Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer’s Disease
Abstract
1. Introduction
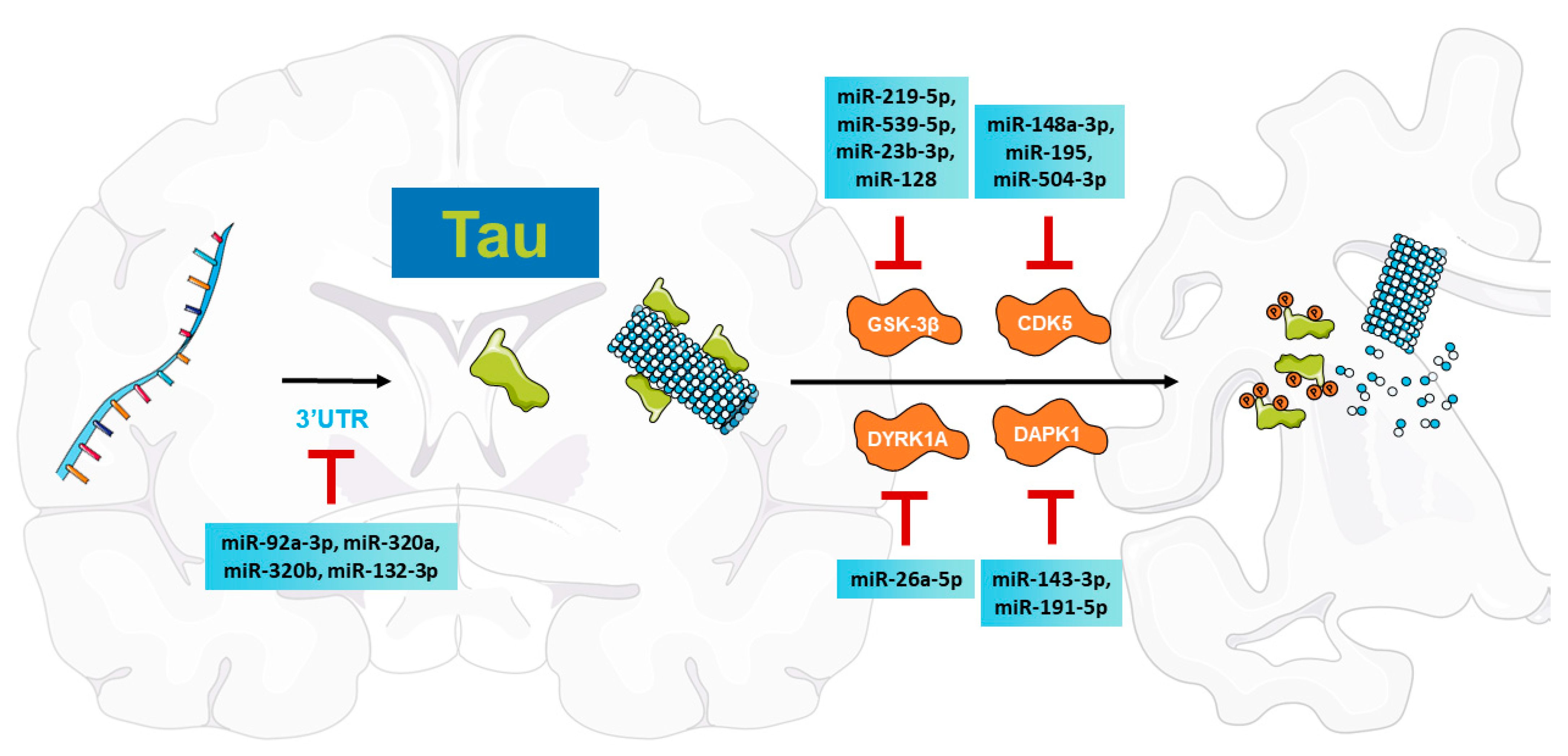
2. MiRNA-Mediated Regulation of Tau Pathologies in AD
2.1. Tau Pathologies in AD
2.2. MiRNAs Regulate MAPT Expression
2.2.1. MiRNAs Directly Modulate MAPT Transcript Levels
2.2.2. MiRNAs Regulate Tau Alternative Splicing
2.3. MiRNAs Regulate Kinases That Phosphorylate Tau
2.3.1. MiRNAs Modulate GSK-3β to Inhibit Tau Phosphorylation
2.3.2. MiRNAs Are Involved in CDK5 Regulation to Reduce Tau-Related Pathologies
2.3.3. MiRNAs Regulate DYRK1A to Suppress Tau Phosphorylation
2.3.4. MiRNAs Target DAPK1 to Attenuate Tau Pathologies
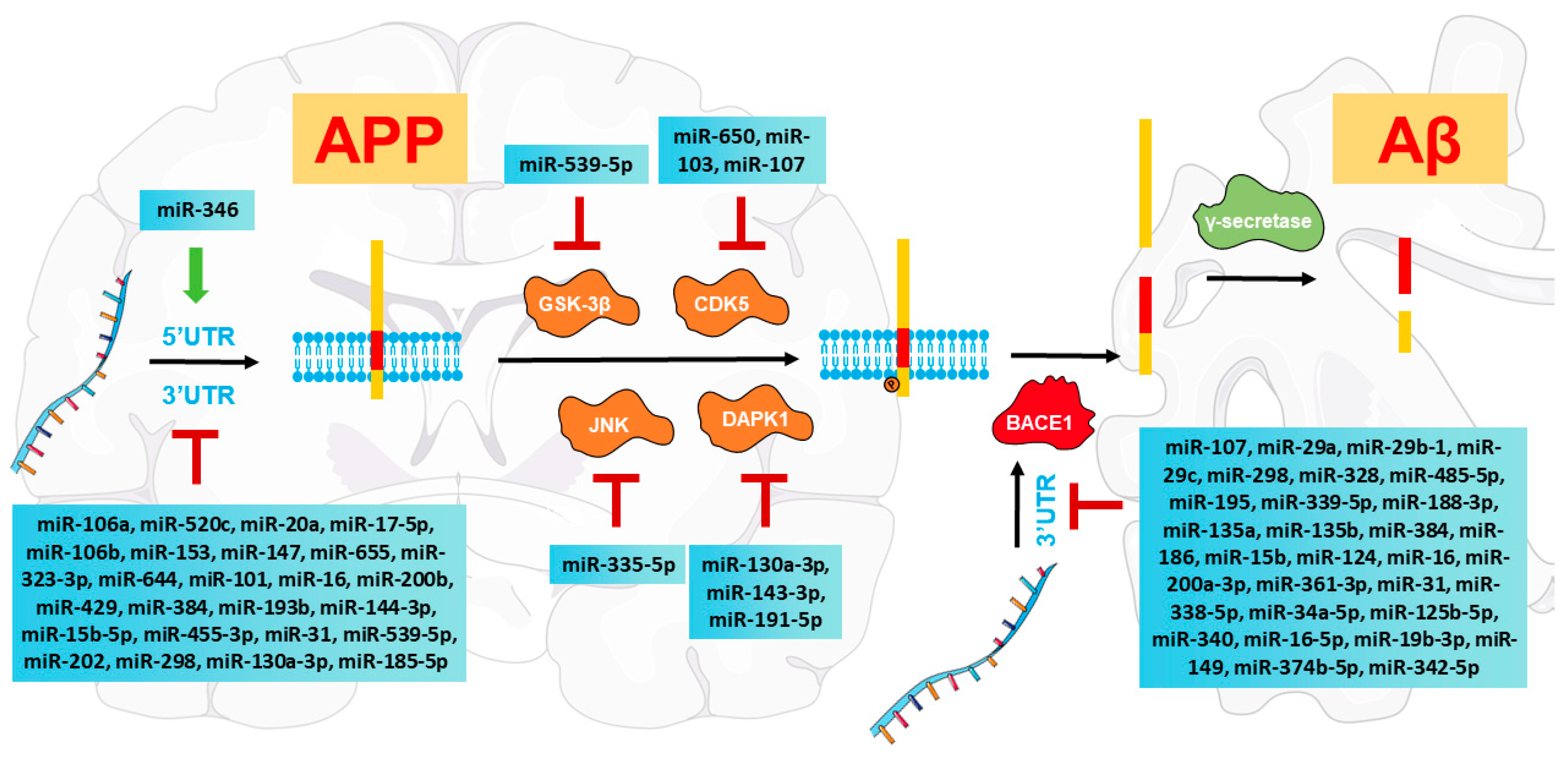
3. MiRNA-Mediated Modulation of Aβ Pathologies in AD
3.1. Amyloidogenic APP Processing in AD
3.2. MiRNAs Regulate Amyloidogenic APP Processing
3.2.1. MiRNAs Directly Modulate APP mRNA Expression
3.2.2. MiRNAs Regulate the Alternative Splicing of APP
3.2.3. MiRNAs Directly Regulate BACE1 Transcripts
3.3. MiRNAs Regulate Kinases That Phosphorylate APP at Thr668
3.3.1. MiRNAs Are Involved in GSK-3β Regulation
3.3.2. MiRNAs Modulate CDK5 to Reduce Aβ Pathologies
3.3.3. MiRNAs Target JNK to Inhibit Aβ Production
3.3.4. MiRNAs Regulate DAPK1 to Alleviate Amyloidogenic APP Processing
3.4. MiRNAs Regulate Apolipoprotein E (APOE)-Mediated Aβ Pathologies
4. MiRNA-Mediated Modulation of Inflammation in AD
4.1. MiRNAs Induce Pro-Inflammatory Responses
4.2. MiRNAs Promote Anti-Inflammatory Responses
5. Diagnostic Potential of miRNAs in AD
6. Therapeutic Potential of miRNAs in AD
7. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Partridge, L.; Deelen, J.; Slagboom, P.E. Facing up to the global challenges of ageing. Nature 2018, 561, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Pietrocola, F.; Roiz-Valle, D.; Galluzzi, L.; Kroemer, G. Meta-hallmarks of aging and cancer. Cell Metab. 2023, 35, 12–35. [Google Scholar] [CrossRef]
- Querfurth, H.W.; LaFerla, F.M. Alzheimer’s disease. N. Engl. J. Med. 2010, 362, 329–344. [Google Scholar] [CrossRef]
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chetelat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.; Chen, D.; Lee, T.H. Peptidyl-Prolyl Cis/Trans Isomerase Pin1 and Alzheimer’s Disease. Front. Cell Dev. Biol. 2020, 8, 355. [Google Scholar] [CrossRef]
- Grobler, C.; van Tongeren, M.; Gettemans, J.; Kell, D.B.; Pretorius, E. Alzheimer’s Disease: A Systems View Provides a Unifying Explanation of Its Development. J. Alzheimers Dis. 2023, 91, 43–70. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. A century of Alzheimer’s disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef]
- Ittner, L.M.; Gotz, J. Amyloid-beta and tau—A toxic pas de deux in Alzheimer’s disease. Nat. Rev. Neurosci. 2011, 12, 65–72. [Google Scholar] [CrossRef] [PubMed]
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grasser, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stahler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Iacomino, G. miRNAs: The Road from Bench to Bedside. Genes 2023, 14, 314. [Google Scholar] [CrossRef]
- Nowak, J.S.; Michlewski, G. miRNAs in development and pathogenesis of the nervous system. Biochem. Soc. Trans. 2013, 41, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Eacker, S.M.; Dawson, T.M.; Dawson, V.L. Understanding microRNAs in neurodegeneration. Nat. Rev. Neurosci. 2009, 10, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Swarbrick, S.; Wragg, N.; Ghosh, S.; Stolzing, A. Systematic Review of miRNA as Biomarkers in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 6156–6167. [Google Scholar] [CrossRef]
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [Google Scholar] [CrossRef]
- Iqbal, K.; Liu, F.; Gong, C.X. Tau and neurodegenerative disease: The story so far. Nat. Rev. Neurol. 2016, 12, 15–27. [Google Scholar] [CrossRef]
- Martin, L.; Latypova, X.; Wilson, C.M.; Magnaudeix, A.; Perrin, M.L.; Yardin, C.; Terro, F. Tau protein kinases: Involvement in Alzheimer’s disease. Ageing Res. Rev. 2013, 12, 289–309. [Google Scholar] [CrossRef] [PubMed]
- Almansoub, H.; Tang, H.; Wu, Y.; Wang, D.Q.; Mahaman, Y.A.R.; Wei, N.; Almansob, Y.A.M.; He, W.; Liu, D. Tau Abnormalities and the Potential Therapy in Alzheimer’s Disease. J. Alzheimers Dis. 2019, 67, 13–33. [Google Scholar] [CrossRef]
- Montalto, G.; Ricciarelli, R. Tau, tau kinases, and tauopathies: An updated overview. Biofactors 2023, 49, 502–511. [Google Scholar] [CrossRef]
- Lindwall, G.; Cole, R.D. Phosphorylation affects the ability of tau protein to promote microtubule assembly. J. Biol. Chem. 1984, 259, 5301–5305. [Google Scholar] [CrossRef]
- Avila, J.; Lucas, J.J.; Perez, M.; Hernandez, F. Role of tau protein in both physiological and pathological conditions. Physiol. Rev. 2004, 84, 361–384. [Google Scholar] [CrossRef]
- Alonso, A.C.; Zaidi, T.; Grundke-Iqbal, I.; Iqbal, K. Role of abnormally phosphorylated tau in the breakdown of microtubules in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1994, 91, 5562–5566. [Google Scholar] [CrossRef]
- Lee, V.M.; Balin, B.J.; Otvos, L., Jr.; Trojanowski, J.Q. A68: A major subunit of paired helical filaments and derivatized forms of normal Tau. Science 1991, 251, 675–678. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Cairns, N.J.; Crowther, R.A. Tau proteins of Alzheimer paired helical filaments: Abnormal phosphorylation of all six brain isoforms. Neuron 1992, 8, 159–168. [Google Scholar] [CrossRef]
- Matsuo, E.S.; Shin, R.W.; Billingsley, M.L.; Van deVoorde, A.; O’Connor, M.; Trojanowski, J.Q.; Lee, V.M. Biopsy-derived adult human brain tau is phosphorylated at many of the same sites as Alzheimer’s disease paired helical filament tau. Neuron 1994, 13, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Ballatore, C.; Lee, V.M.; Trojanowski, J.Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 2007, 8, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Alafuzoff, I.; Bigio, E.H.; Bouras, C.; Braak, H.; Cairns, N.J.; Castellani, R.J.; Crain, B.J.; Davies, P.; Del Tredici, K.; et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J. Neuropathol. Exp. Neurol. 2012, 71, 362–381. [Google Scholar] [CrossRef]
- Arnsten, A.F.T.; Datta, D.; Del Tredici, K.; Braak, H. Hypothesis: Tau pathology is an initiating factor in sporadic Alzheimer’s disease. Alzheimers Dement. 2021, 17, 115–124. [Google Scholar] [CrossRef]
- Piscopo, P.; Grasso, M.; Manzini, V.; Zeni, A.; Castelluzzo, M.; Fontana, F.; Talarico, G.; Castellano, A.E.; Rivabene, R.; Crestini, A.; et al. Identification of miRNAs regulating MAPT expression and their analysis in plasma of patients with dementia. Front. Mol. Neurosci. 2023, 16, 1127163. [Google Scholar] [CrossRef] [PubMed]
- Denk, J.; Oberhauser, F.; Kornhuber, J.; Wiltfang, J.; Fassbender, K.; Schroeter, M.L.; Volk, A.E.; Diehl-Schmid, J.; Prudlo, J.; Danek, A.; et al. Specific serum and CSF microRNA profiles distinguish sporadic behavioural variant of frontotemporal dementia compared with Alzheimer patients and cognitively healthy controls. PLoS ONE 2018, 13, e0197329. [Google Scholar] [CrossRef]
- Pena-Bautista, C.; Tarazona-Sanchez, A.; Braza-Boils, A.; Balaguer, A.; Ferre-Gonzalez, L.; Canada-Martinez, A.J.; Baquero, M.; Chafer-Pericas, C. Plasma microRNAs as potential biomarkers in early Alzheimer disease expression. Sci. Rep. 2022, 12, 15589. [Google Scholar] [CrossRef]
- Siedlecki-Wullich, D.; Catala-Solsona, J.; Fabregas, C.; Hernandez, I.; Clarimon, J.; Lleo, A.; Boada, M.; Saura, C.A.; Rodriguez-Alvarez, J.; Minano-Molina, A.J. Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer’s disease. Alzheimers Res. Ther. 2019, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.J.; Wong, B.Y.X.; Vaidyanathan, R.; Sreejith, S.; Chia, S.Y.; Kandiah, N.; Ng, A.S.L.; Zeng, L. Altered Cerebrospinal Fluid Exosomal microRNA Levels in Young-Onset Alzheimer’s Disease and Frontotemporal Dementia. J. Alzheimers Dis. Rep. 2021, 5, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, S.; Laskowska-Kaszub, K.; Debski, K.J.; Wojsiat, J.; Dabrowski, M.; Gabryelewicz, T.; Kuznicki, J.; Wojda, U. Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer’s disease patients from non-demented subjects. Oncotarget 2017, 8, 16122–16143. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.Y.; Hernandez-Rapp, J.; Jolivette, F.; Lecours, C.; Bisht, K.; Goupil, C.; Dorval, V.; Parsi, S.; Morin, F.; Planel, E.; et al. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 2015, 24, 6721–6735. [Google Scholar] [CrossRef]
- Wu, H.; Huang, M.; Lu, M.; Zhu, W.; Shu, Y.; Cao, P.; Liu, P. Regulation of microtubule-associated protein tau (MAPT) by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel. Cancer Chemother. Pharmacol. 2013, 71, 1159–1171. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Z.; Sun, L.; Fang, Y.; Xu, X.; Zhou, G. miR-186 regulates chemo-sensitivity to paclitaxel via targeting MAPT in non-small cell lung cancer (NSCLC). Mol. Biosyst. 2016, 12, 3417–3424. [Google Scholar] [CrossRef]
- Li, J.W.; Ren, S.H.; Ren, J.R.; Zhen, Z.G.; Li, L.R.; Hao, X.D.; Ji, H.M. Nimodipine Improves Cognitive Impairment After Subarachnoid Hemorrhage in Rats Through IncRNA NEAT1/miR-27a/MAPT Axis. Drug Des. Devel. Ther. 2020, 14, 2295–2306. [Google Scholar] [CrossRef]
- Qian, W.; Liu, F. Regulation of alternative splicing of tau exon 10. Neurosci. Bull. 2014, 30, 367–377. [Google Scholar] [CrossRef]
- Liu, F.; Gong, C.X. Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 2008, 3, 8. [Google Scholar] [CrossRef]
- Smith, P.Y.; Delay, C.; Girard, J.; Papon, M.A.; Planel, E.; Sergeant, N.; Buee, L.; Hebert, S.S. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum. Mol. Genet. 2011, 20, 4016–4024. [Google Scholar] [CrossRef]
- Salta, E.; Sierksma, A.; Vanden Eynden, E.; De Strooper, B. miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer’s brain. EMBO Mol. Med. 2016, 8, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- El Fatimy, R.; Li, S.; Chen, Z.; Mushannen, T.; Gongala, S.; Wei, Z.; Balu, D.T.; Rabinovsky, R.; Cantlon, A.; Elkhal, A.; et al. MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol. 2018, 136, 537–555. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, D.; Huang, H.Z.; Wang, Z.H.; Hou, T.Y.; Yang, X.; Pang, P.; Wei, N.; Zhou, Y.F.; Dupras, M.J.; et al. A Novel MicroRNA-124/PTPN1 Signal Pathway Mediates Synaptic and Memory Deficits in Alzheimer’s Disease. Biol. Psychiatry 2018, 83, 395–405. [Google Scholar] [CrossRef]
- Hou, T.Y.; Zhou, Y.; Zhu, L.S.; Wang, X.; Pang, P.; Wang, D.Q.; Liuyang, Z.Y.; Man, H.; Lu, Y.; Zhu, L.Q.; et al. Correcting abnormalities in miR-124/PTPN1 signaling rescues tau pathology in Alzheimer’s disease. J. Neurochem. 2020, 154, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef]
- Subramanian, M.; Hyeon, S.J.; Das, T.; Suh, Y.S.; Kim, Y.K.; Lee, J.S.; Song, E.J.; Ryu, H.; Yu, K. UBE4B, a microRNA-9 target gene, promotes autophagy-mediated Tau degradation. Nat. Commun. 2021, 12, 3291. [Google Scholar] [CrossRef]
- Geekiyanage, H.; Chan, C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. J. Neurosci. 2011, 31, 14820–14830. [Google Scholar] [CrossRef]
- Jiang, Y.; Xu, B.; Chen, J.; Sui, Y.; Ren, L.; Li, J.; Zhang, H.; Guo, L.; Sun, X. Micro-RNA-137 Inhibits Tau Hyperphosphorylation in Alzheimer’s Disease and Targets the CACNA1C Gene in Transgenic Mice and Human Neuroblastoma SH-SY5Y Cells. Med. Sci. Monit. 2018, 24, 5635–5644. [Google Scholar] [CrossRef]
- Sayas, C.L.; Avila, J. GSK-3 and Tau: A Key Duet in Alzheimer’s Disease. Cells 2021, 10, 721. [Google Scholar] [CrossRef]
- Leroy, K.; Yilmaz, Z.; Brion, J.P. Increased level of active GSK-3beta in Alzheimer’s disease and accumulation in argyrophilic grains and in neurones at different stages of neurofibrillary degeneration. Neuropathol. Appl. Neurobiol. 2007, 33, 43–55. [Google Scholar] [CrossRef]
- Li, J.; Chen, W.; Yi, Y.; Tong, Q. miR-219-5p inhibits tau phosphorylation by targeting TTBK1 and GSK-3beta in Alzheimer’s disease. J. Cell. Biochem. 2019, 120, 9936–9946. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, Y.; Su, L. MiR-539-5p Decreases amyloid beta-protein production, hyperphosphorylation of Tau and Memory Impairment by Regulating PI3K/Akt/GSK-3beta Pathways in APP/PS1 Double Transgenic Mice. Neurotox. Res. 2020, 38, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Liu, J.; Guo, S.; Zeng, L.; Cai, Z.; Zhang, J.; Wang, L.; Li, Z.; Liu, R. miR-23b-3p rescues cognition in Alzheimer’s disease by reducing tau phosphorylation and apoptosis via GSK-3beta signaling pathways. Mol. Ther. Nucleic Acids 2022, 28, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Poon, C.H.; Zhang, Z.; Yue, M.; Chen, R.; Zhang, Y.; Hossain, M.F.; Pan, Y.; Zhao, J.; Rong, L.; et al. MicroRNA-128 suppresses tau phosphorylation and reduces amyloid-beta accumulation by inhibiting the expression of GSK3beta, APPBP2, and mTOR in Alzheimer’s disease. CNS Neurosci. Ther. 2023, 29, 1848–1864. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L.; Wang, C.; Jiang, T.; Tan, L.; Xing, A.; Yu, J.T. The Role of Cdk5 in Alzheimer’s Disease. Mol. Neurobiol. 2016, 53, 4328–4342. [Google Scholar] [CrossRef]
- Maitra, S.; Vincent, B. Cdk5-p25 as a key element linking amyloid and tau pathologies in Alzheimer’s disease: Mechanisms and possible therapeutic interventions. Life Sci. 2022, 308, 120986. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.; Skuntz, S.; Pant, H.C. Deregulated Cdk5 activity is involved in inducing Alzheimer’s disease. Arch. Med. Res. 2012, 43, 655–662. [Google Scholar] [CrossRef]
- Noble, W.; Olm, V.; Takata, K.; Casey, E.; Mary, O.; Meyerson, J.; Gaynor, K.; LaFrancois, J.; Wang, L.; Kondo, T.; et al. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron 2003, 38, 555–565. [Google Scholar] [CrossRef]
- Zeng, L.; Jiang, H.; Ashraf, G.M.; Liu, J.; Wang, L.; Zhao, K.; Liu, M.; Li, Z.; Liu, R. Implications of miR-148a-3p/p35/PTEN signaling in tau hyperphosphorylation and autoregulatory feedforward of Akt/CREB in Alzheimer’s disease. Mol. Ther. Nucleic Acids 2022, 27, 256–275. [Google Scholar] [CrossRef]
- Sun, L.H.; Ban, T.; Liu, C.D.; Chen, Q.X.; Wang, X.; Yan, M.L.; Hu, X.L.; Su, X.L.; Bao, Y.N.; Sun, L.L.; et al. Activation of Cdk5/p25 and tau phosphorylation following chronic brain hypoperfusion in rats involves microRNA-195 down-regulation. J. Neurochem. 2015, 134, 1139–1151. [Google Scholar] [CrossRef]
- Chen, D.; Lan, G.; Li, R.; Mei, Y.; Shui, X.; Gu, X.; Wang, L.; Zhang, T.; Gan, C.L.; Xia, Y.; et al. Melatonin ameliorates tau-related pathology via the miR-504-3p and CDK5 axis in Alzheimer’s disease. Transl. Neurodegener. 2022, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Wegiel, J.; Gong, C.X.; Hwang, Y.W. The role of DYRK1A in neurodegenerative diseases. FEBS J. 2011, 278, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Kimura, R.; Kamino, K.; Yamamoto, M.; Nuripa, A.; Kida, T.; Kazui, H.; Hashimoto, R.; Tanaka, T.; Kudo, T.; Yamagata, H.; et al. The DYRK1A gene, encoded in chromosome 21 Down syndrome critical region, bridges between beta-amyloid production and tau phosphorylation in Alzheimer disease. Hum. Mol. Genet. 2007, 16, 15–23. [Google Scholar] [CrossRef]
- Ryoo, S.R.; Jeong, H.K.; Radnaabazar, C.; Yoo, J.J.; Cho, H.J.; Lee, H.W.; Kim, I.S.; Cheon, Y.H.; Ahn, Y.S.; Chung, S.H.; et al. DYRK1A-mediated hyperphosphorylation of Tau. A functional link between Down syndrome and Alzheimer disease. J. Biol. Chem. 2007, 282, 34850–34857. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Xie, F.; Wang, X.; Hou, Y.; Wang, X.; Liu, J. Overexpression of miR-26a-5p Suppresses Tau Phosphorylation and Abeta Accumulation in the Alzheimer’s Disease Mice by Targeting DYRK1A. Curr. Neurovasc Res. 2020, 17, 241–248. [Google Scholar] [CrossRef]
- Chen, D.; Zhou, X.Z.; Lee, T.H. Death-Associated Protein Kinase 1 as a Promising Drug Target in Cancer and Alzheimer’s Disease. Recent. Pat. Anticancer. Drug Discov. 2019, 14, 144–157. [Google Scholar] [CrossRef]
- Kim, N.; Chen, D.; Zhou, X.Z.; Lee, T.H. Death-Associated Protein Kinase 1 Phosphorylation in Neuronal Cell Death and Neurodegenerative Disease. Int. J. Mol. Sci. 2019, 20, 3131. [Google Scholar] [CrossRef]
- Li, Y.; Grupe, A.; Rowland, C.; Nowotny, P.; Kauwe, J.S.; Smemo, S.; Hinrichs, A.; Tacey, K.; Toombs, T.A.; Kwok, S.; et al. DAPK1 variants are associated with Alzheimer’s disease and allele-specific expression. Hum. Mol. Genet. 2006, 15, 2560–2568. [Google Scholar] [CrossRef]
- Li, H.; Wetten, S.; Li, L.; St Jean, P.L.; Upmanyu, R.; Surh, L.; Hosford, D.; Barnes, M.R.; Briley, J.D.; Borrie, M.; et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch. Neurol. 2008, 65, 45–53. [Google Scholar] [CrossRef]
- Laumet, G.; Chouraki, V.; Grenier-Boley, B.; Legry, V.; Heath, S.; Zelenika, D.; Fievet, N.; Hannequin, D.; Delepine, M.; Pasquier, F.; et al. Systematic analysis of candidate genes for Alzheimer’s disease in a French, genome-wide association study. J. Alzheimers Dis. 2010, 20, 1181–1188. [Google Scholar] [CrossRef]
- Gaj, P.; Paziewska, A.; Bik, W.; Dabrowska, M.; Baranowska-Bik, A.; Styczynska, M.; Chodakowska-Zebrowska, M.; Pfeffer-Baczuk, A.; Barcikowska, M.; Baranowska, B.; et al. Identification of a late onset Alzheimer’s disease candidate risk variant at 9q21.33 in Polish patients. J. Alzheimers Dis. 2012, 32, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.M.; You, M.H.; Chen, C.H.; Lee, S.; Hong, Y.; Hong, Y.; Kimchi, A.; Zhou, X.Z.; Lee, T.H. Death-associated protein kinase 1 has a critical role in aberrant tau protein regulation and function. Cell Death Dis. 2014, 5, e1237. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.M.; You, M.H.; Chen, C.H.; Suh, J.; Tanzi, R.E.; Ho Lee, T. Inhibition of death-associated protein kinase 1 attenuates the phosphorylation and amyloidogenic processing of amyloid precursor protein. Hum. Mol. Genet. 2016, 25, 2498–2513. [Google Scholar] [CrossRef]
- You, M.H.; Kim, B.M.; Chen, C.H.; Begley, M.J.; Cantley, L.C.; Lee, T.H. Death-associated protein kinase 1 phosphorylates NDRG2 and induces neuronal cell death. Cell Death Differ. 2017, 24, 238–250. [Google Scholar] [CrossRef]
- Nakamura, K.; Greenwood, A.; Binder, L.; Bigio, E.H.; Denial, S.; Nicholson, L.; Zhou, X.Z.; Lu, K.P. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer’s disease. Cell 2012, 149, 232–244. [Google Scholar] [CrossRef]
- Kondo, A.; Shahpasand, K.; Mannix, R.; Qiu, J.; Moncaster, J.; Chen, C.H.; Yao, Y.; Lin, Y.M.; Driver, J.A.; Sun, Y.; et al. Antibody against early driver of neurodegeneration cis P-tau blocks brain injury and tauopathy. Nature 2015, 523, 431–436. [Google Scholar] [CrossRef]
- Wang, R.; Lu, K.P.; Zhou, X.Z. Function and regulation of cis P-tau in the pathogenesis and treatment of conventional and nonconventional tauopathies. J. Neurochem. 2023, 166, 904–914. [Google Scholar] [CrossRef]
- Wang, L.; Shui, X.; Mei, Y.; Xia, Y.; Lan, G.; Hu, L.; Zhang, M.; Gan, C.L.; Li, R.; Tian, Y.; et al. miR-143-3p Inhibits Aberrant Tau Phosphorylation and Amyloidogenic Processing of APP by Directly Targeting DAPK1 in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 7992. [Google Scholar] [CrossRef]
- Wang, L.; Shui, X.; Zhang, M.; Mei, Y.; Xia, Y.; Lan, G.; Hu, L.; Gan, C.L.; Tian, Y.; Li, R.; et al. MiR-191-5p Attenuates Tau Phosphorylation, Abeta Generation, and Neuronal Cell Death by Regulating Death-Associated Protein Kinase 1. ACS Chem. Neurosci. 2022, 13, 3554–3566. [Google Scholar] [CrossRef]
- Hardy, J.; Allsop, D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 1991, 12, 383–388. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Goldgaber, D.; Lerman, M.I.; McBride, O.W.; Saffiotti, U.; Gajdusek, D.C. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer’s disease. Science 1987, 235, 877–880. [Google Scholar] [CrossRef] [PubMed]
- Tanzi, R.E.; Gusella, J.F.; Watkins, P.C.; Bruns, G.A.; St George-Hyslop, P.; Van Keuren, M.L.; Patterson, D.; Pagan, S.; Kurnit, D.M.; Neve, R.L. Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science 1987, 235, 880–884. [Google Scholar] [CrossRef]
- Zheng, H.; Koo, E.H. The amyloid precursor protein: Beyond amyloid. Mol. Neurodegener. 2006, 1, 5. [Google Scholar] [CrossRef][Green Version]
- Rovelet-Lecrux, A.; Hannequin, D.; Raux, G.; Le Meur, N.; Laquerriere, A.; Vital, A.; Dumanchin, C.; Feuillette, S.; Brice, A.; Vercelletto, M.; et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006, 38, 24–26. [Google Scholar] [CrossRef]
- Sleegers, K.; Brouwers, N.; Gijselinck, I.; Theuns, J.; Goossens, D.; Wauters, J.; Del-Favero, J.; Cruts, M.; van Duijn, C.M.; Van Broeckhoven, C. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain 2006, 129, 2977–2983. [Google Scholar] [CrossRef]
- Vetrivel, K.S.; Thinakaran, G. Amyloidogenic processing of beta-amyloid precursor protein in intracellular compartments. Neurology 2006, 66, S69–S73. [Google Scholar] [CrossRef]
- Thinakaran, G.; Koo, E.H. Amyloid precursor protein trafficking, processing, and function. J. Biol. Chem. 2008, 283, 29615–29619. [Google Scholar] [CrossRef]
- Cohen, M.L.; Golde, T.E.; Usiak, M.F.; Younkin, L.H.; Younkin, S.G. In situ hybridization of nucleus basalis neurons shows increased beta-amyloid mRNA in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1988, 85, 1227–1231. [Google Scholar] [CrossRef]
- Higgins, G.A.; Lewis, D.A.; Bahmanyar, S.; Goldgaber, D.; Gajdusek, D.C.; Young, W.G.; Morrison, J.H.; Wilson, M.C. Differential regulation of amyloid-beta-protein mRNA expression within hippocampal neuronal subpopulations in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1988, 85, 1297–1301. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, D.; Lee, T.H. Phosphorylation Signaling in APP Processing in Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 21, 209. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Kao, S.C.; Lemere, C.A.; Xia, W.; Tseng, H.C.; Zhou, Y.; Neve, R.; Ahlijanian, M.K.; Tsai, L.H. APP processing is regulated by cytoplasmic phosphorylation. J. Cell Biol. 2003, 163, 83–95. [Google Scholar] [CrossRef]
- Suzuki, T.; Nakaya, T. Regulation of amyloid beta-protein precursor by phosphorylation and protein interactions. J. Biol. Chem. 2008, 283, 29633–29637. [Google Scholar] [CrossRef] [PubMed]
- Tamayev, R.; Zhou, D.; D’Adamio, L. The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol. Neurodegener. 2009, 4, 28. [Google Scholar] [CrossRef]
- Patel, N.; Hoang, D.; Miller, N.; Ansaloni, S.; Huang, Q.; Rogers, J.T.; Lee, J.C.; Saunders, A.J. MicroRNAs can regulate human APP levels. Mol. Neurodegener. 2008, 3, 10. [Google Scholar] [CrossRef]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Bergmans, B.; Papadopoulou, A.S.; Delacourte, A.; De Strooper, B. MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiol. Dis. 2009, 33, 422–428. [Google Scholar] [CrossRef]
- Liang, C.; Zhu, H.; Xu, Y.; Huang, L.; Ma, C.; Deng, W.; Liu, Y.; Qin, C. MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 2012, 1455, 103–113. [Google Scholar] [CrossRef]
- Long, J.M.; Ray, B.; Lahiri, D.K. MicroRNA-153 physiologically inhibits expression of amyloid-beta precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 2012, 287, 31298–31310. [Google Scholar] [CrossRef]
- Delay, C.; Calon, F.; Mathews, P.; Hebert, S.S. Alzheimer-specific variants in the 3’UTR of Amyloid precursor protein affect microRNA function. Mol. Neurodegener. 2011, 6, 70. [Google Scholar] [CrossRef]
- Vilardo, E.; Barbato, C.; Ciotti, M.; Cogoni, C.; Ruberti, F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J. Biol. Chem. 2010, 285, 18344–18351. [Google Scholar] [CrossRef] [PubMed]
- Long, J.M.; Lahiri, D.K. MicroRNA-101 downregulates Alzheimer’s amyloid-beta precursor protein levels in human cell cultures and is differentially expressed. Biochem. Biophys. Res. Commun. 2011, 404, 889–895. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.; Zhu, J.; Shu, P.; Yin, B.; Gong, Y.; Qiang, B.; Yuan, J.; Peng, X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 2012, 33, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Parsi, S.; Smith, P.Y.; Goupil, C.; Dorval, V.; Hebert, S.S. Preclinical Evaluation of miR-15/107 Family Members as Multifactorial Drug Targets for Alzheimer’s Disease. Mol. Ther. Nucleic Acids 2015, 4, e256. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, C.F.; Wang, A.H.; Lin, Q.F. MiR-16 regulates cell death in Alzheimer’s disease by targeting amyloid precursor protein. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 4020–4027. [Google Scholar]
- Liu, C.G.; Wang, J.L.; Li, L.; Xue, L.X.; Zhang, Y.Q.; Wang, P.C. MicroRNA-135a and -200b, potential Biomarkers for Alzheimer׳s disease, regulate beta secretase and amyloid precursor protein. Brain Res. 2014, 1583, 55–64. [Google Scholar] [CrossRef]
- Liu, C.G.; Wang, J.L.; Li, L.; Wang, P.C. MicroRNA-384 regulates both amyloid precursor protein and beta-secretase expression and is a potential biomarker for Alzheimer’s disease. Int. J. Mol. Med. 2014, 34, 160–166. [Google Scholar] [CrossRef]
- Liu, C.G.; Song, J.; Zhang, Y.Q.; Wang, P.C. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol. Med. Rep. 2014, 10, 2395–2400. [Google Scholar] [CrossRef]
- Li, K.; Zhang, J.; Ji, C.; Wang, L. MiR-144-3p and Its Target Gene beta-Amyloid Precursor Protein Regulate 1-Methyl-4-Phenyl-1,2-3,6-Tetrahydropyridine-Induced Mitochondrial Dysfunction. Mol. Cells 2016, 39, 543–549. [Google Scholar] [CrossRef]
- Liu, H.Y.; Fu, X.; Li, Y.F.; Li, X.L.; Ma, Z.Y.; Zhang, Y.; Gao, Q.C. miR-15b-5p targeting amyloid precursor protein is involved in the anti-amyloid eflect of curcumin in swAPP695-HEK293 cells. Neural Regen. Res. 2019, 14, 1603–1609. [Google Scholar]
- Kumar, S.; Reddy, A.P.; Yin, X.; Reddy, P.H. Novel MicroRNA-455-3p and its protective effects against abnormal APP processing and amyloid beta toxicity in Alzheimer’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2428–2440. [Google Scholar] [CrossRef] [PubMed]
- Barros-Viegas, A.T.; Carmona, V.; Ferreiro, E.; Guedes, J.; Cardoso, A.M.; Cunha, P.; Pereira de Almeida, L.; Resende de Oliveira, C.; Pedro de Magalhaes, J.; Peca, J.; et al. miRNA-31 Improves Cognition and Abolishes Amyloid-beta Pathology by Targeting APP and BACE1 in an Animal Model of Alzheimer’s Disease. Mol. Ther. Nucleic Acids 2020, 19, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.H.; Sun, L.; Zhang, W.J.; Wang, X.Y.; Li, J.M. Reduced serum miR-202 may promote the progression of Alzheimer’s disease patients via targeting amyloid precursor protein. Kaohsiung J. Med. Sci. 2021, 37, 730–738. [Google Scholar] [CrossRef]
- Chopra, N.; Wang, R.; Maloney, B.; Nho, K.; Beck, J.S.; Pourshafie, N.; Niculescu, A.; Saykin, A.J.; Rinaldi, C.; Counts, S.E.; et al. MicroRNA-298 reduces levels of human amyloid-beta precursor protein (APP), beta-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties. Mol. Psychiatry 2021, 26, 5636–5657. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhu, L.; Tan, J.; Chen, K.; Yu, B. Suppression of miR-130a-3p Attenuates Oxygen-Glucose Deprivation/Reoxygenation-Induced Dendritic Spine Loss by Promoting APP. Front. Neurosci. 2021, 15, 601850. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Yang, X.; Xia, X.; Li, Y.; Wang, Y.; Li, C.; Sun, Y.; Gao, G.; Zhao, S.; Sheng, S.; et al. Exosomes Mediate APP Dysregulation via APP-miR-185-5p Axis. Front. Cell Dev. Biol. 2022, 10, 793388. [Google Scholar] [CrossRef]
- Fan, X.; Xu, S.; Yang, C. miR-373-3p promotes lung adenocarcinoma cell proliferation via APP. Oncol. Lett. 2018, 15, 1046–1050. [Google Scholar] [CrossRef]
- Zhou, Q.; Luo, L.; Wang, X.; Li, X. Relationship between single nucleotide polymorphisms in the 3’UTR of amyloid precursor protein and risk of Alzheimer’s disease and its mechanism. Biosci. Rep. 2019, 39, BSR20182485. [Google Scholar] [CrossRef]
- Long, J.M.; Maloney, B.; Rogers, J.T.; Lahiri, D.K. Novel upregulation of amyloid-beta precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5’-untranslated region: Implications in Alzheimer’s disease. Mol. Psychiatry 2019, 24, 345–363. [Google Scholar] [CrossRef]
- Nikom, D.; Zheng, S. Alternative splicing in neurodegenerative disease and the promise of RNA therapies. Nat. Rev. Neurosci. 2023, 24, 457–473. [Google Scholar] [CrossRef]
- Smith, P.; Al Hashimi, A.; Girard, J.; Delay, C.; Hebert, S.S. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J. Neurochem. 2011, 116, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Vassar, R. Targeting the beta secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol. 2014, 13, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Vassar, R.; De Strooper, B.; Hardy, J.; Willem, M.; Singh, N.; Zhou, J.; Yan, R.; Vanmechelen, E.; De Vos, A.; et al. The beta-Secretase BACE1 in Alzheimer’s Disease. Biol. Psychiatry 2021, 89, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Rajeev, B.W.; Stromberg, A.J.; Ren, N.; Tang, G.; Huang, Q.; Rigoutsos, I.; Nelson, P.T. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008, 28, 1213–1223. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, H.; Dong, W.; Quan, X.; Zhu, H.; Xu, Y.; Huang, L.; Ma, C.; Qin, C. miR-29c regulates BACE1 protein expression. Brain Res. 2011, 1395, 108–115. [Google Scholar] [CrossRef]
- Lei, X.; Lei, L.; Zhang, Z.; Zhang, Z.; Cheng, Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 2015, 8, 1565–1574. [Google Scholar]
- Yang, G.; Song, Y.; Zhou, X.; Deng, Y.; Liu, T.; Weng, G.; Yu, D.; Pan, S. MicroRNA-29c targets beta-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol. Med. Rep. 2015, 12, 3081–3088. [Google Scholar] [CrossRef]
- Boissonneault, V.; Plante, I.; Rivest, S.; Provost, P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J. Biol. Chem. 2009, 284, 1971–1981. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Zhang, M.; Huang, J.; Modarresi, F.; Van der Brug, M.P.; Nalls, M.A.; Cookson, M.R.; St-Laurent, G., 3rd; Wahlestedt, C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010, 11, R56. [Google Scholar] [CrossRef]
- Zhu, H.C.; Wang, L.M.; Wang, M.; Song, B.; Tan, S.; Teng, J.F.; Duan, D.X. MicroRNA-195 downregulates Alzheimer’s disease amyloid-beta production by targeting BACE1. Brain Res. Bull. 2012, 88, 596–601. [Google Scholar] [CrossRef]
- Long, J.M.; Ray, B.; Lahiri, D.K. MicroRNA-339-5p down-regulates protein expression of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J. Biol. Chem. 2014, 289, 5184–5198. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, M.; Teng, Z.; Tang, Y.P.; Chen, C. Synaptic and cognitive improvements by inhibition of 2-AG metabolism are through upregulation of microRNA-188-3p in a mouse model of Alzheimer’s disease. J. Neurosci. 2014, 34, 14919–14933. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xing, H.; Guo, S.; Zheng, Z.; Wang, H.; Xu, D. MicroRNA-135b has a neuroprotective role via targeting of beta-site APP-cleaving enzyme 1. Exp. Ther. Med. 2016, 12, 809–814. [Google Scholar] [CrossRef]
- Kim, J.; Yoon, H.; Chung, D.E.; Brown, J.L.; Belmonte, K.C.; Kim, J. miR-186 is decreased in aged brain and suppresses BACE1 expression. J. Neurochem. 2016, 137, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; An, F.; Wang, Y.; Bian, M.; Yu, L.J.; Wei, C. miR-15b represses BACE1 expression in sporadic Alzheimer’s disease. Oncotarget 2017, 8, 91551–91557. [Google Scholar] [CrossRef]
- Li, J.; Wang, H. miR-15b reduces amyloid-beta accumulation in SH-SY5Y cell line through targetting NF-kappaB signaling and BACE1. Biosci. Rep. 2018, 38, BSR20180051. [Google Scholar]
- An, F.; Gong, G.; Wang, Y.; Bian, M.; Yu, L.; Wei, C. MiR-124 acts as a target for Alzheimer’s disease by regulating BACE1. Oncotarget 2017, 8, 114065–114071. [Google Scholar] [CrossRef]
- Du, X.; Huo, X.; Yang, Y.; Hu, Z.; Botchway, B.O.A.; Jiang, Y.; Fang, M. miR-124 downregulates BACE 1 and alters autophagy in APP/PS1 transgenic mice. Toxicol. Lett. 2017, 280, 195–205. [Google Scholar] [CrossRef]
- Zhong, Z.; Yuan, K.; Tong, X.; Hu, J.; Song, Z.; Zhang, G.; Fang, X.; Zhang, W. MiR-16 attenuates beta-amyloid-induced neurotoxicity through targeting beta-site amyloid precursor protein-cleaving enzyme 1 in an Alzheimer’s disease cell model. Neuroreport 2018, 29, 1365–1372. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Wang, Q.; Jiang, H.; Zeng, L.; Li, Z.; Liu, R. MicroRNA-200a-3p Mediates Neuroprotection in Alzheimer-Related Deficits and Attenuates Amyloid-Beta Overproduction and Tau Hyperphosphorylation via Coregulating BACE1 and PRKACB. Front. Pharmacol. 2019, 10, 806. [Google Scholar] [CrossRef]
- Ji, Y.; Wang, D.; Zhang, B.; Lu, H. MiR-361-3p inhibits beta-amyloid accumulation and attenuates cognitive deficits through targeting BACE1 in Alzheimer’s disease. J. Integr. Neurosci. 2019, 18, 285–291. [Google Scholar]
- Qian, Q.; Zhang, J.; He, F.P.; Bao, W.X.; Zheng, T.T.; Zhou, D.M.; Pan, H.Y.; Zhang, H.; Zhang, X.Q.; He, X.; et al. Down-regulated expression of microRNA-338-5p contributes to neuropathology in Alzheimer’s disease. FASEB J. 2019, 33, 4404–4417. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xu, Y.; Wang, B.; Huang, J.; Li, Q. miR-34a-5p and miR-125b-5p attenuate Abeta-induced neurotoxicity through targeting BACE1. J. Neurol. Sci. 2020, 413, 116793. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Luo, Y.; Pi, D.; Xia, L.; Li, Z.; Tu, Q. MiR-340 Reduces the Accumulation of Amyloid-beta Through Targeting BACE1 (beta-site Amyloid Precursor Protein Cleaving Enzyme 1) in Alzheimer’s Disease. Curr. Neurovasc Res. 2020, 17, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Li, W.W.; Lv, C.M.; Gao, Y.W.; Liu, X.L.; Zhao, L. miR-16-5p and miR-19b-3p prevent amyloid beta-induced injury by targeting BACE1 in SH-SY5Y cells. Neuroreport 2020, 31, 205–212. [Google Scholar] [CrossRef]
- Du, W.; Lei, C.; Dong, Y. MicroRNA-149 is downregulated in Alzheimer’s disease and inhibits beta-amyloid accumulation and ameliorates neuronal viability through targeting BACE1. Genet. Mol. Biol. 2021, 44, e20200064. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, R. Deregulated lncRNA MAGI2-AS3 in Alzheimer’s disease attenuates amyloid-beta induced neurotoxicity and neuroinflammation by sponging miR-374b-5p. Exp. Gerontol. 2021, 144, 111180. [Google Scholar] [CrossRef]
- Dong, Z.; Gu, H.; Guo, Q.; Liu, X.; Li, F.; Liu, H.; Sun, L.; Ma, H.; Zhao, K. Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer’s Disease. Cells 2022, 11, 3830. [Google Scholar] [CrossRef]
- Aplin, A.E.; Gibb, G.M.; Jacobsen, J.S.; Gallo, J.M.; Anderton, B.H. In vitro phosphorylation of the cytoplasmic domain of the amyloid precursor protein by glycogen synthase kinase-3beta. J. Neurochem. 1996, 67, 699–707. [Google Scholar] [CrossRef]
- Acevedo, K.M.; Opazo, C.M.; Norrish, D.; Challis, L.M.; Li, Q.X.; White, A.R.; Bush, A.I.; Camakaris, J. Phosphorylation of amyloid precursor protein at threonine 668 is essential for its copper-responsive trafficking in SH-SY5Y neuroblastoma cells. J. Biol. Chem. 2014, 289, 11007–11019. [Google Scholar] [CrossRef]
- Iijima, K.; Ando, K.; Takeda, S.; Satoh, Y.; Seki, T.; Itohara, S.; Greengard, P.; Kirino, Y.; Nairn, A.C.; Suzuki, T. Neuron-specific phosphorylation of Alzheimer’s beta-amyloid precursor protein by cyclin-dependent kinase 5. J. Neurochem. 2000, 75, 1085–1091. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Su, Y.; Li, B.; Zhou, Y.; Ryder, J.; Gonzalez-DeWhitt, P.; May, P.C.; Ni, B. Regulation of amyloid precursor protein (APP) phosphorylation and processing by p35/Cdk5 and p25/Cdk5. FEBS Lett. 2003, 547, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Liu, X.; Cheng, X.; Li, Y.; Gearing, M.; Levey, A.; Huang, X.; Li, Y.; Jin, P.; Li, X. MicroRNA-650 Regulates the Pathogenesis of Alzheimer’s Disease Through Targeting Cyclin-Dependent Kinase 5. Mol. Neurobiol. 2023, 60, 2426–2441. [Google Scholar] [CrossRef] [PubMed]
- Moncini, S.; Lunghi, M.; Valmadre, A.; Grasso, M.; Del Vescovo, V.; Riva, P.; Denti, M.A.; Venturin, M. The miR-15/107 Family of microRNA Genes Regulates CDK5R1/p35 with Implications for Alzheimer’s Disease Pathogenesis. Mol. Neurobiol. 2017, 54, 4329–4342. [Google Scholar] [CrossRef] [PubMed]
- Standen, C.L.; Brownlees, J.; Grierson, A.J.; Kesavapany, S.; Lau, K.F.; McLoughlin, D.M.; Miller, C.C. Phosphorylation of thr(668) in the cytoplasmic domain of the Alzheimer’s disease amyloid precursor protein by stress-activated protein kinase 1b (Jun N-terminal kinase-3). J. Neurochem. 2001, 76, 316–320. [Google Scholar] [CrossRef]
- Colombo, A.; Bastone, A.; Ploia, C.; Sclip, A.; Salmona, M.; Forloni, G.; Borsello, T. JNK regulates APP cleavage and degradation in a model of Alzheimer’s disease. Neurobiol. Dis. 2009, 33, 518–525. [Google Scholar] [CrossRef]
- Zhu, X.; Raina, A.K.; Rottkamp, C.A.; Aliev, G.; Perry, G.; Boux, H.; Smith, M.A. Activation and redistribution of c-jun N-terminal kinase/stress activated protein kinase in degenerating neurons in Alzheimer’s disease. J. Neurochem. 2001, 76, 435–441. [Google Scholar] [CrossRef]
- Wang, D.; Fei, Z.; Luo, S.; Wang, H. MiR-335-5p Inhibits beta-Amyloid (Abeta) Accumulation to Attenuate Cognitive Deficits Through Targeting c-jun-N-terminal Kinase 3 in Alzheimer’s Disease. Curr. Neurovasc Res. 2020, 17, 93–101. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, M.; Hong, Z.; Kang, J.; Pan, H.; Yan, C. MiR-130a-3p Has Protective Effects in Alzheimer’s Disease via Targeting DAPK1. Am. J. Alzheimers Dis. Other Demen 2021, 36, 15333175211020572. [Google Scholar] [CrossRef]
- Raulin, A.C.; Doss, S.V.; Trottier, Z.A.; Ikezu, T.C.; Bu, G.; Liu, C.C. ApoE in Alzheimer’s disease: Pathophysiology and therapeutic strategies. Mol. Neurodegener. 2022, 17, 72. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Zhao, N.; Caulfield, T.R.; Liu, C.C.; Bu, G. Apolipoprotein E and Alzheimer disease: Pathobiology and targeting strategies. Nat. Rev. Neurol. 2019, 15, 501–518. [Google Scholar] [CrossRef] [PubMed]
- Souza, V.C.; Morais, G.S., Jr.; Henriques, A.D.; Machado-Silva, W.; Perez, D.I.V.; Brito, C.J.; Camargos, E.F.; Moraes, C.F.; Nobrega, O.T. Whole-Blood Levels of MicroRNA-9 Are Decreased in Patients With Late-Onset Alzheimer Disease. Am. J. Alzheimers Dis. Other Demen 2020, 35, 1533317520911573. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qin, W.; Wei, C.B.; Tang, Y.; Zhao, L.N.; Jin, H.M.; Li, Y.; Wang, Q.; Luan, X.Q.; He, J.C.; et al. The microRNA-1908 up-regulation in the peripheral blood cells impairs amyloid clearance by targeting ApoE. Int. J. Geriatr. Psychiatry 2018, 33, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Boccardi, V.; Poli, G.; Cecchetti, R.; Bastiani, P.; Scamosci, M.; Febo, M.; Mazzon, E.; Bruscoli, S.; Brancorsini, S.; Mecocci, P. miRNAs and Alzheimer’s Disease: Exploring the Role of Inflammation and Vitamin E in an Old-Age Population. Nutrients 2023, 15, 634. [Google Scholar] [CrossRef]
- Pascoal, T.A.; Benedet, A.L.; Ashton, N.J.; Kang, M.S.; Therriault, J.; Chamoun, M.; Savard, M.; Lussier, F.Z.; Tissot, C.; Karikari, T.K.; et al. Microglial activation and tau propagate jointly across Braak stages. Nat. Med. 2021, 27, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Long, Y.F.; Xu, P.; Guo, H.D.; Cui, G.H. Pathogenesis of miR-155 on nonmodifiable and modifiable risk factors in Alzheimer’s disease. Alzheimers Res. Ther. 2023, 15, 122. [Google Scholar] [CrossRef]
- Rastegar-Moghaddam, S.H.; Ebrahimzadeh-Bideskan, A.; Shahba, S.; Malvandi, A.M.; Mohammadipour, A. Roles of the miR-155 in Neuroinflammation and Neurological Disorders: A Potent Biological and Therapeutic Target. Cell Mol. Neurobiol. 2023, 43, 455–467. [Google Scholar] [CrossRef]
- Readhead, B.; Haure-Mirande, J.V.; Mastroeni, D.; Audrain, M.; Fanutza, T.; Kim, S.H.; Blitzer, R.D.; Gandy, S.; Dudley, J.T.; Ehrlich, M.E. miR155 regulation of behavior, neuropathology, and cortical transcriptomics in Alzheimer’s disease. Acta Neuropathol. 2020, 140, 295–315. [Google Scholar] [CrossRef]
- Aloi, M.S.; Prater, K.E.; Sopher, B.; Davidson, S.; Jayadev, S.; Garden, G.A. The pro-inflammatory microRNA miR-155 influences fibrillar beta-Amyloid(1)(-42) catabolism by microglia. Glia 2021, 69, 1736–1748. [Google Scholar] [CrossRef]
- Li, Y.Y.; Cui, J.G.; Dua, P.; Pogue, A.I.; Bhattacharjee, S.; Lukiw, W.J. Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci. Lett. 2011, 499, 109–113. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Su, W.; Aloi, M.S.; Garden, G.A. MicroRNAs mediating CNS inflammation: Small regulators with powerful potential. Brain Behav. Immun. 2016, 52, 1–8. [Google Scholar] [CrossRef]
- Long, J.M.; Holtzman, D.M. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019, 179, 312–339. [Google Scholar] [CrossRef] [PubMed]
- Vermunt, L.; Sikkes, S.A.M.; van den Hout, A.; Handels, R.; Bos, I.; van der Flier, W.M.; Kern, S.; Ousset, P.J.; Maruff, P.; Skoog, I.; et al. Duration of preclinical, prodromal, and dementia stages of Alzheimer’s disease in relation to age, sex, and APOE genotype. Alzheimers Dement. 2019, 15, 888–898. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Sperling, R.A.; Aisen, P.S.; Beckett, L.A.; Bennett, D.A.; Craft, S.; Fagan, A.M.; Iwatsubo, T.; Jack, C.R., Jr.; Kaye, J.; Montine, T.J.; et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 280–292. [Google Scholar] [CrossRef]
- Zetterberg, H.; Skillback, T.; Mattsson, N.; Trojanowski, J.Q.; Portelius, E.; Shaw, L.M.; Weiner, M.W.; Blennow, K.; Alzheimer’s Disease Neuroimaging, I. Association of Cerebrospinal Fluid Neurofilament Light Concentration With Alzheimer Disease Progression. JAMA Neurol. 2016, 73, 60–67. [Google Scholar] [CrossRef]
- Hampel, H.; O’Bryant, S.E.; Molinuevo, J.L.; Zetterberg, H.; Masters, C.L.; Lista, S.; Kiddle, S.J.; Batrla, R.; Blennow, K. Blood-based biomarkers for Alzheimer disease: Mapping the road to the clinic. Nat. Rev. Neurol. 2018, 14, 639–652. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kaab, S.; Wakili, R.; et al. Stability of Circulating Blood-Based MicroRNAs—Pre-Analytic Methodological Considerations. PLoS ONE 2017, 12, e0167969. [Google Scholar] [CrossRef]
- Coenen-Stass, A.M.L.; Pauwels, M.J.; Hanson, B.; Martin Perez, C.; Conceicao, M.; Wood, M.J.A.; Mager, I.; Roberts, T.C. Extracellular microRNAs exhibit sequence-dependent stability and cellular release kinetics. RNA Biol. 2019, 16, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Dezso, Z.; MacKenzie, C.; Oestreicher, J.; Agoulnik, S.; Byrne, M.; Bernier, F.; Yanagimachi, M.; Aoshima, K.; Oda, Y. Circulating miRNA biomarkers for Alzheimer’s disease. PLoS ONE 2013, 8, e69807. [Google Scholar] [CrossRef] [PubMed]
- Leidinger, P.; Backes, C.; Deutscher, S.; Schmitt, K.; Mueller, S.C.; Frese, K.; Haas, J.; Ruprecht, K.; Paul, F.; Stahler, C.; et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013, 14, R78. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Yu, J.T.; Liu, Q.Y.; Tan, M.S.; Zhang, W.; Hu, N.; Wang, Y.L.; Sun, L.; Jiang, T.; Tan, L. Circulating miR-125b as a biomarker of Alzheimer’s disease. J. Neurol. Sci. 2014, 336, 52–56. [Google Scholar] [CrossRef]
- Tan, L.; Yu, J.T.; Tan, M.S.; Liu, Q.Y.; Wang, H.F.; Zhang, W.; Jiang, T.; Tan, L. Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer’s disease. J. Alzheimers Dis. 2014, 40, 1017–1027. [Google Scholar] [CrossRef]
- Galimberti, D.; Villa, C.; Fenoglio, C.; Serpente, M.; Ghezzi, L.; Cioffi, S.M.; Arighi, A.; Fumagalli, G.; Scarpini, E. Circulating miRNAs as potential biomarkers in Alzheimer’s disease. J. Alzheimers Dis. 2014, 42, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Li, J.; Huang, L.; Chen, X.; Li, D.; Wang, T.; Hu, C.; Xu, J.; Zhang, C.; Zen, K.; et al. Serum MicroRNA Profiles Serve as Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Dis. Markers 2015, 2015, 625659. [Google Scholar] [CrossRef]
- Satoh, J.; Kino, Y.; Niida, S. MicroRNA-Seq Data Analysis Pipeline to Identify Blood Biomarkers for Alzheimer’s Disease from Public Data. Biomark. Insights 2015, 10, 21–31. [Google Scholar] [CrossRef]
- Cheng, L.; Doecke, J.D.; Sharples, R.A.; Villemagne, V.L.; Fowler, C.J.; Rembach, A.; Martins, R.N.; Rowe, C.C.; Macaulay, S.L.; Masters, C.L.; et al. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 2015, 20, 1188–1196. [Google Scholar] [CrossRef]
- Lugli, G.; Cohen, A.M.; Bennett, D.A.; Shah, R.C.; Fields, C.J.; Hernandez, A.G.; Smalheiser, N.R. Plasma Exosomal miRNAs in Persons with and without Alzheimer Disease: Altered Expression and Prospects for Biomarkers. PLoS ONE 2015, 10, e0139233. [Google Scholar] [CrossRef]
- Yilmaz, S.G.; Erdal, M.E.; Ozge, A.A.; Sungur, M.A. Can Peripheral MicroRNA Expression Data Serve as Epigenomic (Upstream) Biomarkers of Alzheimer’s Disease? OMICS 2016, 20, 456–461. [Google Scholar] [CrossRef]
- Hara, N.; Kikuchi, M.; Miyashita, A.; Hatsuta, H.; Saito, Y.; Kasuga, K.; Murayama, S.; Ikeuchi, T.; Kuwano, R. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 2017, 5, 10. [Google Scholar] [CrossRef]
- Cosin-Tomas, M.; Antonell, A.; Llado, A.; Alcolea, D.; Fortea, J.; Ezquerra, M.; Lleo, A.; Marti, M.J.; Pallas, M.; Sanchez-Valle, R.; et al. Plasma miR-34a-5p and miR-545-3p as Early Biomarkers of Alzheimer’s Disease: Potential and Limitations. Mol. Neurobiol. 2017, 54, 5550–5562. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Vijayan, M.; Reddy, P.H. MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum. Mol. Genet. 2017, 26, 3808–3822. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Fan, G.; Zhang, J.; Wu, C.; Du, Y.; Ye, H.; Li, Z.; Wang, L.; Zhang, Z.; Zhang, L.; et al. A 9-microRNA Signature in Serum Serves as a Noninvasive Biomarker in Early Diagnosis of Alzheimer’s Disease. J. Alzheimers Dis. 2017, 60, 1365–1377. [Google Scholar] [CrossRef]
- Kenny, A.; McArdle, H.; Calero, M.; Rabano, A.; Madden, S.F.; Adamson, K.; Forster, R.; Spain, E.; Prehn, J.H.M.; Henshall, D.C.; et al. Elevated Plasma microRNA-206 Levels Predict Cognitive Decline and Progression to Dementia from Mild Cognitive Impairment. Biomolecules 2019, 9, 734. [Google Scholar] [CrossRef]
- Zhao, X.; Kang, J.; Svetnik, V.; Warden, D.; Wilcock, G.; David Smith, A.; Savage, M.J.; Laterza, O.F. A Machine Learning Approach to Identify a Circulating MicroRNA Signature for Alzheimer Disease. J. Appl. Lab. Med. 2020, 5, 15–28. [Google Scholar] [CrossRef]
- Jia, L.; Zhu, M.; Yang, J.; Pang, Y.; Wang, Q.; Li, T.; Li, F.; Wang, Q.; Li, Y.; Wei, Y. Exosomal MicroRNA-Based Predictive Model for Preclinical Alzheimer’s Disease: A Multicenter Study. Biol. Psychiatry 2022, 92, 44–53. [Google Scholar] [CrossRef]
- Geekiyanage, H.; Jicha, G.A.; Nelson, P.T.; Chan, C. Blood serum miRNA: Non-invasive biomarkers for Alzheimer’s disease. Exp. Neurol. 2012, 235, 491–496. [Google Scholar] [CrossRef]
- Kiko, T.; Nakagawa, K.; Tsuduki, T.; Furukawa, K.; Arai, H.; Miyazawa, T. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J. Alzheimers Dis. 2014, 39, 253–259. [Google Scholar] [CrossRef]
- Bhatnagar, S.; Chertkow, H.; Schipper, H.M.; Yuan, Z.; Shetty, V.; Jenkins, S.; Jones, T.; Wang, E. Increased microRNA-34c abundance in Alzheimer’s disease circulating blood plasma. Front. Mol. Neurosci. 2014, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, C.; Sun, A.; Wang, Y.; Zhou, S. Quantification of microRNA-210 in the cerebrospinal fluid and serum: Implications for Alzheimer’s disease. Exp. Ther. Med. 2015, 9, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.H.; Liu, Y.N. Downregulated serum miR-223 servers as biomarker in Alzheimer’s disease. Cell Biochem. Funct. 2016, 34, 233–237. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, J.; Xu, J.; Cheng, J.; Jiao, D.; Zhou, C.; Dai, Y.; Chen, Q. Lower Serum Levels of miR-29c-3p and miR-19b-3p as Biomarkers for Alzheimer’s Disease. Tohoku J. Exp. Med. 2017, 242, 129–136. [Google Scholar] [CrossRef]
- Yang, T.T.; Liu, C.G.; Gao, S.C.; Zhang, Y.; Wang, P.C. The Serum Exosome Derived MicroRNA-135a, -193b, and -384 Were Potential Alzheimer’s Disease Biomarkers. Biomed. Environ. Sci. 2018, 31, 87–96. [Google Scholar]
- Yang, Q.; Zhao, Q.; Yin, Y. miR-133b is a potential diagnostic biomarker for Alzheimer’s disease and has a neuroprotective role. Exp. Ther. Med. 2019, 18, 2711–2718. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C.; Zhang, Y. An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer’s disease. J. Clin. Lab. Anal. 2020, 34, e23006. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, S.; Sun, W. Expression of miR-28-3p in patients with Alzheimer’s disease before and after treatment and its clinical value. Exp. Ther. Med. 2020, 20, 2218–2226. [Google Scholar] [CrossRef]
- Sabry, R.; El Sharkawy, R.E.; Gad, N.M. MiRNA -483-5p as a Potential Noninvasive Biomarker for Early Detection of Alzheimer’s Disease. Egypt. J. Immunol. 2020, 27, 59–72. [Google Scholar]
- Liu, Q.; Lei, C. Neuroprotective effects of miR-331-3p through improved cell viability and inflammatory marker expression: Correlation of serum miR-331-3p levels with diagnosis and severity of Alzheimer’s disease. Exp. Gerontol. 2021, 144, 111187. [Google Scholar] [CrossRef]
- Zhang, M.; Han, W.; Xu, Y.; Li, D.; Xue, Q. Serum miR-128 Serves as a Potential Diagnostic Biomarker for Alzheimer’s Disease. Neuropsychiatr. Dis. Treat. 2021, 17, 269–275. [Google Scholar] [CrossRef]
- Madadi, S.; Saidijam, M.; Yavari, B.; Soleimani, M. Downregulation of serum miR-106b: A potential biomarker for Alzheimer disease. Arch. Physiol. Biochem. 2022, 128, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Sheinerman, K.S.; Tsivinsky, V.G.; Crawford, F.; Mullan, M.J.; Abdullah, L.; Umansky, S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging 2012, 4, 590–605. [Google Scholar] [CrossRef]
- Sheinerman, K.S.; Tsivinsky, V.G.; Abdullah, L.; Crawford, F.; Umansky, S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment: Biomarker validation study. Aging 2013, 5, 925–938. [Google Scholar] [CrossRef]
- Wang, T.; Chen, K.; Li, H.; Dong, S.; Su, N.; Liu, Y.; Cheng, Y.; Dai, J.; Yang, C.; Xiao, S. The feasibility of utilizing plasma MiRNA107 and BACE1 messenger RNA gene expression for clinical diagnosis of amnestic mild cognitive impairment. J. Clin. Psychiatry 2015, 76, 135–141. [Google Scholar] [CrossRef]
- Xie, B.; Liu, Z.; Jiang, L.; Liu, W.; Song, M.; Zhang, Q.; Zhang, R.; Cui, D.; Wang, X.; Xu, S. Increased Serum miR-206 Level Predicts Conversion from Amnestic Mild Cognitive Impairment to Alzheimer’s Disease: A 5-Year Follow-up Study. J. Alzheimers Dis. 2017, 55, 509–520. [Google Scholar] [CrossRef]
- Salama, I.I.; Salama, S.I.; Elmosalami, D.M.; Saleh, R.M.; Rasmy, H.; Ibrahim, M.H.; Kamel, S.A.; Ganem, M.M.F.; Raslan, H.M. Risk Factors Associated with Mild Cognitive Impairment among Apparently Healthy People and the Role of MicroRNAs. Open Access Maced. J. Med. Sci. 2019, 7, 3253–3261. [Google Scholar] [CrossRef]
- He, H.; Liu, A.; Zhang, W.; Yang, H.; Zhang, M.; Xu, H.; Liu, Y.; Hong, B.; Yan, F.; Yue, L.; et al. Novel Plasma miRNAs as Biomarkers and Therapeutic Targets of Alzheimer’s Disease at the Prodromal Stage. J. Alzheimers Dis. 2021, 83, 779–790. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, P.H. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim. Biophys. Acta 2016, 1862, 1617–1627. [Google Scholar] [CrossRef]
- Piscopo, P.; Bellenghi, M.; Manzini, V.; Crestini, A.; Pontecorvi, G.; Corbo, M.; Ortona, E.; Care, A.; Confaloni, A. A Sex Perspective in Neurodegenerative Diseases: microRNAs as Possible Peripheral Biomarkers. Int. J. Mol. Sci. 2021, 22, 4423. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Chen, N. Resveratrol as a Natural Autophagy Regulator for Prevention and Treatment of Alzheimer’s Disease. Nutrients 2017, 9, 927. [Google Scholar] [CrossRef]
- Chakravarthy, M.; Chen, S.; Dodd, P.R.; Veedu, R.N. Nucleic Acid-Based Theranostics for Tackling Alzheimer’s Disease. Theranostics 2017, 7, 3933–3947. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Taniguchi, H.; Suzuki, Y.; Imai, K.; Adachi, Y. Antitumoral RNA-targeted oligonucleotide therapeutics: The third pillar after small molecule inhibitors and antibodies. Cancer Sci. 2022, 113, 2952–2961. [Google Scholar] [CrossRef]
- Padmakumar, S.; D’Souza, A.; Parayath, N.N.; Bleier, B.S.; Amiji, M.M. Nucleic acid therapies for CNS diseases: Pathophysiology, targets, barriers, and delivery strategies. J. Control Release 2022, 352, 121–145. [Google Scholar] [CrossRef]
- Egli, M.; Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef]
- Rodgers, G.; Austin, C.; Anderson, J.; Pawlyk, A.; Colvis, C.; Margolis, R.; Baker, J. Glimmers in illuminating the druggable genome. Nat. Rev. Drug Discov. 2018, 17, 301–302. [Google Scholar] [CrossRef]
- Dammes, N.; Peer, D. Paving the Road for RNA Therapeutics. Trends Pharmacol. Sci. 2020, 41, 755–775. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.M.; Choi, Y.H.; Tu, M.J. RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 2020, 72, 862–898. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Guo, Y.; Chen, L.; Zhang, X.; Wu, W.; Yang, Z.; Li, X.; Wang, Y.; Hu, Z.; Wang, Z. Co-delivery of phagocytosis checkpoint and STING agonist by a Trojan horse nanocapsule for orthotopic glioma immunotherapy. Theranostics 2022, 12, 5488–5503. [Google Scholar] [CrossRef]
- Ouyang, Q.; Meng, Y.; Zhou, W.; Tong, J.; Cheng, Z.; Zhu, Q. New advances in brain-targeting nano-drug delivery systems for Alzheimer’s disease. J. Drug Target. 2022, 30, 61–81. [Google Scholar] [CrossRef] [PubMed]
- Taliyan, R.; Kakoty, V.; Sarathlal, K.C.; Kharavtekar, S.S.; Karennanavar, C.R.; Choudhary, Y.K.; Singhvi, G.; Riadi, Y.; Dubey, S.K.; Kesharwani, P. Nanocarrier mediated drug delivery as an impeccable therapeutic approach against Alzheimer’s disease. J. Control Release 2022, 343, 528–550. [Google Scholar] [CrossRef]
- Hu, L.; Tao, Y.; Jiang, Y.; Qin, F. Recent progress of nanomedicine in the treatment of Alzheimer’s disease. Front. Cell Dev. Biol. 2023, 11, 1228679. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, L.; Chen, L.F.; Wu, W.; Yang, Z.M.; Wang, Y.Z.; Wang, A.Q.; Jiang, S.J.; Qin, X.Z.; Ye, Z.C.; et al. Glioblastoma cell-derived exosomes functionalized with peptides as efficient nanocarriers for synergistic chemotherapy of glioblastoma with improved biosafety. Nano Res. 2023, 1–11. [Google Scholar] [CrossRef]
- Soares Martins, T.; Trindade, D.; Vaz, M.; Campelo, I.; Almeida, M.; Trigo, G.; da Cruz, E.S.O.A.B.; Henriques, A.G. Diagnostic and therapeutic potential of exosomes in Alzheimer’s disease. J. Neurochem. 2021, 156, 162–181. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, F.; Wang, K.; Zhong, Y.; Wei, X.; Wang, Q.; Zhang, H. Engineered Exosomes: A Promising Drug Delivery Strategy for Brain Diseases. Curr. Med. Chem. 2022, 29, 3111–3124. [Google Scholar] [CrossRef]
- Joo, H.S.; Jeon, H.Y.; Hong, E.B.; Kim, H.Y.; Lee, J.M. Exosomes for the diagnosis and treatment of dementia. Curr. Opin. Psychiatry 2023, 36, 119–125. [Google Scholar] [CrossRef] [PubMed]
| miRNA Profile | Sample | Project Description | References |
|---|---|---|---|
| miR-let-7d-5p, miR-let-7g-5p, miR-15b-5p, miR-142-3p, miR-191-5p, miR-301a-3p, and miR-545-3p | plasma | These 7 signature miRNAs are downregulated in the plasma of AD patients and can discriminate AD individuals from healthy controls with >95% accuracy (AUC of 0.953). | [193] |
| miR-112, miR-161, miR-let-7d-3p, miR-5010-3p, miR-26a-5p, miR-1285-5p, and miR-151a-3p, miR-103a-3p, miR-107, miR-532-5p, miR-26b-5p, and miR-let-7f-5p | blood | These 12 signature miRNAs can discriminate AD patients from healthy controls with an accuracy of 93%, a specificity of 95%, and a sensitivity of 92%. | [194] |
| miR-125b | serum | The level of this miRNA is downregulated in the serum of AD patients, distinguishing AD individuals from healthy controls with a specificity up to 68.3% and a sensitivity of 80.8% and is correlated with the MMSE in AD patients. | [195] |
| miR-98-5p, miR-885-5p, miR-483-3p, miR-342-3p, miR-191-5p, and miR-let-7d-5p | serum | These miRNAs are downregulated in AD patients, while miR-342-3p has the best sensitivity (81.5%) and specificity (70.1%) and is correlated to MMSE score. | [196] |
| miR-125b, miR-23a, and miR-26b | serum | The levels of these miRNAs are decreased in the serum of AD patients, and serum miR-125 levels can distinguish AD individuals from healthy controls with an accuracy of 82%. | [197] |
| miR-31, miR-93, miR-143, and miR-146a | serum | The levels of these miRNAs are decreased in the serum of AD patients, and this panel can be used to distinguish AD individuals from healthy controls with an AUC more than 0.7. | [198] |
| miR-26b-3p, miR-28-3p, miR-30c-5p, miR-30d-5p, miR-148b-5p, miR-151a-3p, miR-186-5p, miR-425-5p, miR-550a-5p, miR-1468, miR-4781-3p, miR-5001-3p, miR-6513-3p, miR-let-7a-5p, miR-let-7e-5p, miR-let-7f-5p, miR-let-7g-5p, miR-15a-5p, miR-17-3p, miR-29b-3p, miR-98-5p, miR-144-5p, miR-148a-3p, miR-502-3p, miR-660-5p, miR-1294, and miR-3200-3p | blood | These miRNAs are differentially expressed between AD and control groups. The entire 27 miRNA panel can distinguish several AD subgroups from controls with an accuracy of 0.801, a sensitivity of 70.8%, and a specificity of 81.8%. | [199] |
| miR-1306-5p, miR-342-3p, miR-18b-5p, miR-20a-5p, miR-30e-5p, miR-582-5p, miR-106a-5p, miR-361-5p, miR-143-3p, miR-424-5p, miR-93-5p, miR-106b-5p, miR-101-3p, miR-15b-3p, miR-335-5p, and miR-15a-5p | serum | These miRNAs can distinguish AD participants from healthy controls with a sensitivity of 87% and a specificity of 77%. | [200] |
| miR-185-5p, miR-342-3p, miR-141-3p, miR-342-5p, miR-23b-3p, miR-338-3p, and miR-3613-3p | plasma | These miRNAs can predict AD status with an accuracy of 83–89% in a machine learning model. | [201] |
| miR-9-5p, miR-106a-5p, miR-106b-5p, and miR-107 | blood | These miRNAs are associated with the risk of AD, and miR-106a-5p as a predictor variable shows a specificity of 93% and a sensitivity of 68%. | [202] |
| miR-501-3p | serum | The level of this miRNA is decreased in the serum of AD patients and shows a sensitivity of 53% and a specificity of 100% with an AUC of 0.82. Its lower levels are associated with lower MMSE scores. | [203] |
| miR-483-5p, miR-486-5p, miR-30a-5p, miR-200a-3p, miR-502-3p, and miR-142-3p | plasma | These miRNAs can distinguish AD patients from healthy controls with specificities from 0.78 to 1 and sensitivities from 0.75 to 1. | [43] |
| miR-34a-5p and miR-545-3p | plasma | These miRNAs are downregulated in AD samples and show suitable diagnostic accuracy to distinguish AD patients from healthy controls. The AUC for miR-34a-5p was 0.77 with a sensitivity of 76.19% and a specificity of 71.43%. The AUC for miR-545-3p was 0.75 with a sensitivity of 94.12% and a specificity of 76.01%. | [204] |
| miR-455-3p | serum | The level of this miRNA is increased in the serum of AD individuals and can distinguish AD patients from healthy controls with an AUC of 0.79. | [205] |
| miR-26a-5p, miR-181c-3p, miR-126-5p, miR-22-3p, miR-148b-5p, miR-106b-3p, miR-6119-5p, miR-1246, and miR-660-5p | serum | These miRNAs can distinguish AD patients from controls with the AUC between 70.0% and 85.3%. Among the 9 miRNAs, miR-22-3p has the best sensitivity of 81.8% and a specificity of 70.9%. | [206] |
| miR-92a-3p, miR-181c-5p, and miR-210-3p | plasma | The levels of these miRNAs are increased in the plasma of AD patients and can distinguish AD individuals from healthy controls with an AUC value of 0.855, a sensitivity of 92.86%, and a specificity of 71.43%. | [41] |
| miR-206 | plasma | The level of this miRNA is increased in the plasma of AD patients and can predict cognitive decline using the MMSE test with a sensitivity of 87.50% and a specificity of 77.78%. | [207] |
| miR-346, miR-345-5p, miR-122-3p, miR-208b-3p, miR-1291, miR-640, miR-499a-5p, miR-650, miR-1285-3p, miR-1299, miR-1267, and miR-206 | serum | The levels of these miRNAs in the serum can distinguish AD patients from healthy controls with an accuracy of 76.0%, a sensitivity of 90.0%, and a specificity of 66.7%. | [208] |
| miR-29c-5p, miR-143-3p, miR-335-5p, miR-485-5p, miR-138-5p, and miR-342-3p | blood | These miRNAs can predict preclinical AD at the asymptomatic stage 5 to 7 years prior to cognitive impairment onset with an AUC of 0.852. | [209] |
| miR-92a-3p and miR-320a | plasma | These miRNAs can directly bind to the MAPT mRNA and distinguish AD patients from healthy controls. MiR-92a-3p can distinguish AD individuals from healthy controls with an AUC of 0.76 and a sensitivity of 63%. The miR-320a can distinguish AD subjects from controls with an AUC of 0.73 and a sensitivity of 84%. | [38] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Shui, X.; Diao, Y.; Chen, D.; Zhou, Y.; Lee, T.H. Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 16259. https://doi.org/10.3390/ijms242216259
Wang L, Shui X, Diao Y, Chen D, Zhou Y, Lee TH. Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer’s Disease. International Journal of Molecular Sciences. 2023; 24(22):16259. https://doi.org/10.3390/ijms242216259
Chicago/Turabian StyleWang, Long, Xindong Shui, Yuelin Diao, Duoting Chen, Ying Zhou, and Tae Ho Lee. 2023. "Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer’s Disease" International Journal of Molecular Sciences 24, no. 22: 16259. https://doi.org/10.3390/ijms242216259
APA StyleWang, L., Shui, X., Diao, Y., Chen, D., Zhou, Y., & Lee, T. H. (2023). Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer’s Disease. International Journal of Molecular Sciences, 24(22), 16259. https://doi.org/10.3390/ijms242216259





