Strain Variation Can Significantly Modulate the miRNA Response to Zika Virus Infection
Abstract
:1. Introduction
2. Results
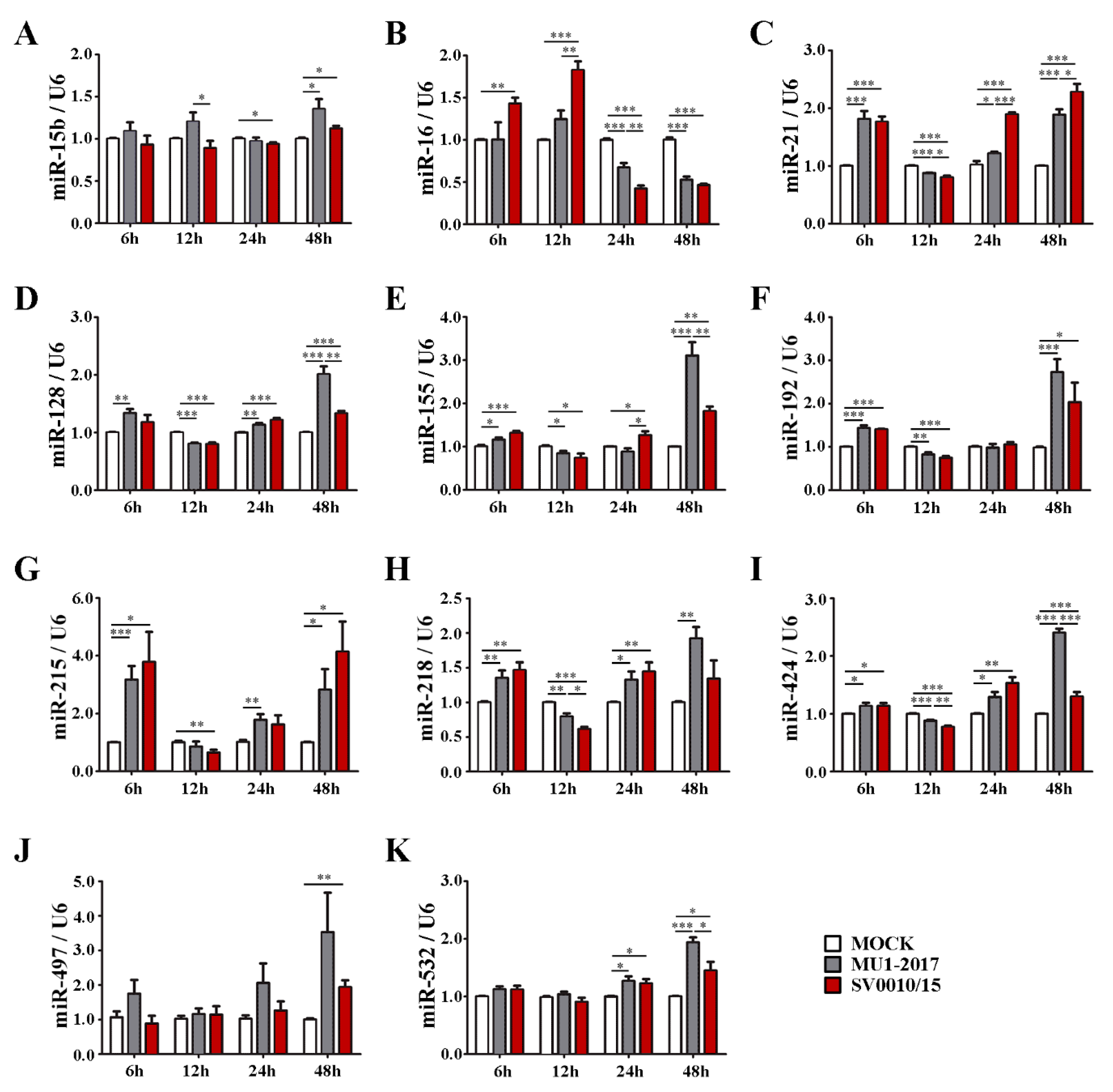
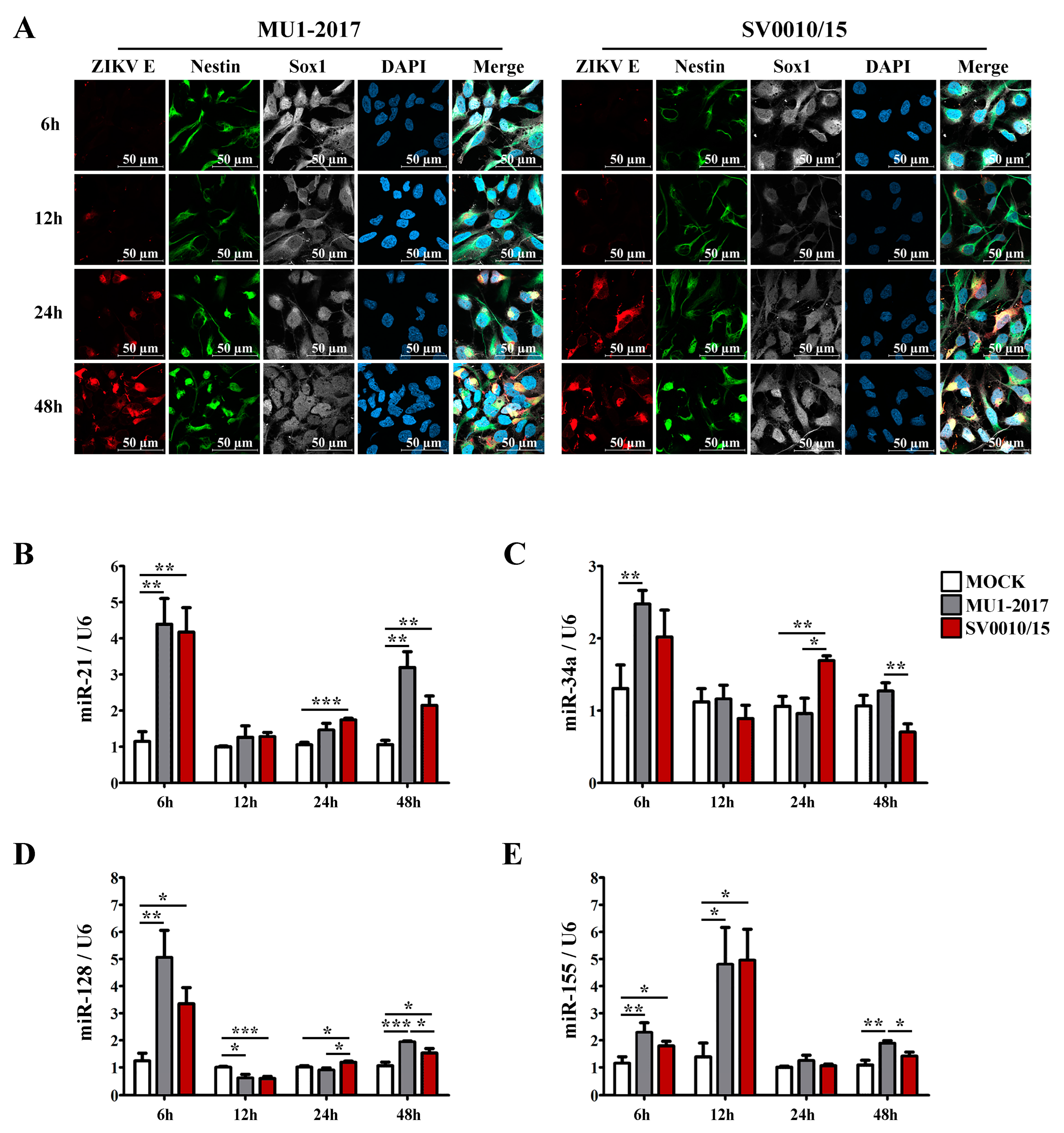
2.1. Validation of miRNA Expression Level in ZIKV-Infected Cells
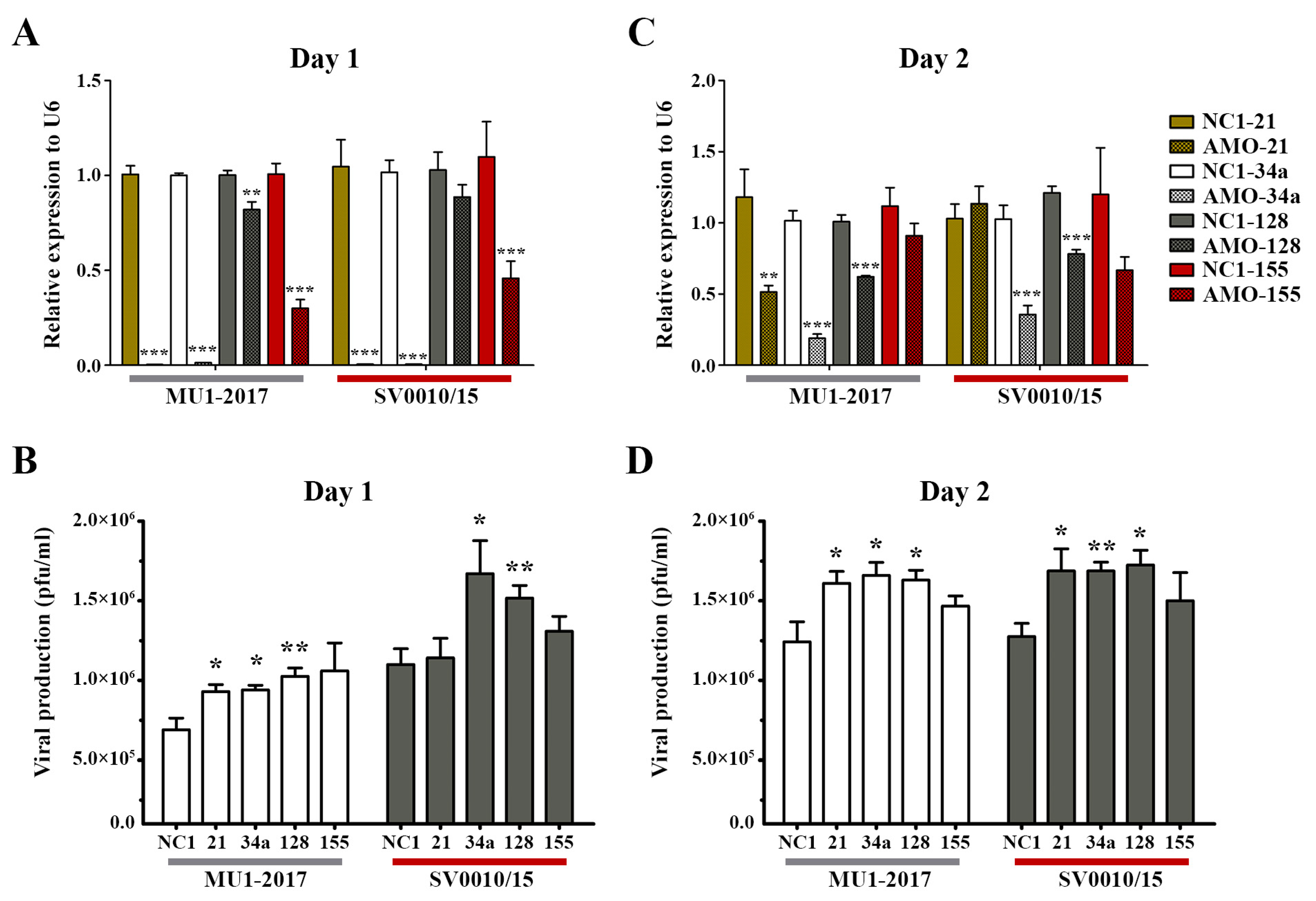
2.2. Inhibition of miRNA Expression
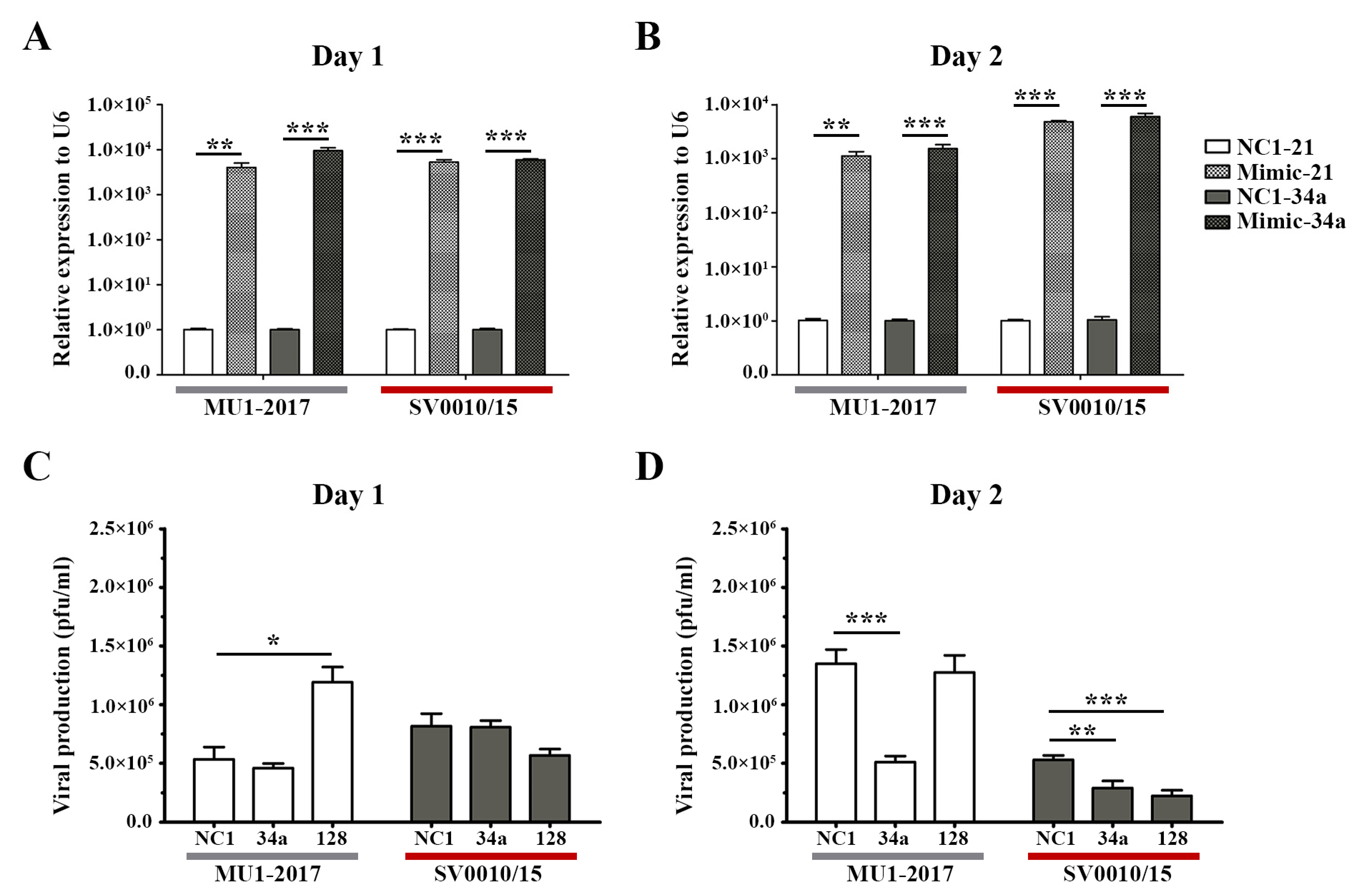
2.3. Treatment with Mimic miRNAs
2.4. Predicted Targets of miR-34a and miR-128
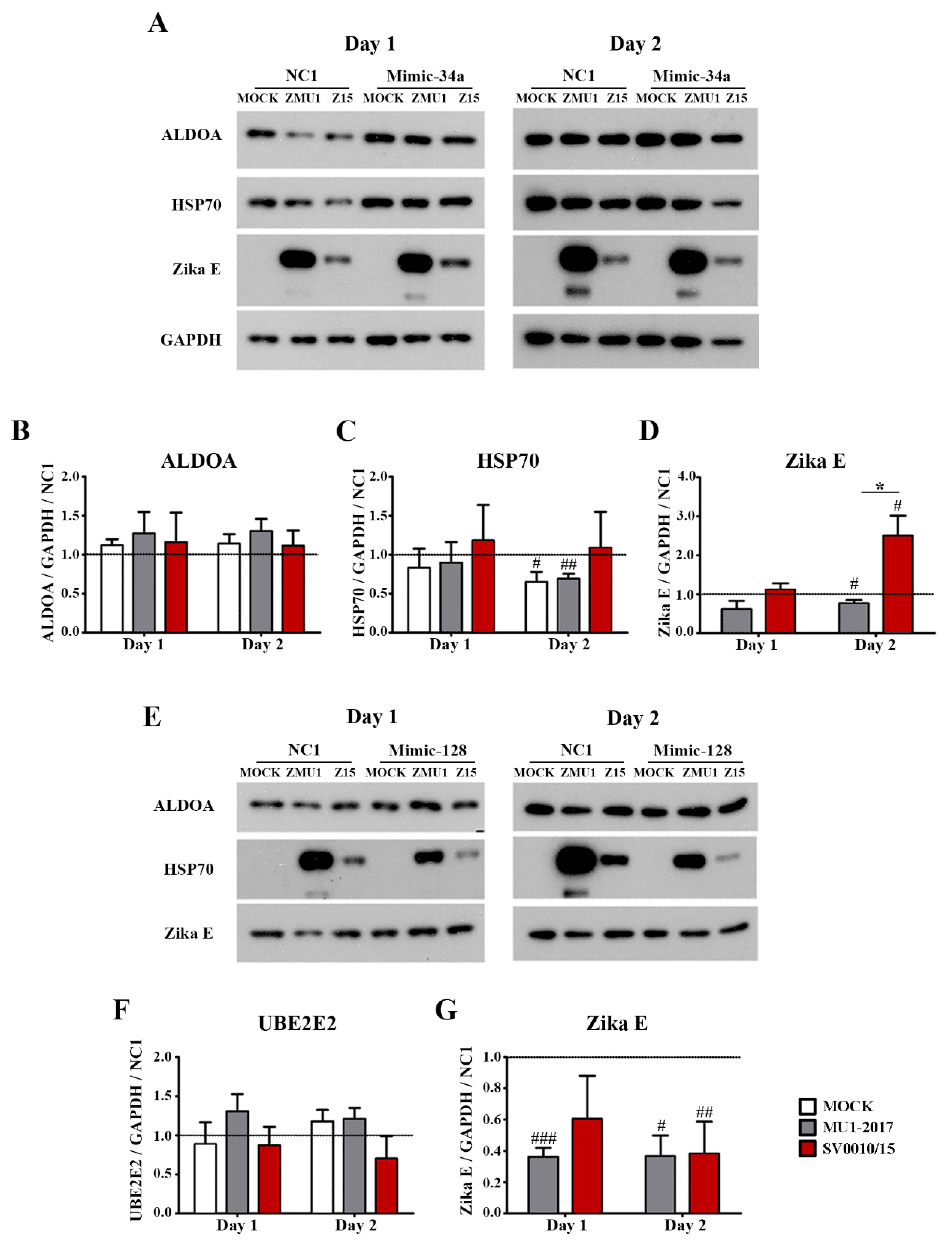
2.5. Effect of miR-34a or miR-128 Inhibition on the Predicted Targets
2.6. Effects of AMO-34a and AMO-128 on Target Protein Expression during Infection
2.7. Effect of Mimic miR-34a and miR-128 on Predicted Targets
2.8. Determination of miRNA Expression Levels in ZIKV-Infected NPCs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Neural Progenitor Cells Differentiation and Culture
4.3. Virus Propagation
4.4. Plaque Assay
4.5. Virus Infection
4.6. Flow Cytometry
4.7. Inhibition of miRNA Using Anti-miRNA Oligonucleotide (AMO) in ZIKV-Infected Cells
4.8. Mimic miRNA Transfection in ZIKV-Infected Cells
4.9. RNA Extraction and Quantitative Reverse-Transcription PCR
4.10. Protein Extraction and Western Blot Analysis
4.11. Immunofluorescence Assay
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, J.J.; Pinsky, B.A. Zika virus: Diagnostics for an Emerging Pandemic Threat. J. Clin. Microbiol. 2016, 54, 860–867. [Google Scholar] [CrossRef]
- Wikan, N.; Smith, D.R. Zika virus: History of a newly emerging arbovirus. Lancet Infect. Dis. 2016, 16, e119–e126. [Google Scholar] [CrossRef]
- Haddow, A.D.; Schuh, A.J.; Yasuda, C.Y.; Kasper, M.R.; Heang, V.; Huy, R.; Guzman, H.; Tesh, R.B.; Weaver, S.C. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLoS Negl. Trop. Dis. 2012, 6, e1477. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.M.; Roche, C.; Teissier, A.; Robin, E.; Berry, A.L.; Mallet, H.P.; Sall, A.A.; Musso, D. Zika virus, French polynesia, South pacific, 2013. Emerg. Infect. Dis. 2014, 20, 1085–1086. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Jing, W.; Liu, J.; Liu, M. The global trends and regional differences in incidence of Zika virus infection and implications for Zika virus infection prevention. PLoS Negl. Trop. Dis. 2022, 16, e0010812. [Google Scholar] [CrossRef]
- Paixao, E.S.; Barreto, F.; Teixeira Mda, G.; Costa Mda, C.; Rodrigues, L.C. History, Epidemiology, and Clinical Manifestations of Zika: A Systematic Review. Am. J. Public Health 2016, 106, 606–612. [Google Scholar] [CrossRef]
- Brasil, P.; Sequeira, P.C.; Freitas, A.D.; Zogbi, H.E.; Calvet, G.A.; de Souza, R.V.; Siqueira, A.M.; de Mendonca, M.C.; Nogueira, R.M.; de Filippis, A.M.; et al. Guillain-Barre syndrome associated with Zika virus infection. Lancet 2016, 387, 1482. [Google Scholar] [CrossRef]
- Moore, C.A.; Staples, J.E.; Dobyns, W.B.; Pessoa, A.; Ventura, C.V.; Fonseca, E.B.; Ribeiro, E.M.; Ventura, L.O.; Neto, N.N.; Arena, J.F.; et al. Characterizing the Pattern of Anomalies in Congenital Zika Syndrome for Pediatric Clinicians. JAMA Pediatr. 2017, 171, 288–295. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.J. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch. Virol. 2007, 152, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Sherman, K.E.; Rouster, S.D.; Kong, L.X.; Aliota, M.T.; Blackard, J.T.; Dean, G.E. Zika virus replication and cytopathic effects in liver cells. PLoS ONE 2019, 14, e0214016. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, E.D.; Peters, K.N.; Connor, J.H.; Bullitt, E. Zika virus induced cellular remodelling. Cell Microbiol. 2017, 19, e12740. [Google Scholar] [CrossRef]
- Serman, T.M.; Gack, M.U. Evasion of Innate and Intrinsic Antiviral Pathways by the Zika virus. Viruses 2019, 11, 970. [Google Scholar] [CrossRef]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Shivdasani, R.A. MicroRNAs: Regulators of gene expression and cell differentiation. Blood 2006, 108, 3646–3653. [Google Scholar] [CrossRef]
- Bueno, M.J.; Perez de Castro, I.; Malumbres, M. Control of cell proliferation pathways by microRNAs. Cell Cycle 2008, 7, 3143–3148. [Google Scholar] [CrossRef]
- Subramanian, S.; Steer, C.J. MicroRNAs as gatekeepers of apoptosis. J. Cell. Physiol. 2010, 223, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Escalera-Cueto, M.; Medina-Martinez, I.; del Angel, R.M.; Berumen-Campos, J.; Gutierrez-Escolano, A.L.; Yocupicio-Monroy, M. Let-7c overexpression inhibits dengue virus replication in human hepatoma Huh-7 cells. Virus Res. 2015, 196, 105–112. [Google Scholar] [CrossRef]
- Wu, N.; Gao, N.; Fan, D.; Wei, J.; Zhang, J.; An, J. miR-223 inhibits dengue virus replication by negatively regulating the microtubule-destabilizing protein STMN1 in EAhy926 cells. Microbes Infect. 2014, 16, 911–922. [Google Scholar] [CrossRef]
- Smith, J.L.; Jeng, S.; McWeeney, S.K.; Hirsch, A.J. A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection. J. Virol. 2017, 91, e02388-16. [Google Scholar] [CrossRef]
- Kanokudom, S.; Vilaivan, T.; Wikan, N.; Thepparit, C.; Smith, D.R.; Assavalapsakul, W. miR-21 promotes dengue virus serotype 2 replication in HepG2 cells. Antivir. Res. 2017, 142, 169–177. [Google Scholar] [CrossRef]
- Zhang, H.; Chang, Y.; Zhang, L.; Kim, S.N.; Otaegi, G.; Zhang, Z.; Nie, Y.; Mubarak, T.; Li, C.; Qin, C.F.; et al. Upregulation of MicroRNA miR-9 Is Associated with Microcephaly and Zika virus Infection in Mice. Mol. Neurobiol. 2019, 56, 4072–4085. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Gubler, D.J. Zika virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef] [PubMed]
- Buathong, R.; Hermann, L.; Thaisomboonsuk, B.; Rutvisuttinunt, W.; Klungthong, C.; Chinnawirotpisan, P.; Manasatienkij, W.; Nisalak, A.; Fernandez, S.; Yoon, I.K.; et al. Detection of Zika virus Infection in Thailand, 2012–2014. Am. J. Trop. Med. Hyg. 2015, 93, 380–383. [Google Scholar] [CrossRef]
- Jitsatja, A.; Ramphan, S.; Promma, P.; Kuadkitkan, A.; Wikan, N.; Uiprasertkul, M.; Phatihattakorn, C.; Smith, D.R. Comparative analysis of a Thai congenital-Zika-syndrome-associated virus with a Thai Zika-fever-associated virus. Arch. Virol. 2020, 165, 1791–1801. [Google Scholar] [CrossRef]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Bochnakian, A.; Zhen, A.; Zisoulis, D.G.; Idica, A.; KewalRamani, V.N.; Neel, N.; Daugaard, I.; Hamdorf, M.; Kitchen, S.; Lee, K.; et al. Interferon-Inducible MicroRNA miR-128 Modulates HIV-1 Replication by Targeting TNPO3 mRNA. J. Virol. 2019, 93, e00364-19. [Google Scholar] [CrossRef]
- Bondanese, V.P.; Francisco-Garcia, A.; Bedke, N.; Davies, D.E.; Sanchez-Elsner, T. Identification of host miRNAs that may limit human rhinovirus replication. World J. Biol. Chem. 2014, 5, 437–456. [Google Scholar] [CrossRef]
- Su, Y.C.; Huang, Y.F.; Wu, Y.W.; Chen, H.F.; Wu, Y.H.; Hsu, C.C.; Hsu, Y.C.; Lee, J.C. MicroRNA-155 inhibits dengue virus replication by inducing heme oxygenase-1-mediated antiviral interferon responses. FASEB J. 2020, 34, 7283–7294. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ling, Y.; Yao, Y.; Zheng, G.; Chen, W. Luteolin inhibits respiratory syncytial virus replication by regulating the MiR-155/SOCS1/STAT1 signaling pathway. Virol. J. 2020, 17, 187. [Google Scholar] [CrossRef]
- Yang, D.; Wang, X.; Gao, H.; Chen, B.; Si, C.; Wang, S. Downregulation of miR-155-5p facilitates enterovirus 71 replication through suppression of type I IFN response by targeting FOXO3/IRF7 pathway. Cell Cycle 2020, 19, 179–192. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Beshai, R.; Bibawy, D.; Bibawy, J. Guillain-Barre Syndrome Secondary to West Nile Virus in New York City. Case Rep. Infect. Dis. 2020, 2020, 6501658. [Google Scholar] [CrossRef]
- Payus, A.O.; Ibrahim, A.; Liew Sat Lin, C.; Hui Jan, T. Sensory Predominant Guillain-Barre Syndrome Concomitant with Dengue Infection: A Case Report. Case Rep. Neurol. 2022, 14, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Matos, A.M.B.; Maia Carvalho, F.M.; Malta, D.L.; Rodrigues, C.L.; Felix, A.C.; Pannuti, C.S.; Lima, A.; Esposito, D.L.A.; Dos Santos, L.M.B.; von Glehn, F.; et al. High proportion of Guillain-Barre syndrome associated with chikungunya in Northeast Brazil. Neurol. Neuroimmunol. Neuroinflamm. 2020, 7, e833. [Google Scholar] [CrossRef]
- Russo, F.B.; Jungmann, P.; Beltrao-Braga, P.C.B. Zika infection and the development of neurological defects. Cell Microbiol. 2017, 19, e12744. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007, 8, 166. [Google Scholar] [CrossRef]
- Peng, K.J.; Wang, J.H.; Su, W.T.; Wang, X.C.; Yang, F.T.; Nie, W.H. Characterization of two human lung adenocarcinoma cell lines by reciprocal chromosome painting. Dongwuxue Yanjiu 2010, 31, 113–121. [Google Scholar] [CrossRef]
- Damania, P.; Sen, B.; Dar, S.B.; Kumar, S.; Kumari, A.; Gupta, E.; Sarin, S.K.; Venugopal, S.K. Hepatitis B virus induces cell proliferation via HBx-induced microRNA-21 in hepatocellular carcinoma by targeting programmed cell death protein4 (PDCD4) and phosphatase and tensin homologue (PTEN). PLoS ONE 2014, 9, e91745. [Google Scholar] [CrossRef]
- Nasci, V.L.; Chuppa, S.; Griswold, L.; Goodreau, K.A.; Dash, R.K.; Kriegel, A.J. miR-21-5p regulates mitochondrial respiration and lipid content in H9C2 cells. Am. J. Physiol. Heart Circ. Physiol. 2019, 316, H710–H721. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Li, A.; Deng, J.; Yang, Y.; Dang, L.; Ye, Y.; Li, Y.; Zhang, W. miR-21 attenuates lipopolysaccharide-induced lipid accumulation and inflammatory response: Potential role in cerebrovascular disease. Lipids Health Dis. 2014, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cappello, T.; Wang, L. Emerging role of microRNAs in lipid metabolism. Acta Pharm. Sin. B 2015, 5, 145–150. [Google Scholar] [CrossRef]
- Villar-Palasi, C.; Larner, J. Glycogen metabolism and glycolytic enzymes. Annu. Rev. Biochem. 1970, 39, 639–672. [Google Scholar] [CrossRef]
- Schwarz, J.D.; Lukassen, S.; Bhandare, P.; Eing, L.; Snaebjornsson, M.T.; Garcia, Y.C.; Kisker, J.P.; Schulze, A.; Wolf, E. The glycolytic enzyme ALDOA and the exon junction complex protein RBM8A are regulators of ribosomal biogenesis. Front. Cell Dev. Biol. 2022, 10, 954358. [Google Scholar] [CrossRef]
- Tangsongcharoen, C.; Roytrakul, S.; Smith, D.R. Analysis of cellular proteome changes in response to ZIKV NS2B-NS3 protease expression. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef]
- Vostakolaei, M.A.; Hatami-Baroogh, L.; Babaei, G.; Molavi, O.; Kordi, S.; Abdolalizadeh, J. Hsp70 in cancer: A double agent in the battle between survival and death. J. Cell. Physiol. 2021, 236, 3420–3444. [Google Scholar] [CrossRef] [PubMed]
- Taguwa, S.; Yeh, M.T.; Rainbolt, T.K.; Nayak, A.; Shao, H.; Gestwicki, J.E.; Andino, R.; Frydman, J. Zika virus Dependence on Host Hsp70 Provides a Protective Strategy against Infection and Disease. Cell Rep. 2019, 26, 906–920.e3. [Google Scholar] [CrossRef]
- Ye, J.; Chen, Z.; Zhang, B.; Miao, H.; Zohaib, A.; Xu, Q.; Chen, H.; Cao, S. Heat shock protein 70 is associated with replicase complex of Japanese encephalitis virus and positively regulates viral genome replication. PLoS ONE 2013, 8, e75188. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Del Valle, J.; Chavez-Salinas, S.; Medina, F.; Del Angel, R.M. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J. Virol. 2005, 79, 4557–4567. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Laxminarayana, S.V.; Chandra, N.; Ravi, V.; Desai, A. Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology 2009, 385, 47–57. [Google Scholar] [CrossRef]
- Thongtan, T.; Wikan, N.; Wintachai, P.; Rattanarungsan, C.; Srisomsap, C.; Cheepsunthorn, P.; Smith, D.R. Characterization of putative Japanese encephalitis virus receptor molecules on microglial cells. J. Med. Virol. 2012, 84, 615–623. [Google Scholar] [CrossRef]
- Cabrera-Hernandez, A.; Thepparit, C.; Suksanpaisan, L.; Smith, D.R. Dengue virus entry into liver (HepG2) cells is independent of hsp90 and hsp70. J. Med. Virol. 2007, 79, 386–392. [Google Scholar] [CrossRef]
- Neutzner, M.; Neutzner, A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012, 52, 37–50. [Google Scholar] [CrossRef]
- Feng, T.; Deng, L.; Lu, X.; Pan, W.; Wu, Q.; Dai, J. Ubiquitin-conjugating enzyme UBE2J1 negatively modulates interferon pathway and promotes RNA virus infection. Virol. J. 2018, 15, 132. [Google Scholar] [CrossRef]
- Ramphan, S.; Khongwichit, S.; Saisawang, C.; Kovanich, D.; Ketterman, A.J.; Ubol, S.; Auewarakul, P.; Roytrakul, S.; Smith, D.R.; Kuadkitkan, A. Ubiquitin-Conjugating Enzyme E2 L3 is Downregulated by the Chikungunya Virus nsP2 Protease. Proteom. Clin. Appl. 2018, 12, e1700020. [Google Scholar] [CrossRef] [PubMed]
- Alpuche-Lazcano, S.P.; Saliba, J.; Costa, V.V.; Campolina-Silva, G.H.; Marim, F.M.; Ribeiro, L.S.; Blank, V.; Mouland, A.J.; Teixeira, M.M.; Gatignol, A. Profound downregulation of neural transcription factor Npas4 and Nr4a family in fetal mice neurons infected with Zika virus. PLoS Negl. Trop. Dis. 2021, 15, e0009425. [Google Scholar] [CrossRef]
- Azouz, F.; Arora, K.; Krause, K.; Nerurkar, V.R.; Kumar, M. Integrated MicroRNA and mRNA Profiling in Zika virus-Infected Neurons. Viruses 2019, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Bagasra, O.; Shamabadi, N.S.; Pandey, P.; Desoky, A.; McLean, E. Differential expression of miRNAs in a human developing neuronal cell line chronically infected with Zika virus. Libyan J. Med. 2021, 16, 1909902. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, R.; Prajapati, B.; Narwal, S.; Agnihotri, N.; Adlakha, Y.K.; Sen, J.; Mani, S.; Seth, P. Zika virus E protein alters the properties of human fetal neural stem cells by modulating microRNA circuitry. Cell Death Differ. 2018, 25, 1837–1854. [Google Scholar] [CrossRef]
- Bhagat, R.; Rajpara, P.; Kaur, G.; Gupta, K.; Seth, P. Zika virus E protein dysregulate mir-204/WNT2 signalling in human fetal neural stem cells. Brain Res. Bull. 2021, 176, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, U.; Singh, S.K. Zika virus NS1 Suppresses VE-Cadherin and Claudin-5 via hsa-miR-101-3p in Human Brain Microvascular Endothelial Cells. Mol. Neurobiol. 2021, 58, 6290–6303. [Google Scholar] [CrossRef]
- Castro, F.L.; Geddes, V.E.V.; Monteiro, F.L.L.; Goncalves, R.; Campanati, L.; Pezzuto, P.; Paquin-Proulx, D.; Schamber-Reis, B.L.; Azevedo, G.S.; Goncalves, A.L.; et al. MicroRNAs 145 and 148a Are Upregulated During Congenital Zika virus Infection. ASN Neuro 2019, 11, 1759091419850983. [Google Scholar] [CrossRef]
- Dang, J.W.; Tiwari, S.K.; Qin, Y.; Rana, T.M. Genome-wide Integrative Analysis of Zika-virus-Infected Neuronal Stem Cells Reveals Roles for MicroRNAs in Cell Cycle and Stemness. Cell Rep. 2019, 27, 3618–3628.e5. [Google Scholar] [CrossRef]
- Kozak, R.A.; Majer, A.; Biondi, M.J.; Medina, S.J.; Goneau, L.W.; Sajesh, B.V.; Slota, J.A.; Zubach, V.; Severini, A.; Safronetz, D.; et al. MicroRNA and mRNA Dysregulation in Astrocytes Infected with Zika virus. Viruses 2017, 9, 297. [Google Scholar] [CrossRef]
- Machado, F.C.; Bittar, C.; Rahal, P.; Calmon, M.F. Identification of differentially expressed miRNAs in human cells infected with different Zika virus strains. Arch. Virol. 2021, 166, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, F.B.C.; Freire, V.; Coelho, S.V.A.; Meuren, L.M.; Palmeira, J.D.F.; Cardoso, A.L.; Neves, F.A.R.; Ribeiro, B.M.; Arganaraz, G.A.; Arruda, L.B.; et al. ZIKV Strains Elicit Different Inflammatory and Anti-Viral Responses in Microglia Cells. Viruses 2023, 15, 1250. [Google Scholar] [CrossRef] [PubMed]
- Polonio, C.M.; da Silva, P.; Russo, F.B.; Hyppolito, B.R.N.; Zanluqui, N.G.; Benazzato, C.; Beltrao-Braga, P.C.B.; Muxel, S.M.; Peron, J.P.S. microRNAs Control Antiviral Immune Response, Cell Death and Chemotaxis Pathways in Human Neuronal Precursor Cells (NPCs) during Zika virus Infection. Int. J. Mol. Sci. 2022, 23, 10282. [Google Scholar] [CrossRef]
- Seong, R.K.; Lee, J.K.; Cho, G.J.; Kumar, M.; Shin, O.S. mRNA and miRNA profiling of Zika virus-infected human umbilical cord mesenchymal stem cells identifies miR-142-5p as an antiviral factor. Emerg. Microbes Infect. 2020, 9, 2061–2075. [Google Scholar] [CrossRef]
- Shukla, A.; Rastogi, M.; Singh, S.K. Zika virus NS1 suppresses the innate immune responses via miR-146a in human microglial cells. Int. J. Biol. Macromol. 2021, 193, 2290–2296. [Google Scholar] [CrossRef]
- Tabari, D.; Scholl, C.; Steffens, M.; Weickhardt, S.; Elgner, F.; Bender, D.; Herrlein, M.L.; Sabino, C.; Semkova, V.; Peitz, M.; et al. Impact of Zika virus Infection on Human Neural Stem Cell MicroRNA Signatures. Viruses 2020, 12, 1219. [Google Scholar] [CrossRef]
- Ye, H.; Kang, L.; Yan, X.; Li, S.; Huang, Y.; Mu, R.; Duan, X.; Chen, L. MiR-103a-3p Promotes Zika virus Replication by Targeting OTU Deubiquitinase 4 to Activate p38 Mitogen-Activated Protein Kinase Signaling Pathway. Front. Microbiol. 2022, 13, 862580. [Google Scholar] [CrossRef] [PubMed]
- Netsrithong, R.; Promnakhon, N.; Boonkaew, B.; Vatanashevanopakorn, C.; Pattanapanyasat, K.; Wattanapanitch, M. Generation of two induced pluripotent stem cell lines (MUSIi011-A and MUSIi011-B) from peripheral blood T lymphocytes of a healthy individual. Stem Cell Res. 2019, 39, 101487. [Google Scholar] [CrossRef]
- Wongsurawat, T.; Athipanyasilp, N.; Jenjaroenpun, P.; Jun, S.R.; Kaewnapan, B.; Wassenaar, T.M.; Leelahakorn, N.; Angkasekwinai, N.; Kantakamalakul, W.; Ussery, D.W.; et al. Case of Microcephaly after Congenital Infection with Asian Lineage Zika virus, Thailand. Emerg. Infect. Dis. 2018, 24, 1758–1761. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef]
- Choong, M.L.; Yang, H.H.; McNiece, I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp. Hematol. 2007, 35, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007, 67, 8433–8438. [Google Scholar] [CrossRef] [PubMed]
- Shell, S.; Park, S.M.; Radjabi, A.R.; Schickel, R.; Kistner, E.O.; Jewell, D.A.; Feig, C.; Lengyel, E.; Peter, M.E. Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 11400–11405. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Magelhaes, P.J.; Ram, S.J. Image Processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]


| Predicted Target | Binding Site | Aggregate PCT TargetScanHuman 7.1 | Target Score miRDB |
|---|---|---|---|
| Position 242–249 of HSPA1B 3′ UTR hsa-miR-34a-5p | 5′ CUUUAAAUGAAUCAACACUGCCA | | | | | | | 3′ UGUUGGUCGAUUCUGUGACGGU | 0.87 | 91 |
| Position 268–275 of ALDOA 3′ UTR hsa-miR-34a-5p | 5′ UCACCCUUUCCGGCACACUGCCA | | | | | | | 3′ UGUUGGUCGAUUCUGUGACGGU | 0.89 | 81 |
| Position 2466–2472 of NOTCH2 3′ UTR hsa-miR-34a-5p | 5′ UGAUGAGGAGGACAACACUGCCU | | | | | | | 3′ UGUUGGUCGAUUCUGUGACGGU | 0.73 | 84 |
| Position 92–99 of PHB 3′ UTR hsa-miR-128-3p | 5′ UCCCACCCCAGAAAUCACUGUGA | | | | | | | 3′ UUUCUCUGGCCAAGUGACACU | 0.85 | 95 |
| Position 546–552 of UBE2E2 3′ UTR hsa-miR-128-3p | 5′ AGCUUCAAUCAGAAUCACUGUGC | | | | | | | 3′ UUUCUCUGGCCAAGUGACACU | 0.95 | 99 |
| Position 1606–1613 of MSI2 3′ UTR hsa-miR-128-3p | 5′ GUAUAAACAUCACUGCACUGUGA | | | | | | | 3′ UUUCUCUGGCCAAGUGACACU | 0.86 | 97 |
| Position 4831–4837 of MSI2 3′ UTR hsa-miR-128-3p | 5′ UAAAACUUUCCCUAGCACUGUGG | | | | | | | 3′ UUUCUCUGGCCAAGUGACACU | 0.86 | 97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramphan, S.; Chumchanchira, C.; Sornjai, W.; Chailangkarn, T.; Jongkaewwattana, A.; Assavalapsakul, W.; Smith, D.R. Strain Variation Can Significantly Modulate the miRNA Response to Zika Virus Infection. Int. J. Mol. Sci. 2023, 24, 16216. https://doi.org/10.3390/ijms242216216
Ramphan S, Chumchanchira C, Sornjai W, Chailangkarn T, Jongkaewwattana A, Assavalapsakul W, Smith DR. Strain Variation Can Significantly Modulate the miRNA Response to Zika Virus Infection. International Journal of Molecular Sciences. 2023; 24(22):16216. https://doi.org/10.3390/ijms242216216
Chicago/Turabian StyleRamphan, Suwipa, Chanida Chumchanchira, Wannapa Sornjai, Thanathom Chailangkarn, Anan Jongkaewwattana, Wanchai Assavalapsakul, and Duncan R. Smith. 2023. "Strain Variation Can Significantly Modulate the miRNA Response to Zika Virus Infection" International Journal of Molecular Sciences 24, no. 22: 16216. https://doi.org/10.3390/ijms242216216
APA StyleRamphan, S., Chumchanchira, C., Sornjai, W., Chailangkarn, T., Jongkaewwattana, A., Assavalapsakul, W., & Smith, D. R. (2023). Strain Variation Can Significantly Modulate the miRNA Response to Zika Virus Infection. International Journal of Molecular Sciences, 24(22), 16216. https://doi.org/10.3390/ijms242216216






