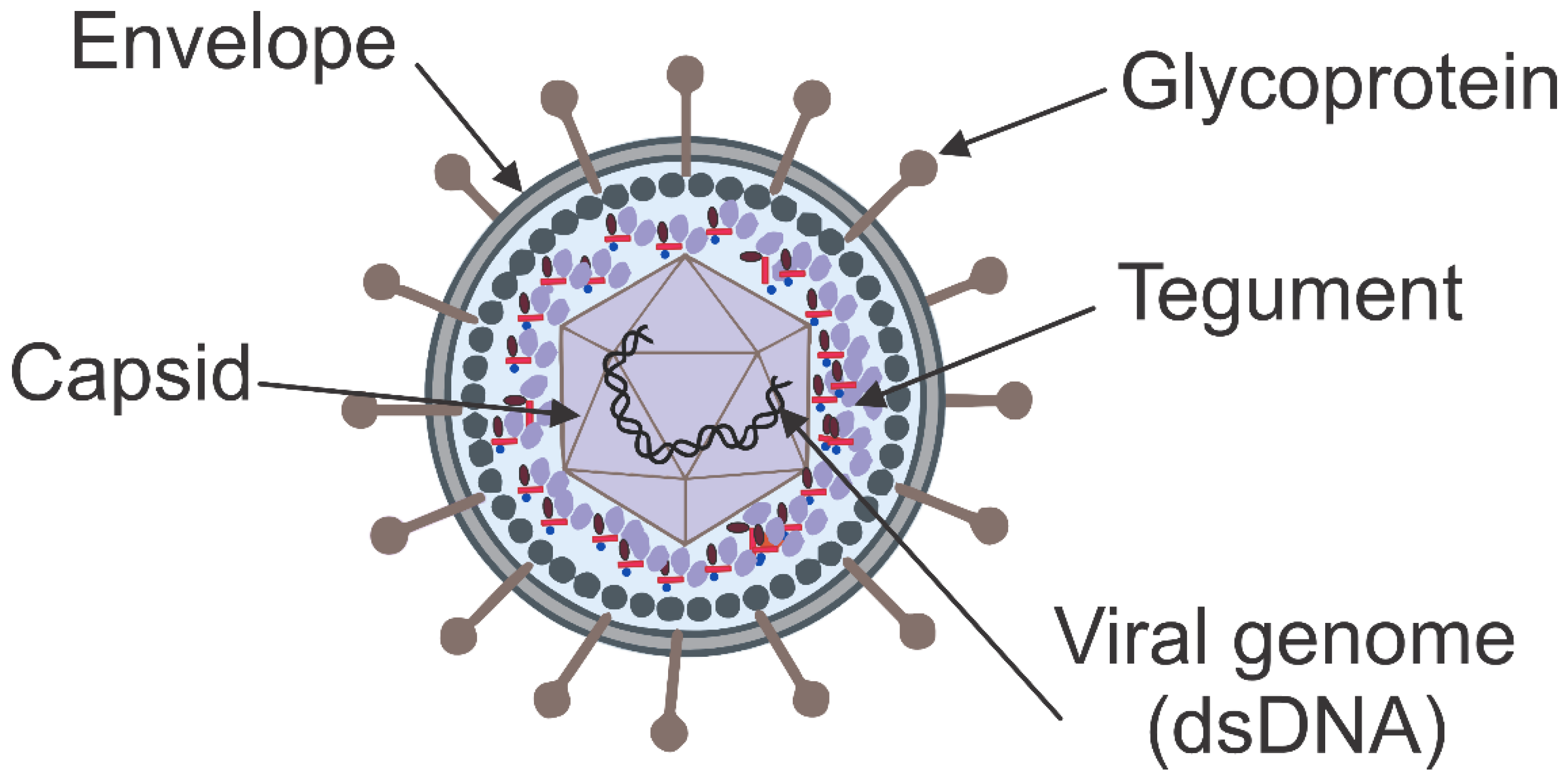
1.1. Herpesvirus Morphology
1.2. Evolutionary Process of Herpesviruses
1.3. Herpesvirus Biology
1.4. Herpesvirus Genomes
1.5. Herpesvirus Taxonomy
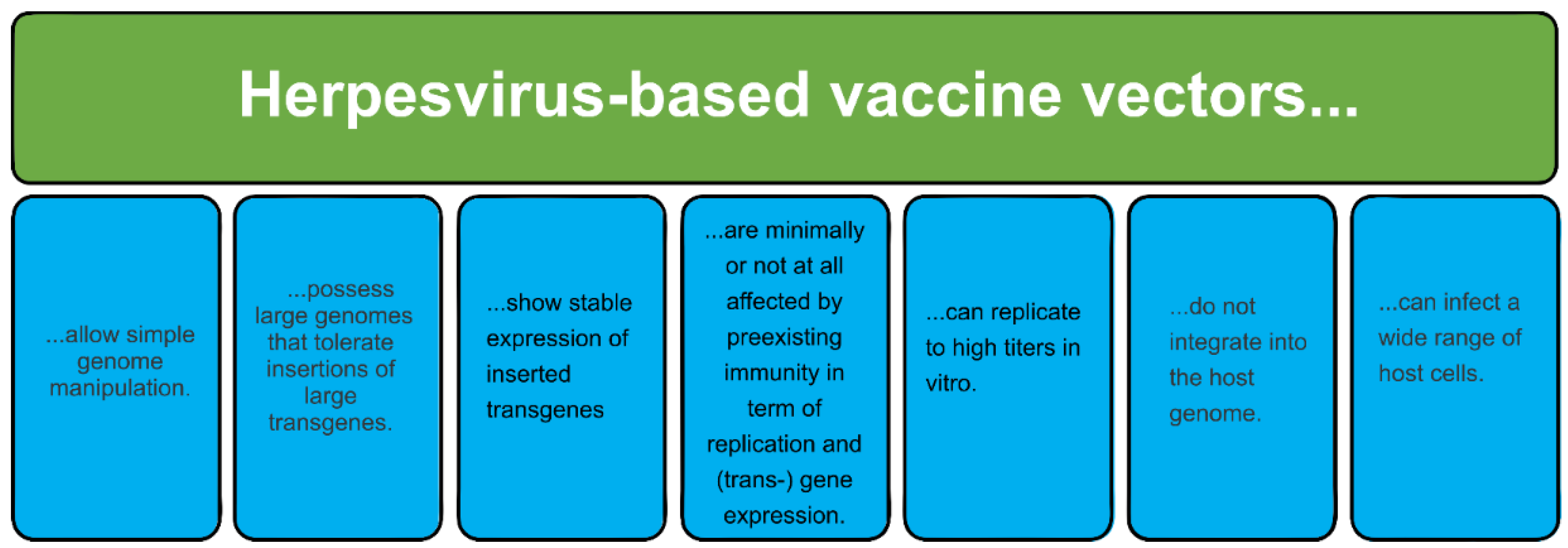
1.6. Replication-Competent Herpesvirus Vectors
1.7. Bacterial Artificial Chromosomes (BACs)
2. Human Herpesviruses as Vectors (HSV, CMV, and VZV)
2.1. Herpes Simplex Viruses
| Viral Infection | Targeted Antigens | Experimental Immunization in | Immune Response | Refs. |
|---|---|---|---|---|
| SIV | Env and Nef | Rhesus macaques | Specific humoral immune response, partial protection | [103] |
| Rotavirus- | VP2, VP6, and VP7 | Mice | Specific humoral immune response, partial protection | [108] |
| HIV | gp120 | Mice | Strong humoral and cell-mediated immune responses | [104] |
| IAV | Modified IAV hemagglutinin (HA) | Mice and guinea pigs | [109] | |
| WNV | Env and prM | Mice | Strong humoral and cell-mediated immune | [106] |
| HSV-1 | gD | Mice | Strong humoral and cell-mediated immune responses | |
| FMDV | P12A3C | Mice | Specific humoral response and partial protection | [110] |
| FMDV | P12A3C | Mice | 75% protection from challenge (Ad/herpes vector, prime-boost) | [111] |
| HIV-1 | HIV-1 gp160 | Adult female BALB/c mice | Antibody responses were measured by ELISA and the induced cell immune responses were measured by intracellular cytokine staining | [112] |
| Malaria | EXP1, UIS3, TMP21, VP26 | BALB/c mice | Potent cellular and humoral immune responses. Protected against challenge in mice | [113] |
| SARS-CoV-1 | spike protein | Balb/C and C57BL/6J mice strains | Antibodies and expression of cytokines were induced | [114] |
| EHV-1 | glycoprotein D | Female BALB/c mice | Strong antiviral humoral and cellular immune responses | [107] |
2.2. Human Cytomegalovirus
| Disease, Bacterial or Viral Infection | Targeted Antigens | Experimental Immunization in | Immune Response | Refs. |
|---|---|---|---|---|
| EBOV | NP CD8+ T cell epitope | Mice | Long-lasting CD8+ T cell response, complete protection | [125] |
| Clostridium tetani | Tetanus toxin fragment C | Mice | Specific humoral response | [143] |
| Cancer | Mouse tyrosinase-related protein 2 | Mice | Prolonged patient survival | [144] |
2.3. Varizella Zoster Virus
3. Molecular Mechanisms and Pathways Used Herpesviruses as Vaccine Vectors
3.1. Immune Response Stimulation
3.2. Genetic Manipulation and Antigen Presentation
3.3. Persistence and Longevity of Immunity
3.4. Immune Evasion Strategies
4. Immune Responses and the Issue of Preexisting Immunity
5. Discussion
6. Challenges, Opportunities, and Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Finco, O.; Rappuoli, R. Designing Vaccines for the Twenty-First Century Society. Front. Immunol. 2014, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, T.; Bhatt, K.; Eggermont, L.J.; O’Hare, N.; Memic, A.; Bencherif, S.A. Memic Supramolecular self-assembled peptide-based vaccines: Current state and future perspectives. Front. Chem. 2020, 8, 598160. [Google Scholar] [CrossRef] [PubMed]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2015, 7, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Faurez, F.; Dory, D.; Le Moigne, V.; Gravier, R.; Jestin, A. Biosafety of DNA vaccines: New generation of DNA vectors and current knowledge on the fate of plasmids after injection. Vaccine 2010, 28, 3888–3895. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, K.; Liljeström, P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccines 2014, 14, 177–194. [Google Scholar] [CrossRef]
- Ura, T.; Okuda, K.; Shimada, M. Developments in Viral Vector-Based Vaccines. Vaccines 2014, 2, 624–641. [Google Scholar] [CrossRef]
- Rollier, C.S.; Reyes-Sandoval, A.; Cottingham, M.G.; Reyes-Sandoval, M.G.; Cottingham, K. Ewer Viral vectors as vaccine platforms: Deployment in sight. Curr. Opin. Immunol. 2011, 23, 377–382. [Google Scholar] [CrossRef]
- Kamel, M.; El-Sayed, A. Utilization of herpesviridae as recombinant viral vectors in vaccine development against animal pathogens. Virus Res. 2019, 270, 197648. [Google Scholar] [CrossRef]
- Ertl, H.C. Viral vectors as vaccine carriers. Curr. Opin. Virol. 2016, 21, 1–8. [Google Scholar] [CrossRef]
- Ewer, K.J.; Lambe, T.; Rollier, C.S.; Spencer, A.J.; Hill, A.V.; Dorrell, L. Viral vectors as vaccine platforms: From immunogenicity to impact. Curr. Opin. Immunol. 2016, 41, 47–54. [Google Scholar] [CrossRef]
- Baron, M.D.; Iqbal, M.; Nair, V. Recent advances in viral vectors in veterinary vaccinology. Curr. Opin. Virol. 2018, 29, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lu, S. Heterologous prime–boost vaccination. Curr. Opin. Immunol. 2009, 21, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Kardani, K.; Bolhassani, A.; Shahbazi, S. Prime-boost vaccine strategy against viral infections: Mechanisms and benefits. Vaccine 2016, 34, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Travieso, T.; Li, J.; Mahesh, S.; Mello, J.D.F.R.E.; Blasi, M. The use of viral vectors in vaccine development. npj Vaccines 2022, 7, 75. [Google Scholar] [CrossRef]
- Tatsis, N.; Ertl, H.C. Adenoviruses as vaccine vectors. Mol. Ther. 2004, 10, 616–629. [Google Scholar] [CrossRef]
- Wagner, M.; Ruzsics, Z.; Koszinowski, U.H. Herpesvirus genetics has come of age. Trends Microbiol. 2002, 10, 318–324. [Google Scholar] [CrossRef]
- Roizman, B.; Baines, J. The diversity and unity of herpesviridae. Comp. Immunol. Microbiol. Infect. Dis. 1991, 14, 63–79. [Google Scholar] [CrossRef]
- Davison, A.J. Evolution of the herpesviruses. Vet. Microbiol. 2001, 86, 69–88. [Google Scholar] [CrossRef]
- Owen, D.J.; Crump, C.M.; Graham, S.C. Tegument Assembly and Secondary Envelopment of Alphaherpesviruses. Viruses 2015, 7, 5084–5114. [Google Scholar] [CrossRef]
- Gruffat, H.; Marchione, R.; Manet, E. Herpesvirus Late Gene Expression: A Viral-Specific Pre-initiation Complex Is Key. Front. Microbiol. 2016, 7, 869. [Google Scholar] [CrossRef]
- Bowman, J.J. Characterization of Expression of the Herpes Simplex Virus Type-1 ICP22 and Us1. 5 Proteins. Ph.D. Thesis, Harvard University, Cambridge, MA, USA, 2008. [Google Scholar]
- Roizman, B.; Carmichael, L.E.; Deinhardt, F.; de-The, G.; Nahmias, A.J.; Plowright, W.; Rapp, F.; Sheldrick, P.; Takahashi, M.; Wolf, K. Herpesviridae: Definition, provisional nomenclature, and taxonomy. Intervirology 1981, 16, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.C. Latent Herpesviruses of Humans. Ann. Intern. Med. 1984, 100, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.C. Alphaherpesvirus Latency: A Dynamic State of Transcription and Reactivation. Adv. Virus Res. 2016, 94, 53–80. [Google Scholar] [CrossRef] [PubMed]
- Crimi, S.; Fiorillo, L.; Bianchi, A.; D’Amico, C.; Amoroso, G.; Gorassini, F.; Mastroieni, R.; Marino, S.; Scoglio, C.; Catalano, F.; et al. Herpes Virus, Oral Clinical Signs and QoL: Systematic Review of Recent Data. Viruses 2019, 11, 463. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.N.; Meers, J.; Fowler, E.; Mahony, T. Back to BAC: The Use of Infectious Clone Technologies for Viral Mutagenesis. Viruses 2012, 4, 211–235. [Google Scholar] [CrossRef]
- Tischer, B.K.; Smith, G.A.; Osterrieder, N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol. Biol. 2010, 634, 421–430. [Google Scholar] [CrossRef]
- Tischer, B.K.; Kaufer, B.B. Viral Bacterial Artificial Chromosomes: Generation, Mutagenesis, and Removal of Mini-F Sequences. J. Biomed. Biotechnol. 2012, 2012, 472537. [Google Scholar] [CrossRef]
- Karstentischer, B.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 2006, 40, 191–197. [Google Scholar] [CrossRef]
- Błażej, P.; Mackiewicz, D.; Wnętrzak, M.; Mackiewicz, P. The Impact of Selection at the Amino Acid Level on the Usage of Synonymous Codons. G3 Genes Genomes Genet. 2017, 7, 967–981. [Google Scholar] [CrossRef]
- Li, G.; Ward, C.; Yeasmin, R.; Skiena, S.; Krug, L.T.; Forrest, J.C. A codon-shuffling method to prevent reversion during production of replication-defective herpesvirus stocks: Implications for herpesvirus vaccines. Sci. Rep. 2017, 7, 44404. [Google Scholar] [CrossRef]
- Manservigi, R.; Argnani, R.; Marconi, P. HSV recombinant vectors for gene therapy. Open Virol. J. 2010, 4, 123. [Google Scholar] [PubMed]
- Marconi, P.; Argnani, R.; Epstein, A.L.; Manservigi, R. HSV as a vector in vaccine development and gene therapy. In Pharmaceutical Biotechnology; Part of the Advances in Experimental Medicine and Biology Book Series (AEMB); Springer: New York, NY, USA, 2009; Volume 655, pp. 118–144. [Google Scholar] [CrossRef]
- Pellet, P.E.; Roizman, B. The Herpesviridae: A brief introduction. In Fields Virology, 5th ed.; Howley, P.M., Ed.; Lippincott: Philadelphia, PA, USA, 2007; pp. 2480–2499. [Google Scholar]
- Lan, K.; Luo, M.-H. Herpesviruses: Epidemiology, pathogenesis, and interventions. Virol. Sin. 2017, 32, 347–348. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J. Herpesviruses: General Features. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Zhang, X.; Efstathiou, S.; Simmons, A. Identification of Novel Herpes Simplex Virus Replicative Intermediates by Field Inversion Gel Electrophoresis: Implications for Viral DNA Amplification Strategies. Virology 1994, 202, 530–539. [Google Scholar] [CrossRef]
- Bataille, D.; Epstein, A. Herpes Simplex Virus Replicative Concatemers Contain L Components in Inverted Orientation. Virology 1994, 203, 384–388. [Google Scholar] [CrossRef]
- Roizman, B.; Pellett, P.E. The family Herpesviridae: A brief introduction. In Fields Virology; Knipe, D.M., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Roizman, B., Straus, S.E., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001; pp. 2381–2398. [Google Scholar]
- Davis-Poynter, N.J.; Farrell, H.E. Masters of deception: A review of herpesvirus immune evasion strategies. Immunol. Cell Biol. 1996, 74, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N.; Wallaschek, N.; Kaufer, B.B. Herpesvirus Genome Integration into Telomeric Repeats of Host Cell Chromosomes. Annu. Rev. Virol. 2014, 1, 215–235. [Google Scholar] [PubMed]
- Wertheim, J.O.; Smith, M.D.; Smith, D.M.; Scheffler, K.; Pond, S.L.K. Evolutionary Origins of Human Herpes Simplex Viruses 1 and 2. Mol. Biol. Evol. 2014, 31, 2356–2364. [Google Scholar] [CrossRef]
- Louten, J. Chapter 13—Herpesviruses. In Essential Human Virology; Louten, J., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 235–256. [Google Scholar]
- Escalera-Zamudio, M.; Rojas-Anaya, E.; Kolokotronis, S.-O.; Taboada, B.; Loza-Rubio, E.; Méndez-Ojeda, M.L.; Arias, C.F.; Osterrieder, N.; Greenwood, A.D. Bats, Primates, and the Evolutionary Origins and Diversification of Mammalian Gammaherpesviruses. mBio 2016, 7, e01425-16. [Google Scholar] [CrossRef]
- Azab, W.; Dayaram, A.; Greenwood, A.D.; Osterrieder, N. How Host Specific Are Herpesviruses? Lessons from Herpesviruses Infecting Wild and Endangered Mammals. Annu. Rev. Virol. 2018, 5, 53–68. [Google Scholar] [CrossRef]
- Grinde, B. Herpesviruses: Latency and reactivation—Viral strategies and host response. J. Oral Microbiol. 2013, 5, 22766. [Google Scholar] [CrossRef]
- Weidner-Glunde, M.; Kruminis-Kaszkiel, E.; Savanagouder, M. Herpesviral Latency—Common Themes. Pathogens 2020, 9, 125. [Google Scholar] [PubMed]
- Bowden, G.H.W. Actinomyces, Propionibacterium propionicus, and Streptomyces. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2. [Google Scholar]
- Tischer, B.K.; Osterrieder, N. Herpesviruses—A zoonotic threat? Vet. Microbiol. 2010, 140, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.E.; Dolin, R.; Blaser, M.J. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Saunders: London, UK, 2019. [Google Scholar]
- Ho, D.Y.; Enriquez, K.; Multani, A. Herpesvirus Infections Potentiated by Biologics. Infect. Dis. Clin. N. Am. 2020, 34, 311–339. [Google Scholar] [CrossRef]
- Rahangdale, R.R.; Tender, T.; Balireddy, S.; Pasupuleti, M.; Hariharapura, R.C. Interplay Between Stress and Immunity Triggers Herpes Zoster Infection in COVID-19 Patients: A Review. Can. J. Microbiol. 2022, 68, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Van Gerwen, O.T.; Muzny, C.A.; Marrazzo, J.M. Muzny Sexually transmitted infections and female reproductive health. Nat. Microbiol. 2022, 7, 1116–1126. [Google Scholar] [PubMed]
- Poole, E.; Sinclair, J. Understanding HCMV Latency Using Unbiased Proteomic Analyses. Pathogens 2020, 9, 590. [Google Scholar] [CrossRef]
- Koyuncu, O.O.; MacGibeny, M.A.; Enquist, L.W. Latent versus productive infection: The alpha herpesvirus switch. Futur. Virol. 2018, 13, 431–443. [Google Scholar] [CrossRef]
- McGeoch, D.J.; Rixon, F.J.; Davison, A.J. Rixon Topics in herpesvirus genomics and evolution. Virus Res. 2006, 117, 90–104. [Google Scholar]
- Arvin, A.; Campadelli-Fiume, G.; Mocarski, E.; Moore, P.S.; Roizman, B.; Whitley, R.; Yamanishi, K. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Cambridge University Press (CUP): Cambridge, UK, 2007; ISBN 9780511545313. [Google Scholar]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order Herpesvirales. Arch. Virol. 2008, 154, 171–177. [Google Scholar] [CrossRef]
- McGeoch, D.J.; Gatherer, D.; Dolan, A. On phylogenetic relationships among major lineages of the Gammaherpesvirinae. J. Gen. Virol. 2005, 86, 307–316. [Google Scholar] [CrossRef]
- Šudomová, M.; Berchová-Bímová, K.; Mazurakova, A.; Šamec, D.; Kubatka, P.; Hassan, S.T.S. Flavonoids Target Human Herpesviruses That Infect the Nervous System: Mechanisms of Action and Therapeutic Insights. Viruses 2022, 14, 592. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.T.S.; Šudomová, M.; Mazurakova, A.; Kubatka, P. Insights into Antiviral Properties and Molecular Mechanisms of Non-Flavonoid Polyphenols against Human Herpesviruses. Int. J. Mol. Sci. 2022, 23, 13891. [Google Scholar] [CrossRef] [PubMed]
- Melnick, W.P. Classification and Nomenclature of Viruses. In Monographs in Virology; Kargerv: Basel, Switzerland, 1971. [Google Scholar]
- Roizman, D.F.B. The Replication of Herpesviruses. In Reproduction; Springer: Berlin/Heidelberg, Germany, 1974; pp. 229–403. [Google Scholar]
- Roizmann, B.; Desrosiers, R.C.; Fleckenstein, B.; Lopez, C.; Minson, A.C.; Studdert, M.J. The family Herpesviridae: An update. The Her-pesvirus Study Group of the International Committee on Taxonomy of Viruses. Arch. Virol. 1992, 123, 425–449. [Google Scholar] [CrossRef]
- Karlin, S.; Mocarski, E.S.; Schachtel, G.A. Molecular evolution of herpesviruses: Genomic and protein sequence comparisons. J. Virol. 1994, 68, 1886–1902. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Mayo, M.A.; Maniloff, J.; Desselberger, U. Virus taxonomy: Eighth report of the International Committee on the taxonomy of viruses. Viruses 2005, 83, 988–992. [Google Scholar]
- Berto, E.; Bozac, A.; Marconi, P. Development and application of replication-incompetent HSV-1-based vectors. Gene Ther. 2005, 12, S98–S102. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Artusi, S.; Miyagawa, Y.; Goins, W.F.; Cohen, J.B.; Glorioso, J.C. Herpes Simplex Virus Vectors for Gene Transfer to the Central Nervous System. Diseases 2018, 6, 74. [Google Scholar] [CrossRef]
- Knop, D.R.; Harrell, H. Bioreactor Production of Recombinant Herpes Simplex Virus Vectors. Biotechnol. Prog. 2008, 23, 715–721. [Google Scholar] [CrossRef]
- Goins, W.F.; Krisky, D.M.; Wechuck, J.B.; Wolfe, D.; Huang, S.; Glorioso, J.C. Generation of replication-competent and -defective HSV vectors. Cold Spring Harb. Protoc. 2011, 2011, prot5615. [Google Scholar] [CrossRef]
- Shah, A.C.; Benos, D.; Gillespie, G.Y.; Markert, J.M. Oncolytic viruses: Clinical applications as vectors for the treatment of malignant gliomas. J. Neuro-Oncol. 2003, 65, 203–226. [Google Scholar] [CrossRef]
- Chiocca, E.A. Oncolytic viruses. Nat. Rev. Cancer 2002, 2, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Knipe, D.M.; Finberg, R.W. Replication-defective mutants of herpes simplex virus (HSV) induce cellular immunity and protect against lethal HSV infection. J. Virol. 1992, 66, 7067–7072. [Google Scholar] [CrossRef] [PubMed]
- Post, D.E.; Fulci, G.; Chiocca, E.A.; Van Meir, E.G. Replicative Oncolytic Herpes Simplex Viruses in Combination Cancer Therapies. Curr. Gene Ther. 2004, 4, 41–51. [Google Scholar] [CrossRef]
- Ward, P.L.; Roizman, B. Herpes simplex genes: The blueprint of a succesful human pathogen. Trends Genet. 1994, 10, 267–274. [Google Scholar] [CrossRef]
- Dogrammatzis, C.; Waisner, H.; Kalamvoki, M. “Non-Essential” Proteins of HSV-1 with Essential Roles In Vivo: A Comprehensive Review. Viruses 2021, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, B. High capacity vectors. In Advances in Biotechnology; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–10. [Google Scholar]
- McGregor, A.; Schleiss, M.R. Recent Advances in Herpesvirus Genetics Using Bacterial Artificial Chromosomes. Mol. Genet. Metab. 2001, 72, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Messerle, M.; Crnkovic, I.; Hammerschmidt, W.; Ziegler, H.; Koszinowski, U.H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 1997, 94, 14759–14763. [Google Scholar] [CrossRef]
- Robert-Guroff, M. Replicating and non-replicating viral vectors for vaccine development. Curr. Opin. Biotechnol. 2007, 18, 546–556. [Google Scholar] [CrossRef]
- Ma, W.; He, H.; Wang, H. Oncolytic herpes simplex virus and immunotherapy. BMC Immunol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Morrison, L.A.; Knipe, D.M. Immunization with replication-defective mutants of herpes simplex virus type 1: Sites of immune intervention in pathogenesis of challenge virus infection. J. Virol. 1994, 68, 689–696. [Google Scholar] [CrossRef]
- Knipe, D.M.; Ruyechan, W.T.; Roizman, B.; Halliburton, I.W. Molecular genetics of herpes simplex virus: Demonstration of regions of obligatory and nonobligatory identity within diploid regions of the genome by sequence replacement and insertion. Proc. Natl. Acad. Sci. USA 1978, 75, 3896–3900. [Google Scholar] [CrossRef] [PubMed]
- Azher, T.N.; Yin, X.T.; Stuart, P.M. Understanding the Role of Chemokines and Cytokines in Experimental Models of Herpes Simplex Keratitis. J. Immunol. Res. 2017, 2017, 7261980. [Google Scholar] [CrossRef] [PubMed]
- Krug, A.; Luker, G.D.; Barchet, W.; Leib, D.A.; Akira, S.; Colonna, M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor. Blood 2004, 103, 1433–1437. [Google Scholar] [CrossRef] [PubMed]
- Kurt-Jones, E.A.; Belko, J.; Yu, C.; Newburger, P.E.; Wang, J.; Chan, M.; Knipe, D.M.; Finberg, R.W. The role of toll-like receptors in herpes simplex infection in neonates. J. Infect. Dis. 2005, 191, 746–748. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, F.; Agostini, S.; Bianchi, A.; Nemni, R.; Piancone, F.; Marventano, I.; Mancuso, R.; Saresella, M.; Clerici, M. Herpes simplex virus-1 (HSV-1) infection induces a potent but ineffective IFN-λ production in immune cells of AD and PD patients. J. Transl. Med. 2019, 17, 286. [Google Scholar] [CrossRef]
- Brockman, M.A.; Knipe, D.M. Herpes Simplex Virus Vectors Elicit Durable Immune Responses in the Presence of Preexisting Host Immunity. J. Virol. 2002, 76, 3678–3687. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Pullum, D.A.; Cardin, R.D.; Bravo, F.J.; Dixon, D.A.; Kousoulas, K.G. The HSV-1 live attenuated VC2 vaccine provides pro-tection against HSV-2 genital infection in the guinea pig model of genital herpes. Vaccine 2019, 37, 61–68. [Google Scholar] [CrossRef]
- Aschner, C.B.; Herold, B.C. Alphaherpesvirus Vaccines. Curr. Issues Mol. Biol. 2021, 41, 469–508. [Google Scholar] [CrossRef]
- Srivastava, R.; Roy, S.; Coulon, P.-G.; Vahed, H.; Prakash, S.; Dhanushkodi, N.; Kim, G.J.; Fouladi, M.A.; Campo, J.; Teng, A.A.; et al. Therapeutic mucosal vaccination of herpes simplex virus 2-infected guinea pigs with ribonucleotide reductase 2 (RR2) protein boosts antiviral neutralizing antibodies and local tissue-resident CD4+ and CD8+ TRM cells associated with protection against recurrent genital herpes. J. Virol. 2019, 93, 2309–2318. [Google Scholar]
- Iyer, A.V.; Pahar, B.; Chouljenko, V.N.; Walker, J.D.; Stanfield, B.; Kousoulas, K.G. Single dose of glycoprotein K (gK)-deleted HSV-1 live-attenuated virus protects mice against lethal vaginal challenge with HSV-1 and HSV-2 and induces lasting T cell memory immune responses. Virol. J. 2013, 10, 317. [Google Scholar] [CrossRef]
- Diaz, F.; Gregory, S.; Nakashima, H.; Viapiano, M.S.; Knipe, D.M. Intramuscular delivery of replication-defective herpes simplex virus gives antigen expression in muscle syncytia and improved protection against pathogenic HSV-2 strains. Virology 2017, 513, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Claiborne, D.T.; Dudek, T.E.; Maldini, C.R.; Power, K.A.; Ghebremichael, M.; Seung, E.; Mellors, E.F.; Vrbanac, V.D.; Krupp, K.; Bisesi, A.; et al. Immunization of BLT Humanized Mice Redirects T Cell Responses to Gag and Reduces Acute HIV-1 Viremia. J. Virol. 2019, 93, e00814-19. [Google Scholar] [CrossRef] [PubMed]
- Duke, C.M.; Maguire, C.A.; Keefer, M.C.; Federoff, H.J.; Bowers, W.J.; Dewhurst, S. HSV-1 amplicon vectors elicit polyfunctional T cell responses to HIV-1 Env, and strongly boost responses to an adenovirus prime. Vaccine 2007, 25, 7410–7421. [Google Scholar] [CrossRef] [PubMed]
- Gorantla, S.; Santos, K.; Meyer, V.; Dewhurst, S.; Bowers, W.J.; Federoff, H.J.; Gendelman, H.E.; Poluektova, L. Human dendritic cells transduced with herpes simplex virus amplicons encoding human immunodeficiency virus type 1 (HIV-1) gp120 elicit adaptive immune responses from human cells engrafted into NOD/SCID mice and confer partial protection against HIV-1 challenge. J. Virol. 2005, 79, 2124–2132. [Google Scholar]
- Santos, K.; Duke, C.M.; Rodriguez-Colon, S.M.; Dakwar, A.; Fan, S.; Keefer, M.C.; Federoff, H.J.; Frelinger, J.G.; Bowers, W.J.; Dewhurst, S. Effect of promoter strength on protein expression and immunogenicity of an HSV-1 amplicon vector encoding HIV-1 Gag. Vaccine 2007, 25, 1634–1646. [Google Scholar] [CrossRef]
- York, I.A.; Roop, C.; Andrews, D.W.; Riddell, S.R.; Graham, F.L.; Johnson, D.C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell 1994, 77, 525–535. [Google Scholar] [CrossRef]
- Barcy, S.; Corey, L. Herpes Simplex Inhibits the Capacity of Lymphoblastoid B Cell Lines to Stimulate CD4+ T Cells. J. Immunol. 2001, 166, 6242–6249. [Google Scholar] [CrossRef]
- Liu, X.; Broberg, E.; Watanabe, D.; Dudek, T.; DeLuca, N.; Knipe, D.M. Genetic engineering of a modified herpes simplex virus 1 vaccine vector. Vaccine 2009, 27, 2760–2767. [Google Scholar] [CrossRef]
- Watanabe, D.; Brockman, M.A.; Ndung, T.; Mathews, L.; Lucas, W.T.; Murphy, C.G.; Felber, B.K.; Pavlakis, G.N.; Deluca, N.A.; Knipe, D.M. Properties of a herpes simplex virus multiple immediate-early gene-deleted recombinant as a vaccine vector. Virology 2007, 357, 186–198. [Google Scholar] [CrossRef]
- Lauterbach, H.; Ried, C.; Epstein, A.L.; Marconi, P.; Brocker, T. Reduced immune responses after vaccination with a recombinant herpes simplex virus type 1 vector in the presence of antiviral immunity. J. Gen. Virol. 2005, 86, 2401–2410. [Google Scholar] [CrossRef]
- Murphy, C.G.; Lucas, W.T.; Means, R.E.; Czajak, S.; Hale, C.L.; Lifson, J.D.; Kaur, A.; Johnson, R.P.; Knipe, D.M.; Desrosiers, R.C. Vaccine Protection against Simian Immunodeficiency Virus by Recombinant Strains of Herpes Simplex Virus. J. Virol. 2000, 74, 7745–7754. [Google Scholar] [CrossRef] [PubMed]
- Hocknell, P.K.; Wiley, R.D.; Wang, X.; Evans, T.G.; Bowers, W.J.; Hanke, T.; Federoff, H.J.; Dewhurst, S. Expression of Human Immunodeficiency Virus Type 1 gp120 from Herpes Simplex Virus Type 1-Derived Amplicons Results in Potent, Specific, and Durable Cellular and Humoral Immune Responses. J. Virol. 2002, 76, 5565–5580. [Google Scholar] [CrossRef] [PubMed]
- Sicurella, M.; Nicoli, F.; Gallerani, E.; Volpi, I.; Berto, E.; Finessi, V.; Destro, F.; Manservigi, R.; Cafaro, A.; Ensoli, B.; et al. An Attenuated Herpes Simplex Virus Type 1 (HSV1) Encoding the HIV-1 Tat Protein Protects Mice from a Deadly Mucosal HSV1 Challenge. PLoS ONE 2014, 9, e100844. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.J.; Diaz, F.; Colgrove, R.C.; Bernard, K.A.; DeLuca, N.A.; Whelan, S.P.; Knipe, D.M. Production of immunogenic West Nile vi-rus-like particles using a herpes simplex virus 1 recombinant vector. Virology 2016, 496, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.A.; Stanfield, B.A.; Chouljenko, V.N.; Naidu, S.; Langohr, I.; Del Piero, F.; Ferracone, J.; Roy, A.A.; Kousoulas, K.G. In-tramuscular Immunization of Mice with the Live-Attenuated Herpes Simplex Virus 1 Vaccine Strain VC2 Expressing Equine Herpesvirus 1 (EHV-1) Glycoprotein D Generates Anti-EHV-1 Immune Responses in Mice. J. Virol. 2017, 91, e02445-16. [Google Scholar] [CrossRef]
- Laimbacher, A.S.; Esteban, L.E.; Castello, A.A.; Cerfoglio, J.C.A.; Argüelles, M.H.; Glikmann, G.; D’Antuono, A.; Mattion, N.; Berois, M.; Arbiza, J.; et al. HSV-1 Amplicon Vectors Launch the Production of Heterologous Rotavirus-like Particles and Induce Rotavirus-specific Immune Responses in Mice. Mol. Ther. 2012, 20, 1810–1820. [Google Scholar] [CrossRef]
- Dardick, J.; Weiss, K.; Lukose, R.; Weinrick, B.; Herold, B.; Jacobs, W.R. A novel vaccine vector: Herpes simplex virus type-2 deleted in glycoprotein D (HSV-2 ΔgD) and expressing modified influenza A (IAV) hemagglutinin (HA) antigens. J. Immunol. 2017, 198, 147.9. [Google Scholar] [CrossRef]
- D’Antuono, A.; Laimbacher, A.S.; La Torre, J.; Tribulatti, V.; Romanutti, C.; Zamorano, P.; Quattrocchi, V.; Schraner, E.M.; Ackermann, M.; Fraefel, C.; et al. HSV-1 amplicon vectors that direct the in situ production of foot-and-mouth disease virus antigens in mammalian cells can be used for genetic immunization. Vaccine 2010, 28, 7363–7372. [Google Scholar] [CrossRef]
- Romanutti, C.; D’antuono, A.; Palacios, C.; Quattrocchi, V.; Zamorano, P.; La Torre, J.; Mattion, N. Evaluation of the immune response elicited by vaccination with viral vectors encoding FMDV capsid proteins and boosted with inactivated virus. Vet. Microbiol. 2013, 165, 333–340. [Google Scholar] [CrossRef]
- Zhang, B.; Mao, H.; Zhu, H.; Guo, J.; Zhou, P.; Ma, Z. Response to HIV-1 gp160-carrying recombinant virus HSV-1 and HIV-1 VLP combined vaccine in BALB/c mice. Front. Microbiol. 2023, 14, 1136664. [Google Scholar] [CrossRef]
- Rider, P.J.F.; Kamil, M.; Yilmaz, I.; Atmaca, H.N.; Kalkan-Yazici, M.; Doymaz, M.Z.; Kousoulas, K.G.; Aly, A.S.I. An Attenuated HSV-1-Derived Malaria Vaccine Expressing Liver-Stage Exported Proteins Induces Sterilizing Protection against Infectious Sporozoite Challenge. Vaccines 2022, 10, 300. [Google Scholar] [CrossRef] [PubMed]
- Kurt-Jones, E.A.; Dudek, T.E.; Watanabe, D.; Mandell, L.; Che, J.; Zhou, S.; Cao, L.; Greenough, T.; Babcock, G.J.; Diaz, F.; et al. Expression of SARS coronavirus 1 spike protein from a herpesviral vector induces innate immune signaling and neutralizing antibody responses. Virology 2021, 559, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Al Mana, H.; Yassine, H.M.; Younes, N.N.; Al-Mohannadi, A.; Al-Sadeq, D.W.; Alhababi, D.; Nasser, E.A.; Nasrallah, G.K.; Al Mana, H.; Al-Mohannadi, A.; et al. The Current Status of Cytomegalovirus (CMV) Prevalence in the MENA Region: A Systematic Review. Pathogens 2019, 8, 213. [Google Scholar] [CrossRef] [PubMed]
- Boeckh, M. Complications, diagnosis, management, and prevention of CMV infections: Current and future. Hematol. Am. Soc. Hematol. Educ. Program 2011, 2011, 305–309. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Erkes, D.A.; Wilski, N.A.; Snyder, C.M. Intratumoral infection by CMV may change the tumor environment by directly interacting with tumor-associated macrophages to promote cancer immunity. Hum. Vaccines Immunother. 2017, 13, 1778–1785. [Google Scholar] [CrossRef] [PubMed]
- Lagenaur, L.A.; Manning, W.C.; Vieira, J.; Martens, C.L.; Mocarski, E.S. Structure and function of the murine cytomegalovirus sgg1 gene: A determinant of viral growth in salivary gland acinar cells. J. Virol. 1994, 68, 7717–7727. [Google Scholar] [CrossRef]
- Manning, W.C.; Mocarski, E.S. Insertional mutagenesis of the murine cytomegalovirus genome: One prominent α gene (ie2) is dispensable for growth. Virology 1988, 167, 477–484. [Google Scholar] [CrossRef]
- Jurak, I.; Brune, W. Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 2006, 25, 2634–2642. [Google Scholar] [CrossRef]
- Abad-Fernandez, M.; Goonetilleke, N. Human cytomegalovirus-vectored vaccines against HIV. Curr. Opin. HIV AIDS 2019, 14, 137–142. [Google Scholar] [CrossRef]
- Ynga-Durand, M.A.; Dekhtiarenko, I.; Cicin-Sain, L. Vaccine Vectors Harnessing the Power of Cytomegaloviruses. Vaccines 2019, 7, 152. [Google Scholar] [CrossRef]
- Fleming, P.; Davis-Poynter, N.; Degli-Esposti, M.; Densley, E.; Papadimitriou, J.; Shellam, G.; Farrell, H. The murine cytomegalovirus chemokine homolog, m131/129, is a determinant of viral pathogenicity. J. Virol. 1999, 73, 6800–6809. [Google Scholar] [CrossRef] [PubMed]
- Balthesen, M.; Messerle, M.; Reddehase, M.J. Lungs are a major organ site of cytomegalovirus latency and recurrence. J. Virol. 1993, 67, 5360–5366. [Google Scholar] [CrossRef] [PubMed]
- Kurz, S.; Steffens, H.P.; Mayer, A.; Harris, J.R.; Reddehase, M.J. Latency versus persistence or intermittent recurrences: Evidence for a latent state of murine cytomegalovirus in the lungs. J. Virol. 1997, 71, 2980–2987. [Google Scholar] [CrossRef]
- Shellam, G. The potential of murine cytomegalovirus as a viral vector for immunocontraception. Reprod. Fertil. Dev. 1994, 6, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Cicin-Sain, L. Cytomegalovirus memory inflation and immune protection. Med. Microbiol. Immunol. 2019, 208, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Gabel, M.; Baumann, N.S.; Oxenius, A.; Graw, F. Investigating the Dynamics of MCMV-Specific CD8+ T Cell Responses in Individual Hosts. Front. Immunol. 2019, 10, 1358. [Google Scholar] [CrossRef]
- Liu, J.; Jaijyan, D.K.; Tang, Q.; Zhu, H. Promising Cytomegalovirus-Based Vaccine Vector Induces Robust CD8+ T-Cell Response. Int. J. Mol. Sci. 2019, 20, 4457. [Google Scholar] [CrossRef]
- Redwood, A.J.; Harvey, N.L.; Lloyd, M.; Lawson, M.A.; Hardy, C.M.; Shellam, G.R. Viral vectored immunocontraception: Screening of multiple fertility antigens using murine cytomegalovirus as a vaccine vector. Vaccine 2007, 25, 698–708. [Google Scholar] [CrossRef]
- Tang, J. Cytomegaloviruses: From Molecular Pathogenesis to Intervention. Emerg. Infect. Dis. 2013, 19, 1906. [Google Scholar] [CrossRef]
- Lawson, C.M.; Grundy, J.E.; Shellam, G.R. Antibody Responses to Murine Cytomegalovirus in Genetically Resistant and Susceptible Strains of Mice. J. Gen. Virol. 1988, 69, 1987–1998. [Google Scholar] [CrossRef]
- Foecking, M.K.; Hofstetter, H. Powerful and versatile enhancer-promoter unit for mammalian expression vectors. Gene 1986, 45, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.; Lloyd, M.; Harvey, N.; Redwood, A.; Lawson, M.; Shellam, G. Species-specificity of a murine immunocontraceptive utilising murine cytomegalovirus as a gene delivery vector. Vaccine 2005, 23, 2959–2969. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, Y.; Caposio, P.; Parkins, C.J.; Botto, S.; Messaoudi, I.; Cicin-Sain, L.; Feldmann, H.; Jarvis, M.A. A replicating cytomegalovirus-based vaccine encoding a single Ebola virus nucleoprotein CTL epitope confers protection against Ebola virus. PLoS Negl. Trop. Dis. 2011, 5, 1275. [Google Scholar] [CrossRef]
- Tsuda, Y.; Parkins, C.J.; Caposio, P.; Feldmann, F.; Botto, S.; Ball, S.; Messaoudi, I.; Cicin-Sain, L.; Feldmann, H.; Jarvis, M.A. A cytomeg-alovirus-based vaccine provides long-lasting protection against lethal Ebola virus challenge after a single dose. Vaccine 2015, 33, 2261–2266. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.; Vieville, C.; Whizin, N.; Coyne-Johnson, L.; Siess, D.; Drummond, D.; Legasse, A.; Axthelm, M.; Oswald, K.; Trubey, C.; et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immuno-deficiency virus challenge. Nat. Med. 2009, 15, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, D.G.; Gold, M.C.; Wagner, M.; Koszinowski, U.H.; Hill, A.B. The Multiple Immune-Evasion Genes of Murine Cytomegalovirus Are Not Redundant. J. Exp. Med. 2001, 194, 967–978. [Google Scholar] [CrossRef]
- Kleijnen, M.F.; Huppa, J.B.; Lucin, P.; Mukherjee, S.; Farrell, H.; Campbell, A.E.; Koszinowski, U.H.; Hill, A.B.; Ploegh, H.L. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 1997, 16, 685–694. [Google Scholar] [CrossRef]
- Reusch, U.; Muranyi, W.; Lucin, P.; Burgert, H.-G.; Hengel, H.; Koszinowski, U.H. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 1999, 18, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, H.; Muranyi, W.; Burgert, H.G.; Kremmer, E.; Koszinowski, U.H. The luminal part of the murine cytomegalovirus glycoprotein gp40 catalyzes the retention of MHC class I molecules. EMBO J. 2000, 19, 870–881. [Google Scholar] [CrossRef]
- Beverley, P.C.L.; Ruzsics, Z.; Hey, A.; Hutchings, C.; Boos, S.; Bolinger, B.; Marchi, E.; O’Hara, G.; Klenerman, P.; Koszinowski, U.H.; et al. A Novel Murine Cytomegalovirus Vaccine Vector Protects against Mycobacterium tuberculosis. J. Immunol. 2014, 193, 2306–2316. [Google Scholar] [CrossRef]
- Tierney, R.; Nakai, T.; Parkins, C.J.; Caposio, P.; Fairweather, N.F.; Sesardic, D.; Jarvis, M.A. A single-dose cytomegalovirus-based vaccine encoding tetanus toxin fragment C induces sustained levels of protective tetanus toxin antibodies in mice. Vaccine 2012, 30, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Smith, T.; Grey, F.; Hill, A.B. Cytomegalovirus-based cancer vaccines expressing TRP2 induce rejection of melanoma in mice. Biochem. Biophys. Res. Commun. 2013, 437, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, P.G.E.; Gershon, A.A. Clinical Features of Varicella-Zoster Virus Infection. Viruses 2018, 10, 609. [Google Scholar] [CrossRef] [PubMed]
- Arvin, A.M. Varicella-zoster virus. Clin. Microbiol. Rev. 1996, 9, 361–381. [Google Scholar] [CrossRef] [PubMed]
- Sorel, O.; Messaoudi, I. Varicella Virus-Host Interactions During Latency and Reactivation: Lessons From Simian Varicella Virus. Front. Microbiol. 2018, 9, 3170. [Google Scholar] [CrossRef]
- Breuer, J. Molecular Genetic Insights Into Varicella Zoster Virus (VZV), the vOka Vaccine Strain, and the Pathogenesis of Latency and Reactivation. J. Infect. Dis. 2018, 218, S75–S80. [Google Scholar] [CrossRef]
- Takahashi, M.; Otsuka, T.; Okuno, Y.; Asano, Y.; Yazaki, T.; Isomura, S. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet 1974, 304, 1288–1290. [Google Scholar] [CrossRef]
- Wu, Q.; Rivailler, P.; Xu, S.; Xu, W. Comparison of the Whole-Genome Sequence of an Oka Varicella Vaccine from China with Other Oka Vaccine Strains Reveals Sites Putatively Critical for Vaccine Efficacy. J. Virol. 2019, 93, 2281. [Google Scholar] [CrossRef]
- Takahashi, M.; Okuno, Y.; Otsuka, T.; Osame, J.; Takamizawa, A. Development of a live attenuated varicella vaccine. Biken J. 1975, 18, 25–33. [Google Scholar]
- Chaves, S.S.; Haber, P.; Walton, K.; Wise, R.P.; Izurieta, H.S.; Schmid, D.S.; Seward, J.F. Safety of varicella vaccine after licensure in the United States: Experience from reports to the vaccine adverse event reporting system. J. Infect. Dis. 2008, 197, 170–177. [Google Scholar] [CrossRef]
- Galea, S.A.; Sweet, A.; Beninger, P.; Steinberg, S.P.; LaRussa, P.S.; Gershon, A.A.; Sharrar, R.G. The safety profile of varicella vaccine: A 10-year review. J. Infect. Dis. 2008, 197, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M. Effectiveness of live varicella vaccine. Expert Opin. Biol. Ther. 2004, 4, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Tombácz, D.; Prazsák, I.; Moldován, N.; Szűcs, A.; Boldogkői, Z. Lytic Transcriptome Dataset of Varicella Zoster Virus Generated by Long-Read Sequencing. Front. Genet. 2018, 9, 460. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.I.; Seidel, K.E. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc. Natl. Acad. Sci. USA 1993, 90, 7376–7380. [Google Scholar] [CrossRef]
- Zhang, Z.; Selariu, A.; Warden, C.; Huang, G.; Huang, Y.; Zaccheus, O.; Cheng, T.; Xia, N.; Zhu, H. Genome-Wide Mutagenesis Reveals That ORF7 Is a Novel VZV Skin-Tropic Factor. PLoS Pathog. 2010, 6, e1000971. [Google Scholar] [CrossRef]
- Nagaike, K.; Mori, Y.; Gomi, Y.; Yoshii, H.; Takahashi, M.; Wagner, M.; Koszinowski, U.; Yamanishi, K. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine 2004, 22, 4069–4074. [Google Scholar] [CrossRef]
- Yoshii, H.; Somboonthum, P.; Takahashi, M.; Yamanishi, K.; Mori, Y. Cloning of full length genome of varicella-zoster virus vaccine strain into a bacterial artificial chromosome and reconstitution of infectious virus. Vaccine 2007, 25, 5006–5012. [Google Scholar] [CrossRef]
- Shiraki, K.; Hayakawa, Y.; Mori, H.; Namazue, J.; Takamizawa, A.; Yoshida, I.; Yamanishi, K.; Takahashi, M. Development of immunogenic recombinant Oka varicella vaccine expressing hepatitis B virus surface antigen. J. Gen. Virol. 1991, 72, 1393–1399. [Google Scholar] [CrossRef]
- Heineman, T.C.; Connelly, B.L.; Bourne, N.; Stanberry, L.R.; Cohen, J. Immunization with recombinant varicella-zoster virus expressing herpes simplex virus type 2 glycoprotein D reduces the severity of genital herpes in guinea pigs. J. Virol. 1995, 69, 8109–8113. [Google Scholar] [CrossRef]
- Murakami, K.; Matsuura, M.; Ota, M.; Gomi, Y.; Yamanishi, K.; Mori, Y. A recombinant varicella vaccine harboring a respiratory syncytial virus gene induces humoral immunity. Vaccine 2015, 33, 6085–6092. [Google Scholar] [CrossRef]
- Lowe, R.S.; Keller, P.M.; Keech, B.J.; Davison, A.J.; Whang, Y.; Morgan, A.J.; Kieff, E.; Ellis, R.W. Varicella-zoster virus as a live vector for the expression of foreign genes. Proc. Natl. Acad. Sci. USA 1987, 84, 3896–3900. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, M.; Somboonthum, P.; Murakami, K.; Ota, M.; Shoji, M.; Kawabata, K.; Mizuguchi, H.; Gomi, Y.; Yamanishi, K.; Mori, Y. Novel polyvalent live vaccine against varicella-zoster and mumps virus infections. Microbiol. Immunol. 2013, 57, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Somboonthum, P.; Koshizuka, T.; Okamoto, S.; Matsuura, M.; Gomi, Y.; Takahashi, M.; Yamanishi, K.; Mori, Y. Rapid and efficient introduction of a foreign gene into bacterial artificial chromosome-cloned varicella vaccine by Tn7-mediated site-specific transposition. Virology 2010, 402, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Somboonthum, P.; Yoshii, H.; Okamoto, S.; Koike, M.; Gomi, Y.; Uchiyama, Y.; Takahashi, M.; Yamanishi, K.; Mori, Y. Generation of a recombinant Oka varicella vaccine expressing mumps virus hemagglutinin-neuraminidase protein as a polyvalent live vaccine. Vaccine 2007, 25, 8741–8755. [Google Scholar] [CrossRef] [PubMed]
- Shiraki, K.; Sato, H.; Yoshida, Y.; Yamamura, J.-I.; Tsurita, M.; Kurokawa, M.; Kageyama, S. Construction of Oka varicella vaccine expressing human immunodeficiency virus env antigen. J. Med. Virol. 2001, 64, 89–95. [Google Scholar] [CrossRef]
- Heineman, T.C.; Pesnicak, L.; Ali, M.A.; Krogmann, T.; Krudwig, N.; Cohen, J.I. Varicella-zoster virus expressing HSV-2 glycoproteins B and D induces protection against HSV-2 challenge. Vaccine 2004, 22, 2558–2565. [Google Scholar] [CrossRef]
- Murakami, K.; Mori, Y. Use of a current varicella vaccine as a live polyvalent vaccine vector. Vaccine 2016, 34, 296–298. [Google Scholar] [CrossRef]
- Kim, Y.; Zheng, X.; Eschke, K.; Chaudhry, M.Z.; Bertoglio, F.; Tomić, A.; Krmpotić, A.; Hoffmann, M.; Bar-On, Y.; Boehme, J.; et al. MCMV-based vaccine vectors expressing full-length viral proteins provide long-term humoral immune protection upon a single-shot vaccination. Cell. Mol. Immunol. 2022, 19, 234–244. [Google Scholar] [CrossRef]
- Humphreys, I.R.; Sebastian, S. Novel viral vectors in infectious diseases. Immunology 2018, 153, 1–9. [Google Scholar] [CrossRef]
- Rider, P.J.F.; Musarrat, F.; Nabi, R.; Naidu, S.; Kousoulas, K.G. First Impressions—The Potential of Altering Initial Host-Virus Interactions for Rational Design of Herpesvirus Vaccine Vectors. Curr. Clin. Microbiol. Rep. 2018, 5, 55–65. [Google Scholar] [CrossRef]
- Stanfield, B.A.; Kousoulas, K.G.; Fernandez, A.; Gershburg, E. Rational Design of Live-Attenuated Vaccines against Herpes Simplex Viruses. Viruses 2021, 13, 1637. [Google Scholar] [CrossRef] [PubMed]
- bin Umair, M.; Akusa, F.N.; Kashif, H.; Fatima, S.E.; Butt, F.; Azhar, M.; Munir, I.; Ahmed, M.; Khalil, W.; Sharyar, H.; et al. Viruses as tools in gene therapy, vaccine development, and cancer treatment. Arch. Virol. 2022, 167, 1387–1404. [Google Scholar] [CrossRef] [PubMed]
- Banks, T.A.; Rouse, B.T. Herpesviruses--Immune Escape Artists? Clin. Infect. Dis. 1992, 14, 933–941. [Google Scholar] [CrossRef] [PubMed]
- York, I.A. Immune evasion strategies of the herpesviruses. Chem. Biol. 1996, 3, 331–335. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Horst, D.; Ressing, M.E.; Wiertz, E.J.H.J. Exploiting human herpesvirus immune evasion for therapeutic gain: Potential and pitfalls. Immunol. Cell Biol. 2011, 89, 359–366. [Google Scholar] [CrossRef]
- Nascimento, I.P.; Leite, L.C.C. Recombinant vaccines and the development of new vaccine strategies. Braz. J. Med. Biol. Res. 2012, 45, 1102–1111. [Google Scholar] [CrossRef]
- Sebastian, S.; Lambe, T. Clinical Advances in Viral-Vectored Influenza Vaccines. Vaccines 2018, 6, 29. [Google Scholar] [CrossRef]
- Nayak, S.; Herzog, R.W. Progress and prospects: Immune responses to viral vectors. Gene Ther. 2010, 17, 295–304. [Google Scholar] [CrossRef]
- Szpara, M.L.; Parsons, L.; Enquist, L.W. Sequence Variability in Clinical and Laboratory Isolates of Herpes Simplex Virus 1 Reveals New Mutations. J. Virol. 2010, 84, 5303–5313. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Z.; Liu, L.; Xiao, Z.; Cao, X.; Cao, Z.; Xue, L.; Miao, L.; He, X.; Li, W. A novel human foamy virus mediated gene transfer of GAD67 reduces neuropathic pain following spinal cord injury. Neurosci. Lett. 2008, 432, 13–18. [Google Scholar] [CrossRef]
- Hao, S.; Mata, M.; Goins, W.; Glorioso, J.C.; Fink, D.J. Transgene-mediated enkephalin release enhances the effect of morphine and evades tolerance to produce a sustained antiallodynic effect in neuropathic pain. Pain 2003, 102, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Guedon, J.-M.G.; Zhang, M.; Glorioso, J.C.; Goins, W.F.; Kinchington, P.R. Relief of pain induced by varicella-zoster virus in a rat model of post-herpetic neuralgia using a herpes simplex virus vector expressing enkephalin. Gene Ther. 2014, 21, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Goss, J.R.; Mata, M.; Goins, W.F.; Wu, H.H.; Glorioso, J.C.; Fink, D.J. Antinociceptive effect of a genomic herpes simplex virus-based vector expressing human proenkephalin in rat dorsal root ganglion. Gene Ther. 2001, 8, 551–556. [Google Scholar] [CrossRef] [PubMed]

| Viral Infection | Targeted Antigens | Experimental Immunization in | Immune Response | Refs. |
|---|---|---|---|---|
| HIV | Env | Guinea pigs | Strong humoral and cell-mediated immune responses | [167] |
| MuV | Hemagglutinin-neuraminidase | Guinea pigs | Specific humoral response | [166] |
| HBV | preS2 and the complete S region | Guinea pigs | Specific humoral response | [160] |
| HSV-2 | gB and gD | Guinea pigs | Significant protection (high antibody titers, reduced lesions, and reduced mortalities) | [168] |
| HSV-2 | gD | Guinea pigs | Significant protection (high antibody titers, reduced lesions, and reduced mortalities) | [161] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamel, M.S.; Munds, R.A.; Verma, M.S. The Quest for Immunity: Exploring Human Herpesviruses as Vaccine Vectors. Int. J. Mol. Sci. 2023, 24, 16112. https://doi.org/10.3390/ijms242216112
Kamel MS, Munds RA, Verma MS. The Quest for Immunity: Exploring Human Herpesviruses as Vaccine Vectors. International Journal of Molecular Sciences. 2023; 24(22):16112. https://doi.org/10.3390/ijms242216112
Chicago/Turabian StyleKamel, Mohamed S., Rachel A. Munds, and Mohit S. Verma. 2023. "The Quest for Immunity: Exploring Human Herpesviruses as Vaccine Vectors" International Journal of Molecular Sciences 24, no. 22: 16112. https://doi.org/10.3390/ijms242216112
APA StyleKamel, M. S., Munds, R. A., & Verma, M. S. (2023). The Quest for Immunity: Exploring Human Herpesviruses as Vaccine Vectors. International Journal of Molecular Sciences, 24(22), 16112. https://doi.org/10.3390/ijms242216112





