Maternal Pre-Existing Diabetes: A Non-Inherited Risk Factor for Congenital Cardiopathies
Abstract
1. Introduction
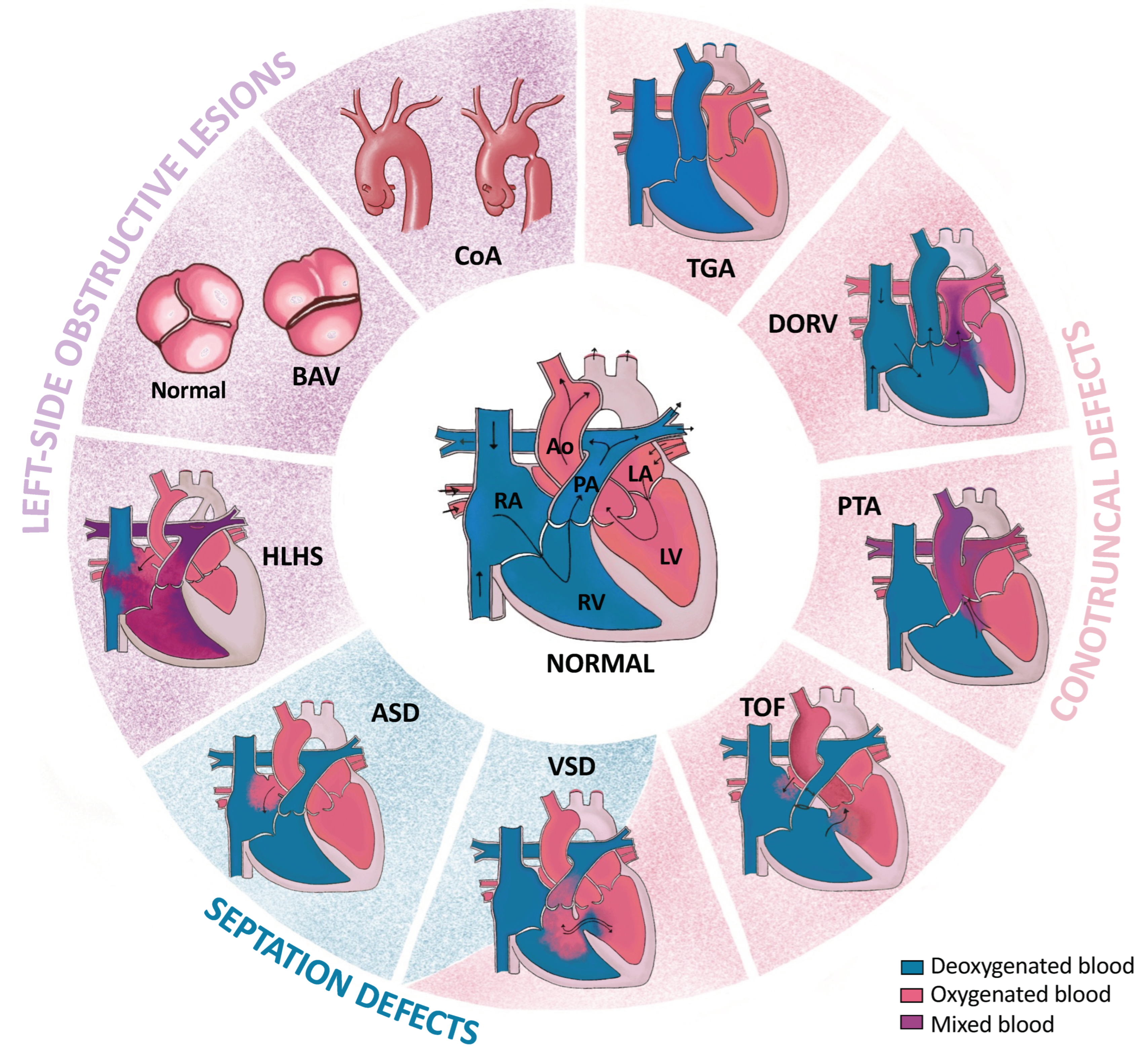
2. Congenital Heart Defects
3. CHD Risk Factors
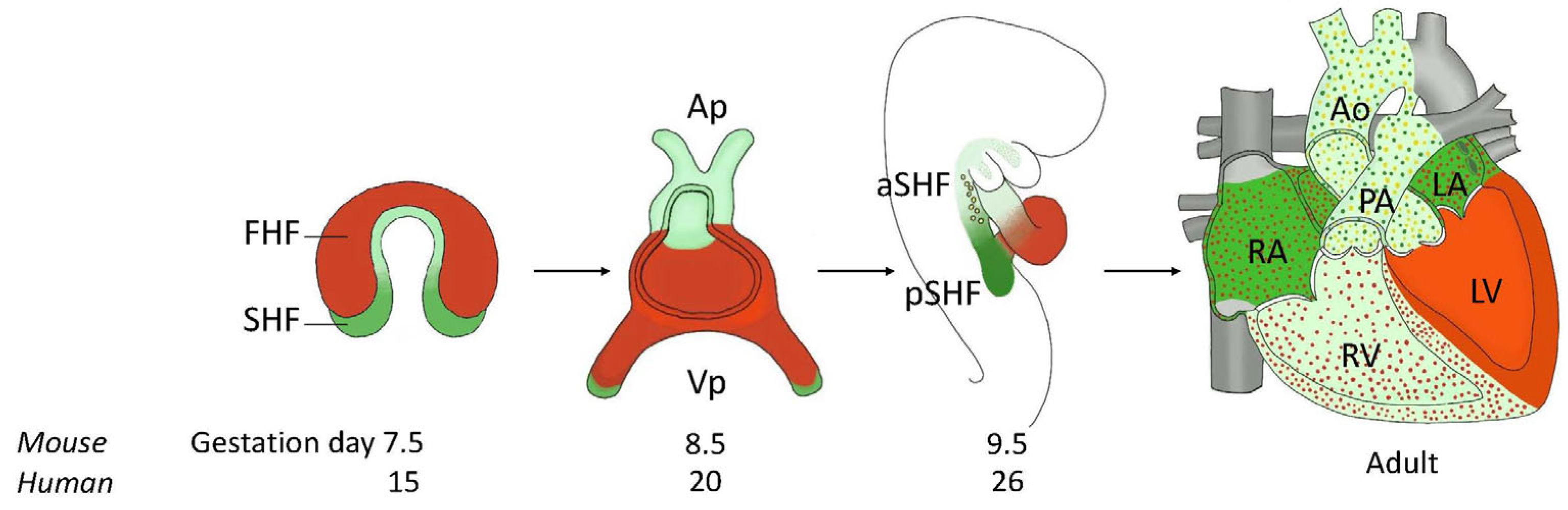
4. Morphogenesis of the Normal Heart
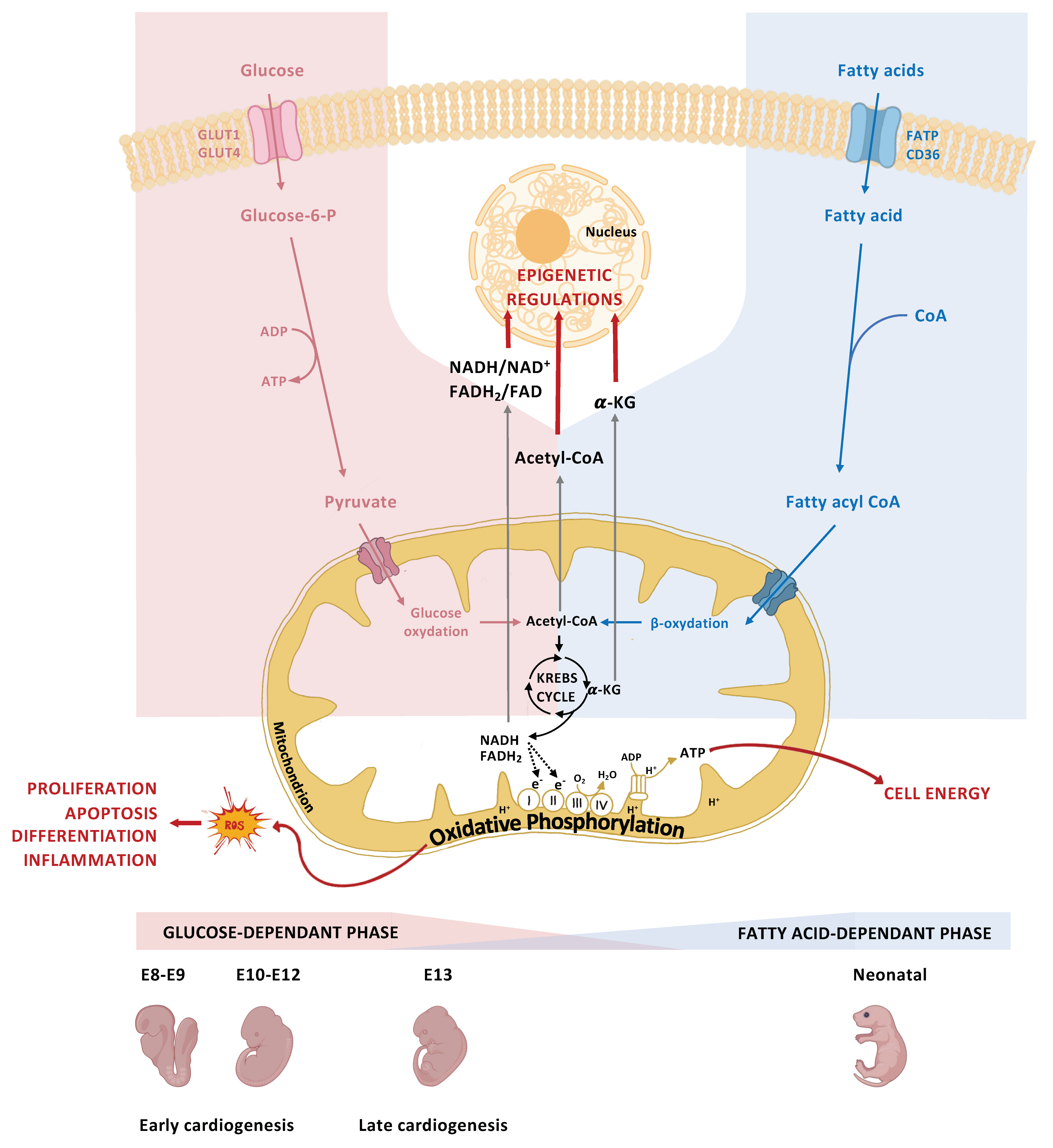
5. The Role of Glucose Metabolism in Normal Heart Development
6. Hyperglycemia Teratogenicity during Heart Development
7. Modeling Pregestational Diabetes
- (a)
- in vivo models
- (b)
- in vitro models
8. Molecular Mechanisms Underlying the Teratogenic Effect of Maternal Pregestational Diabetes
- (a)
- Oxidative stress, endoplasmic reticulum stress, and apoptosis
- (b)
- Dysregulated signaling pathways
- (c)
- Dysregulated transcription factors encoding gene expression
- (d)
- Dysregulated epigenetic regulation
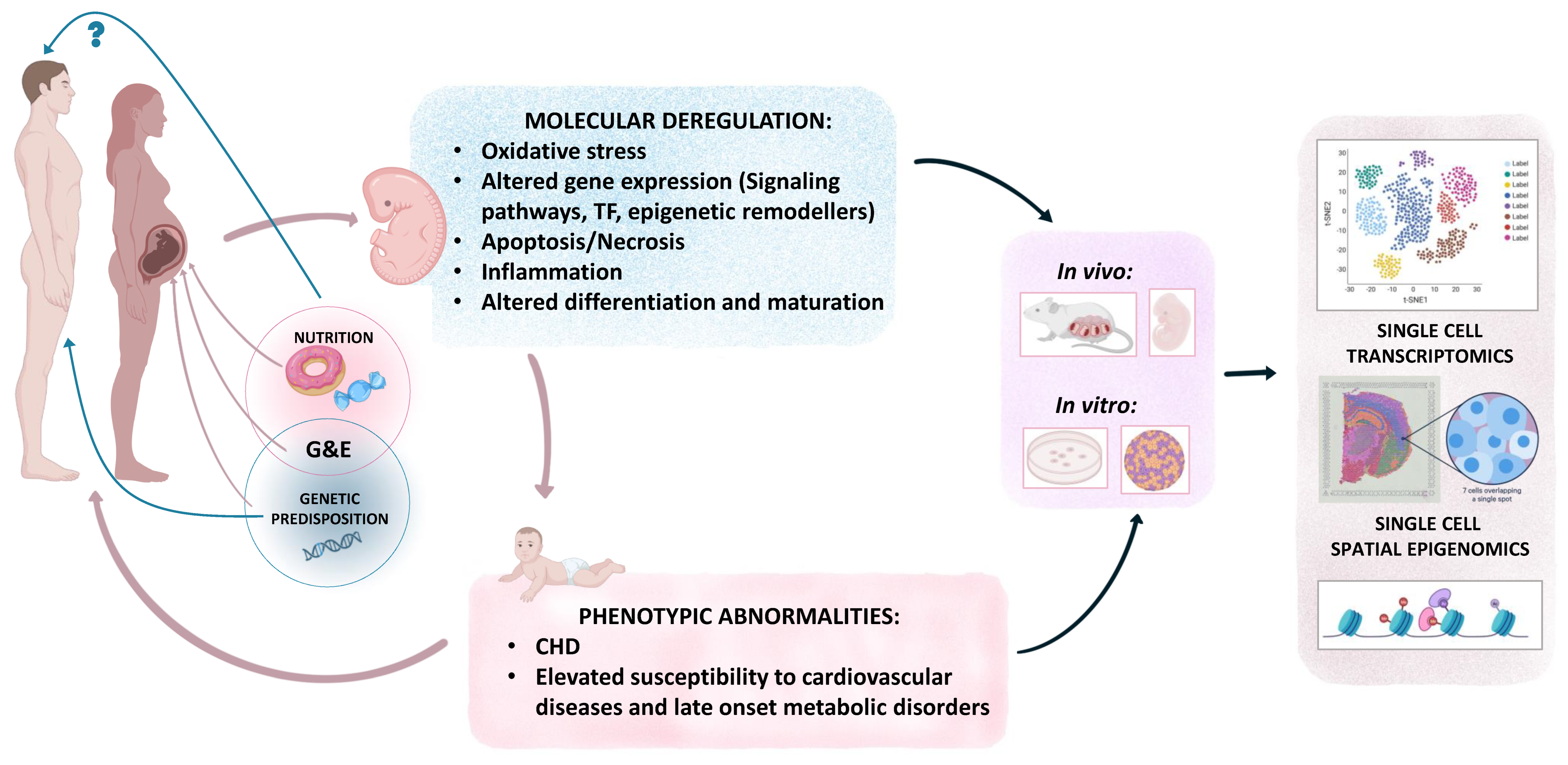
9. Heightened CHD Risk Due to Combined Effect of Genetic Predisposition and Hyperglycemia
10. Combined Teratogenicity of Diabetes and Obesity in CHDs
11. Future Directions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ogurtsova, K.; da Rocha Fernandes, J.D.; Huang, Y.; Linnenkamp, U.; Guariguata, L.; Cho, N.H.; Cavan, D.; Shaw, J.E.; Makaroff, L.E. IDF Diabetes Atlas: Global Estimates for the Prevalence of Diabetes for 2015 and 2040. Diabetes Res. Clin. Pract. 2017, 128, 40–50. [Google Scholar] [CrossRef] [PubMed]
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Hilliard, M.E.; Isaacs, D.; Johnson, E.L.; et al. 2. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes-2023. Diabetes Care 2023, 46, S19–S40. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Reece, E.A. New Concepts in Diabetic Embryopathy. Clin. Lab. Med. 2013, 33, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Kc, K.; Shakya, S.; Zhang, H. Gestational Diabetes Mellitus and Macrosomia: A Literature Review. Ann. Nutr. Metab. 2015, 66, 14–20. [Google Scholar] [CrossRef]
- van der Linde, D.; Konings, E.E.M.; Slager, M.A.; Witsenburg, M.; Helbing, W.A.; Takkenberg, J.J.M.; Roos-Hesselink, J.W. Birth Prevalence of Congenital Heart Disease Worldwide. J. Am. Coll. Cardiol. 2011, 58, 2241–2247. [Google Scholar] [CrossRef]
- Helle, E.; Priest, J.R. Maternal Obesity and Diabetes Mellitus as Risk Factors for Congenital Heart Disease in the Offspring. J. Am. Heart Assoc. 2020, 9, e011541. [Google Scholar] [CrossRef]
- Mackie, A.S.; Tran, D.T.; Marelli, A.J.; Kaul, P. Cost of Congenital Heart Disease Hospitalizations in Canada: A Population-Based Study. Can. J. Cardiol. 2017, 33, 792–798. [Google Scholar] [CrossRef]
- Zaidi, S.; Brueckner, M. Genetics and Genomics of Congenital Heart Disease. Circ. Res. 2017, 120, 923–940. [Google Scholar] [CrossRef]
- Øyen, N.; Diaz, L.J.; Leirgul, E.; Boyd, H.A.; Priest, J.; Mathiesen, E.R.; Quertermous, T.; Wohlfahrt, J.; Melbye, M. Prepregnancy Diabetes and Offspring Risk of Congenital Heart Disease: A Nationwide Cohort Study. Circulation 2016, 133, 2243–2253. [Google Scholar] [CrossRef]
- Bruneau, B.G. The Developmental Genetics of Congenital Heart Disease. Nature 2008, 451, 943–948. [Google Scholar] [CrossRef]
- Kalisch-Smith, J.I.; Ved, N.; Sparrow, D.B. Environmental Risk Factors for Congenital Heart Disease. Cold Spring Harb. Perspect. Biol. 2020, 12, a037234. [Google Scholar] [CrossRef]
- Martins, P.; Castela, E. Transposition of the Great Arteries. Orphanet J. Rare Dis. 2008, 3, 27. [Google Scholar] [CrossRef]
- Obler, D.; Juraszek, A.L.; Smoot, L.B.; Natowicz, M.R. Double Outlet Right Ventricle: Aetiologies and Associations. J. Med. Genet. 2008, 45, 481–497. [Google Scholar] [CrossRef]
- Bartelings, M.M.; Gittenberger-de Groot, A.C. Morphogenetic Considerations on Congenital Malformations of the Outflow Tract. Part 1: Common Arterial Trunk and Tetralogy of Fallot. Int. J. Cardiol. 1991, 32, 213–230. [Google Scholar] [CrossRef] [PubMed]
- Geva, T.; Martins, J.D.; Wald, R.M. Atrial Septal Defects. Lancet 2014, 383, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Calkoen, E.E.; Hazekamp, M.G.; Blom, N.A.; Elders, B.B.L.J.; Gittenberger-de Groot, A.C.; Haak, M.C.; Bartelings, M.M.; Roest, A.A.W.; Jongbloed, M.R.M. Atrioventricular Septal Defect: From Embryonic Development to Long-Term Follow-Up. Int. J. Cardiol. 2016, 202, 784–795. [Google Scholar] [CrossRef] [PubMed]
- Barron, D.J.; Kilby, M.D.; Davies, B.; Wright, J.G.; Jones, T.J.; Brawn, W.J. Hypoplastic Left Heart Syndrome. Lancet 2009, 374, 551–564. [Google Scholar] [CrossRef]
- Siu, S.C.; Silversides, C.K. Bicuspid Aortic Valve Disease. J. Am. Coll. Cardiol. 2010, 55, 2789–2800. [Google Scholar] [CrossRef]
- Franklin, R.C.G.; Béland, M.J.; Colan, S.D.; Walters, H.L.; Aiello, V.D.; Anderson, R.H.; Bailliard, F.; Boris, J.R.; Cohen, M.S.; Gaynor, J.W.; et al. Nomenclature for Congenital and Paediatric Cardiac Disease: The International Paediatric and Congenital Cardiac Code (IPCCC) and the Eleventh Iteration of the International Classification of Diseases (ICD-11). Cardiol. Young 2017, 27, 1872–1938. [Google Scholar] [CrossRef]
- LaHaye, S.; Corsmeier, D.; Basu, M.; Bowman, J.L.; Fitzgerald-Butt, S.; Zender, G.; Bosse, K.; McBride, K.L.; White, P.; Garg, V. Utilization of Whole Exome Sequencing to Identify Causative Mutations in Familial Congenital Heart Disease. Circ. Cardiovasc. Genet. 2016, 9, 320–329. [Google Scholar] [CrossRef]
- Hartman, R.J.; Rasmussen, S.A.; Botto, L.D.; Riehle-Colarusso, T.; Martin, C.L.; Cragan, J.D.; Shin, M.; Correa, A. The Contribution of Chromosomal Abnormalities to Congenital Heart Defects: A Population-Based Study. Pediatr. Cardiol. 2011, 32, 1147–1157. [Google Scholar] [CrossRef]
- McCulley, D.J.; Black, B.L. Transcription Factor Pathways and Congenital Heart Disease. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 100, pp. 253–277. ISBN 978-0-12-387786-4. [Google Scholar]
- Fahed, A.C.; Gelb, B.D.; Seidman, J.G.; Seidman, C.E. Genetics of Congenital Heart Disease: The Glass Half Empty. Circ. Res. 2013, 112, 707–720. [Google Scholar] [CrossRef]
- Bruneau, B.G. Signaling and Transcriptional Networks in Heart Development and Regeneration. Cold Spring Harb. Perspect. Biol. 2013, 5, a008292. [Google Scholar] [CrossRef]
- Lindsay, E.A.; Vitelli, F.; Su, H.; Morishima, M.; Huynh, T.; Pramparo, T.; Jurecic, V.; Ogunrinu, G.; Sutherland, H.F.; Scambler, P.J.; et al. Tbx1 Haploinsufficiency in the DiGeorge Syndrome Region Causes Aortic Arch Defects in Mice. Nature 2001, 410, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Pehlivan, T.; Pober, B.R.; Brueckner, M.; Garrett, S.; Slaugh, R.; Van Rheeden, R.; Wilson, D.B.; Watson, M.S.; Hing, A.V. GATA4 Haploinsufficiency in Patients with Interstitial Deletion of Chromosome Region 8p23.1 and Congenital Heart Disease. Am. J. Med. Genet. 1999, 83, 201–206. [Google Scholar] [CrossRef]
- Cordell, H.J.; Bentham, J.; Topf, A.; Zelenika, D.; Heath, S.; Mamasoula, C.; Cosgrove, C.; Blue, G.; Granados-Riveron, J.; Setchfield, K.; et al. Genome-Wide Association Study of Multiple Congenital Heart Disease Phenotypes Identifies a Susceptibility Locus for Atrial Septal Defect at Chromosome 4p16. Nat. Genet. 2013, 45, 822–824. [Google Scholar] [CrossRef]
- Homsy, J.; Zaidi, S.; Shen, Y.; Ware, J.S.; Samocha, K.E.; Karczewski, K.J.; DePalma, S.R.; McKean, D.; Wakimoto, H.; Gorham, J.; et al. De Novo Mutations in Congenital Heart Disease with Neurodevelopmental and Other Congenital Anomalies. Science 2015, 350, 1262–1266. [Google Scholar] [CrossRef] [PubMed]
- Vargesson, N. Thalidomide-induced Teratogenesis: History and Mechanisms. Birth Defect. Res. C 2015, 105, 140–156. [Google Scholar] [CrossRef]
- Khiali, S.; Gharekhani, A.; Entezari-Maleki, T. Isotretinoin; A Review on the Utilization Pattern in Pregnancy. Adv. Pharm. Bull. 2018, 8, 377–382. [Google Scholar] [CrossRef]
- Nora, J.J.; Nora, A.H.; Toews, W.H. Lithium, Ebstein’s Anomaly, and Other Congenital Heart Defects. Lancet 1974, 304, 594–595. [Google Scholar] [CrossRef]
- Stambolic, V.; Ruel, L.; Woodgett, J.R. Lithium Inhibits Glycogen Synthase Kinase-3 Activity and Mimics Wingless Signalling in Intact Cells. Curr. Biol. 1996, 6, 1664–1669. [Google Scholar] [CrossRef] [PubMed]
- Oster, M.E.; Riehle-Colarusso, T.; Correa, A. An Update on Cardiovascular Malformations in Congenital Rubella Syndrome. Birth Defect. Res. A 2009, 88, 1–18. [Google Scholar] [CrossRef]
- Edwards, M.J. Congenital Defects in Guinea Pigs: Fetal Resorptions, Abortions, and Malformations Following Induced Hyperthermia during Early Gestation. Teratology 1969, 2, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Karatza, A.A.; Giannakopoulos, I.; Dassios, T.G.; Belavgenis, G.; Mantagos, S.P.; Varvarigou, A.A. Periconceptional Tobacco Smoking and Xisolated Congenital Heart Defects in the Neonatal Period. Int. J. Cardiol. 2011, 148, 295–299. [Google Scholar] [CrossRef]
- Bolin, E.H.; Gokun, Y.; Romitti, P.A.; Tinker, S.C.; Summers, A.D.; Roberson, P.K.; Hobbs, C.A.; Malik, S.; Botto, L.D.; Nembhard, W.N. Maternal Smoking and Congenital Heart Defects, National Birth Defects Prevention Study, 1997–2011. J. Pediatr. 2022, 240, 79–86.e1. [Google Scholar] [CrossRef]
- Burd, L.; Deal, E.; Rios, R.; Adickes, E.; Wynne, J.; Klug, M.G. Congenital Heart Defects and Fetal Alcohol Spectrum Disorders. Congenit. Heart Dis. 2007, 2, 250–255. [Google Scholar] [CrossRef]
- Yang, J.; Qiu, H.; Qu, P.; Zhang, R.; Zeng, L.; Yan, H. Prenatal Alcohol Exposure and Congenital Heart Defects: A Meta-Analysis. PLoS ONE 2015, 10, e0130681. [Google Scholar] [CrossRef][Green Version]
- Mao, B.; Qiu, J.; Zhao, N.; Shao, Y.; Dai, W.; He, X.; Cui, H.; Lin, X.; Lv, L.; Tang, Z.; et al. Maternal Folic Acid Supplementation and Dietary Folate Intake and Congenital Heart Defects. PLoS ONE 2017, 12, e0187996. [Google Scholar] [CrossRef]
- Chen, L.; Yang, T.; Chen, L.; Wang, L.; Wang, T.; Zhao, L.; Ye, Z.; Zhang, S.; Luo, L.; Zheng, Z.; et al. Risk of Congenital Heart Defects in Offspring Exposed to Maternal Diabetes Mellitus: An Updated Systematic Review and Meta-Analysis. Arch. Gynecol. Obs. 2019, 300, 1491–1506. [Google Scholar] [CrossRef]
- Hoang, T.T.; Marengo, L.K.; Mitchell, L.E.; Canfield, M.A.; Agopian, A.J. Original Findings and Updated Meta-Analysis for the Association Between Maternal Diabetes and Risk for Congenital Heart Disease Phenotypes. Am. J. Epidemiol. 2017, 186, 118–128. [Google Scholar] [CrossRef]
- Stockman, J.A. Maternal Overweight and Obesity and the Risk of Congenital Anomalies: A Systematic Review and Meta-Analysis. Yearb. Pediatr. 2010, 2010, 451–453. [Google Scholar] [CrossRef]
- Wu, L.; Li, N.; Liu, Y. Association Between Maternal Factors and Risk of Congenital Heart Disease in Offspring: A Systematic Review and Meta-Analysis. Matern. Child. Health J. 2023, 27, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Sylva, M.; van den Hoff, M.J.B.; Moorman, A.F.M. Development of the Human Heart. Am. J. Med. Genet. 2014, 164, 1347–1371. [Google Scholar] [CrossRef] [PubMed]
- Meilhac, S.M.; Buckingham, M.E. The Deployment of Cell Lineages That Form the Mammalian Heart. Nat. Rev. Cardiol. 2018, 15, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Buijtendijk, M.F.J.; Barnett, P.; Hoff, M.J.B. Development of the Human Heart. Am. J. Med. Genet. 2020, 184, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Christoffels, V.M.; Habets, P.E.M.H.; Franco, D.; Campione, M.; de Jong, F.; Lamers, W.H.; Bao, Z.-Z.; Palmer, S.; Biben, C.; Harvey, R.P.; et al. Chamber Formation and Morphogenesis in the Developing Mammalian Heart. Dev. Biol. 2000, 223, 266–278. [Google Scholar] [CrossRef]
- Snarr, B.S.; Kern, C.B.; Wessels, A. Origin and Fate of Cardiac Mesenchyme. Dev. Dyn. 2008, 237, 2804–2819. [Google Scholar] [CrossRef]
- van Weerd, J.H.; Christoffels, V.M. The Formation and Function of the Cardiac Conduction System. Development 2016, 143, 197–210. [Google Scholar] [CrossRef]
- Lager, S.; Powell, T.L. Regulation of Nutrient Transport across the Placenta. J. Pregnancy 2012, 2012, 1–14. [Google Scholar] [CrossRef]
- Depre, C.; Vanoverschelde, J.-L.J.; Taegtmeyer, H. Glucose for the Heart. Circulation 1999, 99, 578–588. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The Key Role of Anaplerosis and Cataplerosis for Citric Acid Cycle Function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef] [PubMed]
- Yaribeygi, H.; Atkin, S.L.; Sahebkar, A. A Review of the Molecular Mechanisms of Hyperglycemia-induced Free Radical Generation Leading to Oxidative Stress. J. Cell. Physiol. 2019, 234, 1300–1312. [Google Scholar] [CrossRef]
- Mackler, B.; Grace, R.; Duncan, H.M. Studies of Mitochondrial Development during Embryogenesis in the Rat. Arch. Biochem. Biophys. 1971, 144, 603–610. [Google Scholar] [CrossRef]
- Cox, S.J.; Gunberg, D.L. Metabolite Utilization by Isolated Embryonic Rat Hearts in Vitro. J. Embryol. Exp. Morphol. 1972, 28, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Minami, I.; Braas, D.; Pappoe, H.; Wu, X.; Sagadevan, A.; Vergnes, L.; Fu, K.; Morselli, M.; Dunham, C.; et al. Glucose Inhibits Cardiac Muscle Maturation through Nucleotide Biosynthesis. eLife 2017, 6, e29330. [Google Scholar] [CrossRef]
- Jansson, T.; Wennergren, M.; Illsley, N.P. Glucose Transporter Protein Expression in Human Placenta throughout Gestation and in Intrauterine Growth Retardation. J. Clin. Endocrinol. Metab. 1993, 77, 1554–1562. [Google Scholar] [CrossRef]
- Abel, E.D. Glucose Transport in the Heart. Front. Biosci. 2004, 9, 201–215. [Google Scholar] [CrossRef]
- Shao, D.; Tian, R. Glucose Transporters in Cardiac Metabolism and Hypertrophy. In Comprehensive Physiology; Terjung, R., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 331–351. ISBN 978-0-470-65071-4. [Google Scholar]
- Baker, L.; Piddington, R.; Goldman, A.; Egler, J.; Moehring, J. Myo-Inositol and Prostaglandins Reverse the Glucose Inhibition of Neural Tube Fusion in Cultured Mouse Embryos. Diabetologia 1990, 33, 593–596. [Google Scholar] [CrossRef]
- Zabihi, S.; Loeken, M.R. Understanding Diabetic Teratogenesis: Where Are We Now and Where Are We Going? Birth Defects Res. A Clin. Mol. Teratol. 2010, 88, 779–790. [Google Scholar] [CrossRef]
- Garnham, E.A.; Beck, F.; Clarke, C.A.; Stanisstreet, M. Effects of Glucose on Rat Embryos in Culture. Diabetologia 1983, 25, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhao, S.; Zhang, Z.; Chen, Y.; Cheng, X.; Chuai, M.; Liu, G.; Lee, K.K.H.; Yang, X. High Glucose Level Induces Cardiovascular Dysplasia During Early Embryo Development. Exp. Clin. Endocrinol. Diabetes 2019, 127, 590–597. [Google Scholar] [CrossRef]
- Rousseau-Ralliard, D.; Couturier-Tarrade, A.; Thieme, R.; Brat, R.; Rolland, A.; Boileau, P.; Aubrière, M.-C.; Daniel, N.; Dahirel, M.; Derisoud, E.; et al. A Short Periconceptional Exposure to Maternal Type-1 Diabetes Is Sufficient to Disrupt the Feto-Placental Phenotype in a Rabbit Model. Mol. Cell. Endocrinol. 2019, 480, 42–53. [Google Scholar] [CrossRef]
- Lock, M.; Botting, K.; Tellam, R.; Brooks, D.; Morrison, J. Adverse Intrauterine Environment and Cardiac MiRNA Expression. Int. J. Mol. Sci. 2017, 18, 2628. [Google Scholar] [CrossRef]
- Rochette, L.; Zeller, M.; Cottin, Y.; Vergely, C. Diabetes, Oxidative Stress and Therapeutic Strategies. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2014, 1840, 2709–2729. [Google Scholar] [CrossRef] [PubMed]
- Lisowski, L.A.; Verheijen, P.M.; Copel, J.A.; Kleinman, C.S.; Wassink, S.; Visser, G.H.A.; Meijboom, E.-J. Congenital Heart Disease in Pregnancies Complicated by Maternal Diabetes Mellitus. An International Clinical Collaboration, Literature Review, and Meta-Analysis. Herz 2010, 35, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Joseph, K.S.; Lisonkova, S.; Rouleau, J.; Van den Hof, M.; Sauve, R.; Kramer, M.S.; Canadian Perinatal Surveillance System (Public Health Agency of Canada). Association between Maternal Chronic Conditions and Congenital Heart Defects: A Population-Based Cohort Study. Circulation 2013, 128, 583–589. [Google Scholar] [CrossRef]
- Stefanovic, S.; Laforest, B.; Desvignes, J.-P.; Lescroart, F.; Argiro, L.; Maurel-Zaffran, C.; Salgado, D.; Plaindoux, E.; De Bono, C.; Pazur, K.; et al. Hox-Dependent Coordination of Mouse Cardiac Progenitor Cell Patterning and Differentiation. eLife 2020, 9, e55124. [Google Scholar] [CrossRef]
- Stefanovic, S.; Etchevers, H.C.; Zaffran, S. Outflow Tract Formation—Embryonic Origins of Conotruncal Congenital Heart Disease. J. Cardiovasc. Dev. Dis. 2021, 8, 42. [Google Scholar] [CrossRef]
- Lescroart, F.; Dumas, C.E.; Adachi, N.; Kelly, R.G. Emergence of Heart and Branchiomeric Muscles in Cardiopharyngeal Mesoderm. Exp. Cell Res. 2022, 410, 112931. [Google Scholar] [CrossRef]
- Adachi, N.; Bilio, M.; Baldini, A.; Kelly, R.G. Cardiopharyngeal Mesoderm Origins of Musculoskeletal and Connective Tissues in the Mammalian Pharynx. Development 2020, 147, dev185256. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.G. Core Issues in Craniofacial Myogenesis. Exp. Cell Res. 2010, 316, 3034–3041. [Google Scholar] [CrossRef] [PubMed]
- Funato, N. Craniofacial Phenotypes and Genetics of DiGeorge Syndrome. J. Dev. Biol. 2022, 10, 18. [Google Scholar] [CrossRef]
- Morton, S.U.; Quiat, D.; Seidman, J.G.; Seidman, C.E. Genomic Frontiers in Congenital Heart Disease. Nat. Rev. Cardiol. 2022, 19, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, S.; Mansfield, C.; Zhang, X.; Kodigepalli, K.M.; Majumdar, U.; Garg, V.; Basu, M. Single-Cell Transcriptomic Profiling Unveils Dysregulation of Cardiac Progenitor Cells and Cardiomyocytes in a Mouse Model of Maternal Hyperglycemia. Commun. Biol. 2022, 5, 820. [Google Scholar] [CrossRef]
- Han, S.; Wang, G.; Jin, Y.; Ma, Z.; Jia, W.; Wu, X.; Wang, X.; He, M.; Cheng, X.; Li, W.; et al. Investigating the Mechanism of Hyperglycemia-Induced Fetal Cardiac Hypertrophy. PLoS ONE 2015, 10, e0139141. [Google Scholar] [CrossRef]
- Hatém, M.A.B.; Zielinsky, P.; Hatém, D.M.; Nicoloso, L.H.; Manica, J.L.; Piccoli, A.L.; Zanettini, J.; Oliveira, V.; Scarpa, F.; Petracco, R. Assessment of Diastolic Ventricular Function in Fetuses of Diabetic Mothers Using Tissue Doppler. Cardiol. Young 2008, 18, 297–302. [Google Scholar] [CrossRef]
- Schierz, I.A.M.; Pinello, G.; Piro, E.; Giuffrè, M.; La Placa, S.; Corsello, G. Transitional Hemodynamics in Infants of Diabetic Mothers by Targeted Neonatal Echocardiography, Electrocardiography and Peripheral Flow Study. J. Matern. Fetal Neonatal Med. 2018, 31, 1578–1585. [Google Scholar] [CrossRef]
- Zielinsky, P.; Piccoli, A.L. Myocardial Hypertrophy and Dysfunction in Maternal Diabetes. Early Hum. Dev. 2012, 88, 273–278. [Google Scholar] [CrossRef]
- Gonzalez, A.B.; Young, L.; Doll, J.A.; Morgan, G.M.; Crawford, S.E.; Plunkett, B.A. Elevated Neonatal Insulin-like Growth Factor I Is Associated with Fetal Hypertrophic Cardiomyopathy in Diabetic Women. Am. J. Obs. Gynecol. 2014, 211, 290.e1–290.e7. [Google Scholar] [CrossRef]
- El-Ganzoury, M.M.; El-Masry, S.A.; El-Farrash, R.A.; Anwar, M.; Abd Ellatife, R.Z. Infants of Diabetic Mothers: Echocardiographic Measurements and Cord Blood IGF-I and IGFBP-1. Pediatr. Diabetes 2012, 13, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Cypryk, K.; Bartyzel, L.; Zurawska-Klis, M.; Mlynarski, W.; Szadkowska, A.; Wilczynski, J.; Nowakowska, D.; Wozniak, L.A.; Fendler, W. Continuous Glucose Monitoring in Type 1 Diabetes Pregnancy Shows That Fetal Heart Rate Correlates with Maternal Glycemia. Diabetes Technol. Ther. 2015, 17, 619–624. [Google Scholar] [CrossRef]
- Bet, B.B.; De Vries, J.M.; Limpens, J.; Van Wely, M.; Van Leeuwen, E.; Clur, S.A.; Pajkrt, E. Implications of Fetal Premature Atrial Contractions: Systematic Review. Ultrasound Obs. Gyne 2022, 60, 721–730. [Google Scholar] [CrossRef]
- Krishnan, A.; Samtani, R.; Dhanantwari, P.; Lee, E.; Yamada, S.; Shiota, K.; Donofrio, M.T.; Leatherbury, L.; Lo, C.W. A Detailed Comparison of Mouse and Human Cardiac Development. Pediatr. Res. 2014, 76, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.D.; Dheen, S.T.; Tay, S.S.W. Maternal Diabetes Induces Congenital Heart Defects in Mice by Altering the Expression of Genes Involved in Cardiovascular Development. Cardiovasc. Diabetol. 2007, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Lehtoranta, L.; Vuolteenaho, O.; Laine, V.J.; Koskinen, A.; Soukka, H.; Kytö, V.; Määttä, J.; Haapsamo, M.; Ekholm, E.; Räsänen, J. Maternal Hyperglycemia Leads to Fetal Cardiac Hyperplasia and Dysfunction in a Rat Model. Am. J. Physiol. -Endocrinol. Metab. 2013, 305, E611–E619. [Google Scholar] [CrossRef]
- Morishima, M.; Yasui, H.; Ando, M.; Nakazawa, M.; Takao, A. Influence of Genetic and Maternal Diabetes in the Pathogenesis of Visceroatrial Heterotaxy in Mice. Teratology 1996, 54, 183–190. [Google Scholar] [CrossRef]
- Eriksson, U.J.; Bone, A.J.; Turnbull, D.M.; Baird, J.D. Timed Interruption of Insulin Therapy in Diabetic BB/E Rat Pregnancy: Effect on Maternal Metabolism and Fetal Outcome. Acta Endocrinol. 1989, 120, 800–810. [Google Scholar] [CrossRef]
- Zangen, S.W.; Yaffe, P.; Shechtman, S.; Zangen, D.H.; Ornoy, A. The Role of Reactive Oxygen Species in Diabetes-Induced Anomalies in Embryos of Cohen Diabetic Rats. Int. J. Exp. Diabetes Res. 2002, 3, 247–255. [Google Scholar] [CrossRef]
- Salbaum, J.M.; Kruger, C.; MacGowan, J.; Herion, N.J.; Burk, D.; Kappen, C. Novel Mode of Defective Neural Tube Closure in the Non-Obese Diabetic (NOD) Mouse Strain. Sci. Rep. 2015, 5, 16917. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, F.; Fu, M.; Wang, C.; Quon, M.J.; Yang, P. Cellular Stress, Excessive Apoptosis, and the Effect of Metformin in a Mouse Model of Type 2 Diabetic Embryopathy. Diabetes 2015, 64, 2526–2536. [Google Scholar] [CrossRef] [PubMed]
- Jaime-Cruz, R.; Sánchez-Gómez, C.; Villavicencio-Guzmán, L.; Lazzarini-Lechuga, R.; Patiño-Morales, C.C.; García-Lorenzana, M.; Ramírez-Fuentes, T.C.; Salazar-García, M. Embryonic Hyperglycemia Disrupts Myocardial Growth, Morphological Development, and Cellular Organization: An In Vivo Experimental Study. Life 2023, 13, 768. [Google Scholar] [CrossRef]
- Liang, J.; Gui, Y.; Wang, W.; Gao, S.; Li, J.; Song, H. Elevated Glucose Induces Congenital Heart Defects by Altering the Expression of Tbx5, Tbx20, and Has2 in Developing Zebrafish Embryos. Birth Defects Res. Part. A Clin. Mol. Teratol. 2010, 88, 480–486. [Google Scholar] [CrossRef]
- Guest, P.C. Characterization of the Goto-Kakizaki (GK) Rat Model of Type 2 Diabetes. Methods Mol. Biol. 2019, 1916, 203–211. [Google Scholar] [CrossRef]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N.; et al. Self-Assembling Human Heart Organoids for the Modeling of Cardiac Development and Congenital Heart Disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.; Zhu, J.-Y.; LaHaye, S.; Majumdar, U.; Jiao, K.; Han, Z.; Garg, V. Epigenetic Mechanisms Underlying Maternal Diabetes-Associated Risk of Congenital Heart Disease. JCI Insight 2017, 2, e95085. [Google Scholar] [CrossRef]
- Yang, P.; Chen, X.; Kaushal, S.; Reece, E.A.; Yang, P. High Glucose Suppresses Embryonic Stem Cell Differentiation into Cardiomyocytes: High Glucose Inhibits ES Cell Cardiogenesis. Stem Cell Res. Ther. 2016, 7, 187. [Google Scholar] [CrossRef]
- Díaz Del Moral, S.; Benaouicha, M.; Muñoz-Chápuli, R.; Carmona, R. The Insulin-like Growth Factor Signalling Pathway in Cardiac Development and Regeneration. Int. J. Mol. Sci. 2021, 23, 234. [Google Scholar] [CrossRef]
- Pavlinkova, G.; Salbaum, J.M.; Kappen, C. Maternal Diabetes Alters Transcriptional Programs in the Developing Embryo. BMC Genom. 2009, 10, 274. [Google Scholar] [CrossRef]
- Vijaya, M.; Manikandan, J.; Parakalan, R.; Dheen, S.T.; Kumar, S.D.; Tay, S.S.W. Differential Gene Expression Profiles during Embryonic Heart Development in Diabetic Mice Pregnancy. Gene 2013, 516, 218–227. [Google Scholar] [CrossRef]
- Alam, M.J.; Uppulapu, S.K.; Tiwari, V.; Varghese, B.; Mohammed, S.A.; Adela, R.; Arava, S.K.; Banerjee, S.K. Pregestational Diabetes Alters Cardiac Structure and Function of Neonatal Rats through Developmental Plasticity. Front. Cardiovasc. Med. 2022, 9, 919293. [Google Scholar] [CrossRef] [PubMed]
- Bohuslavova, R.; Skvorova, L.; Sedmera, D.; Semenza, G.L.; Pavlinkova, G. Increased Susceptibility of HIF-1α Heterozygous-Null Mice to Cardiovascular Malformations Associated with Maternal Diabetes. J. Mol. Cell. Cardiol. 2013, 60, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Engineer, A.; Saiyin, T.; Lu, X.; Kucey, A.S.; Urquhart, B.L.; Drysdale, T.A.; Norozi, K.; Feng, Q. Sapropterin Treatment Prevents Congenital Heart Defects Induced by Pregestational Diabetes Mellitus in Mice. J. Am. Heart Assoc. 2018, 7, e009624. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.C.; Prater, M.R.; Smith, B.J.; Freeman, L.E.; Mallela, M.K.; Holladay, S.D. Late-Gestation Ventricular Myocardial Reduction in Fetuses of Hyperglycemic CD1 Mice Is Associated with Increased Apoptosis. Birth Defect. Res. B 2009, 86, 409–415. [Google Scholar] [CrossRef]
- Kumar, S.D.; Yong, S.-K.; Dheen, S.T.; Bay, B.-H.; Tay, S.S.-W. Cardiac Malformations Are Associated with Altered Expression of Vascular Endothelial Growth Factor and Endothelial Nitric Oxide Synthase Genes in Embryos of Diabetic Mice. Exp. Biol. Med. 2008, 233, 1421–1432. [Google Scholar] [CrossRef]
- Lin, N.; Cai, Y.; Zhang, L.; Chen, Y. Identification of Key Genes Associated with Congenital Heart Defects in Embryos of Diabetic Mice. Mol. Med. Rep. 2017, 17, 3697–3707. [Google Scholar] [CrossRef]
- Moazzen, H.; Lu, X.; Ma, N.L.; Velenosi, T.J.; Urquhart, B.L.; Wisse, L.J.; Gittenberger-de Groot, A.C.; Feng, Q. N-Acetylcysteine Prevents Congenital Heart Defects Induced by Pregestational Diabetes. Cardiovasc. Diabetol. 2014, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Roest, P.A.M.; Molin, D.G.M.; Schalkwijk, C.G.; van Iperen, L.; Wentzel, P.; Eriksson, U.J.; Gittenberger-de Groot, A.C. Specific Local Cardiovascular Changes of Nɛ-(Carboxymethyl)Lysine, Vascular Endothelial Growth Factor, and Smad2 in the Developing Embryos Coincide With Maternal Diabetes–Induced Congenital Heart Defects. Diabetes 2009, 58, 1222–1228. [Google Scholar] [CrossRef]
- Saiyin, T.; Engineer, A.; Greco, E.R.; Kim, M.Y.; Lu, X.; Jones, D.L.; Feng, Q. Maternal Voluntary Exercise Mitigates Oxidative Stress and Incidence of Congenital Heart Defects in Pre-gestational Diabetes. J. Cell. Mol. Med. 2019, 23, 5553–5565. [Google Scholar] [CrossRef]
- Scott-Drechsel, D.E.; Rugonyi, S.; Marks, D.L.; Thornburg, K.L.; Hinds, M.T. Hyperglycemia Slows Embryonic Growth and Suppresses Cell Cycle via Cyclin D1 and P21. Diabetes 2013, 62, 234–242. [Google Scholar] [CrossRef]
- Wang, F.; Wu, Y.; Quon, M.J.; Li, X.; Yang, P. ASK1 Mediates the Teratogenicity of Diabetes in the Developing Heart by Inducing ER Stress and Inhibiting Critical Factors Essential for Cardiac Development. Am. J. Physiol. -Endocrinol. Metab. 2015, 309, E487–E499. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Fisher, S.A.; Zhong, J.; Wu, Y.; Yang, P. Superoxide Dismutase 1 In Vivo Ameliorates Maternal Diabetes Mellitus–Induced Apoptosis and Heart Defects Through Restoration of Impaired Wnt Signaling. Circ. Cardiovasc. Genet. 2015, 8, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Reece, E.A.; Yang, P. Oxidative Stress Is Responsible for Maternal Diabetes-Impaired Transforming Growth Factor Beta Signaling in the Developing Mouse Heart. Am. J. Obstet. Gynecol. 2015, 212, 650.e1–650.e11. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Reece, E.A.; Zhong, J.; Dong, D.; Shen, W.-B.; Harman, C.R.; Yang, P. Type 2 Diabetes Mellitus Induces Congenital Heart Defects in Murine Embryos by Increasing Oxidative Stress, Endoplasmic Reticulum Stress, and Apoptosis. Am. J. Obs. Gynecol. 2016, 215, 366.e1–366.e10. [Google Scholar] [CrossRef]
- Zhao, Z. TGFβ and Wnt in Cardiac Outflow Tract Defects in Offspring of Diabetic Pregnancies: OFT MALFORMATIONS IN DIABETIC EMBRYOPATHY. Birth Defects Res. B 2014, 101, 364–370. [Google Scholar] [CrossRef]
- Zhao, Z. Cardiac Malformations and Alteration of TGFβ Signaling System in Diabetic Embryopathy. Birth Defect. Res. B 2010, 89, 97–105. [Google Scholar] [CrossRef]
- Zhao, J.; Hakvoort, T.B.M.; Willemsen, A.M.; Jongejan, A.; Sokolovic, M.; Bradley, E.J.; de Boer, V.C.J.; Baas, F.; van Kampen, A.H.C.; Lamers, W.H. Effect of Hyperglycemia on Gene Expression during Early Organogenesis in Mice. PLoS ONE 2016, 11, e0158035. [Google Scholar] [CrossRef]
- Morgan, S.C.; Relaix, F.; Sandell, L.L.; Loeken, M.R. Oxidative Stress during Diabetic Pregnancy Disrupts Cardiac Neural Crest Migration and Causes Outflow Tract Defects. Birth Defect. Res. A 2008, 82, 453–463. [Google Scholar] [CrossRef]
- Wentzel, P.; Gäreskog, M.; Eriksson, U.J. Decreased Cardiac Glutathione Peroxidase Levels and Enhanced Mandibular Apoptosis in Malformed Embryos of Diabetic Rats. Diabetes 2008, 57, 3344–3352. [Google Scholar] [CrossRef]
- Herb, M.; Gluschko, A.; Schramm, M. Reactive Oxygen Species: Not Omnipresent but Important in Many Locations. Front. Cell Dev. Biol. 2021, 9, 716406. [Google Scholar] [CrossRef]
- Brownlee, M. Biochemistry and Molecular Cell Biology of Diabetic Complications. Nature 2001, 414, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.D.; Vijaya, M.; Samy, R.P.; Dheen, S.T.; Ren, M.; Watt, F.; Kang, Y.J.; Bay, B.-H.; Tay, S.S.W. Zinc Supplementation Prevents Cardiomyocyte Apoptosis and Congenital Heart Defects in Embryos of Diabetic Mice. Free Radic. Biol. Med. 2012, 53, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zhao, Z.; Reece, E.A. Activation of Oxidative Stress Signaling That Is Implicated in Apoptosis with a Mouse Model of Diabetic Embryopathy. Am. J. Obstet. Gynecol. 2008, 198, 130.e1–130.e7. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Yang, W.W.; Chen, X.; Kaushal, S.; Dong, D.; Shen, W.-B. Maternal Diabetes and High Glucose in Vitro Trigger Sca1+ Cardiac Progenitor Cell Apoptosis through FoxO3a. Biochem. Biophys. Res. Commun. 2017, 482, 575–581. [Google Scholar] [CrossRef]
- Sivan, E.; Lee, Y.-C.; Wu, Y.-K.; Reece, E.A. Free Radical Scavenging Enzymes in Fetal Dysmorphogenesis among Offspring of Diabetic Rats. Teratology 1997, 56, 343–349. [Google Scholar] [CrossRef]
- Sato, H.; Leonardi, M.L.; Roberti, S.L.; Jawerbaum, A.; Higa, R. Maternal Diabetes Increases FOXO1 Activation during Embryonic Cardiac Development. Mol. Cell. Endocrinol. 2023, 575, 111999. [Google Scholar] [CrossRef]
- Kostina, A.; Lewis-Israeli, Y.R.; Abdelhamid, M.; Gabalski, M.A.; Volmert, B.D.; Lankerd, H.; Huang, A.R.; Wasserman, A.H.; Lydic, T.; Chan, C.; et al. ER Stress and Lipid Imbalance Drive Embryonic Cardiomyopathy in a Human Heart Organoid Model of Pregestational Diabetes. bioRxiv 2023. preprint. [Google Scholar] [CrossRef]
- de la Pompa, J.L.; Timmerman, L.A.; Takimoto, H.; Yoshida, H.; Elia, A.J.; Samper, E.; Potter, J.; Wakeham, A.; Marengere, L.; Langille, B.L.; et al. Role of the NF-ATc Transcription Factor in Morphogenesis of Cardiac Valves and Septum. Nature 1998, 392, 182–186. [Google Scholar] [CrossRef]
- Niessen, K.; Fu, Y.; Chang, L.; Hoodless, P.A.; McFadden, D.; Karsan, A. Slug Is a Direct Notch Target Required for Initiation of Cardiac Cushion Cellularization. J. Cell Biol. 2008, 182, 315–325. [Google Scholar] [CrossRef]
- Hachisuga, M.; Oki, S.; Kitajima, K.; Ikuta, S.; Sumi, T.; Kato, K.; Wake, N.; Meno, C. Hyperglycemia Impairs Left–Right Axis Formation and Thereby Disturbs Heart Morphogenesis in Mouse Embryos. Proc. Natl. Acad. Sci. USA 2015, 112, E5300–E5307. [Google Scholar] [CrossRef]
- Feng, Q.; Song, W.; Lu, X.; Hamilton, J.A.; Lei, M.; Peng, T.; Yee, S.-P. Development of Heart Failure and Congenital Septal Defects in Mice Lacking Endothelial Nitric Oxide Synthase. Circulation 2002, 106, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Hamblet, N.S.; Lijam, N.; Ruiz-Lozano, P.; Wang, J.; Yang, Y.; Luo, Z.; Mei, L.; Chien, K.R.; Sussman, D.J.; Wynshaw-Boris, A. Dishevelled 2 Is Essential for Cardiac Outflow Tract Development, Somite Segmentation and Neural Tube Closure. Development 2002, 129, 5827–5838. [Google Scholar] [CrossRef] [PubMed]
- Sanford, L.P.; Ormsby, I.; Gittenberger-de Groot, A.C.; Sariola, H.; Friedman, R.; Boivin, G.P.; Cardell, E.L.; Doetschman, T. TGFbeta2 Knockout Mice Have Multiple Developmental Defects That Are Non-Overlapping with Other TGFbeta Knockout Phenotypes. Development 1997, 124, 2659–2670. [Google Scholar] [CrossRef] [PubMed]
- van Weerd, J.H.; Koshiba-Takeuchi, K.; Kwon, C.; Takeuchi, J.K. Epigenetic Factors and Cardiac Development. Cardiovasc. Res. 2011, 91, 203–211. [Google Scholar] [CrossRef]
- Meier, K.; Brehm, A. Chromatin Regulation: How Complex Does It Get? Epigenetics 2014, 9, 1485–1495. [Google Scholar] [CrossRef]
- Nora, J.J. Multifactorial Inheritance Hypothesis for the Etiology of Congenital Heart Diseases. The Genetic-Environmental Interaction. Circulation 1968, 38, 604–617. [Google Scholar] [CrossRef]
- Zhu, H.; Kartiko, S.; Finnell, R. Importance of Gene-Environment Interactions in the Etiology of Selected Birth Defects. Clin. Genet. 2009, 75, 409–423. [Google Scholar] [CrossRef]
- Moreau, J.L.M.; Kesteven, S.; Martin, E.M.M.A.; Lau, K.S.; Yam, M.X.; O’Reilly, V.C.; del Monte-Nieto, G.; Baldini, A.; Feneley, M.P.; Moon, A.M.; et al. Gene-Environment Interaction Impacts on Heart Development and Embryo Survival. Development 2019, 146, dev172957. [Google Scholar] [CrossRef]
- Teramo, K.; Klemetti, M.; Tikkanen, M.; Nuutila, M. Maternal diabetes and fetal hypoxia. Duodecim 2013, 129, 228–234. [Google Scholar]
- Zhao, M.; Diao, J.; Huang, P.; Li, J.; Li, Y.; Yang, Y.; Luo, L.; Zhang, S.; Chen, L.; Wang, T.; et al. Association of Maternal Diabetes Mellitus and Polymorphisms of the NKX2.5 Gene in Children with Congenital Heart Disease: A Single Centre-Based Case-Control Study. J. Diabetes Res. 2020, 2020, 3854630. [Google Scholar] [CrossRef]
- Gauderman, W.J.; Mukherjee, B.; Aschard, H.; Hsu, L.; Lewinger, J.P.; Patel, C.J.; Witte, J.S.; Amos, C.; Tai, C.G.; Conti, D.; et al. Update on the State of the Science for Analytical Methods for Gene-Environment Interactions. Am. J. Epidemiol. 2017, 186, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Morton, S.U.; Pereira, A.C.; Quiat, D.; Richter, F.; Kitaygorodsky, A.; Hagen, J.; Bernstein, D.; Brueckner, M.; Goldmuntz, E.; Kim, R.W.; et al. Genome-Wide De Novo Variants in Congenital Heart Disease Are Not Associated With Maternal Diabetes or Obesity. Circ. Genom. Precis. Med. 2022, 15, e003500. [Google Scholar] [CrossRef] [PubMed]
- Chapman, G.; Moreau, J.L.M.; Ip, E.; Szot, J.O.; Iyer, K.R.; Shi, H.; Yam, M.X.; O’Reilly, V.C.; Enriquez, A.; Greasby, J.A.; et al. Functional Genomics and Gene-Environment Interaction Highlight the Complexity of Congenital Heart Disease Caused by Notch Pathway Variants. Hum. Mol. Genet. 2020, 29, 566–579. [Google Scholar] [CrossRef]
- Kristensen, J.; Vestergaard, M.; Wisborg, K.; Kesmodel, U.; Secher, N.J. Pre-Pregnancy Weight and the Risk of Stillbirth and Neonatal Death. BJOG Int. J. Obstet. Gynaecol. 2005, 112, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Watkins, M.L.; Rasmussen, S.A.; Honein, M.A.; Botto, L.D.; Moore, C.A. Maternal Obesity and Risk for Birth Defects. Pediatrics 2003, 111, 1152–1158. [Google Scholar] [CrossRef]
- Madsen, N.L.; Schwartz, S.M.; Lewin, M.B.; Mueller, B.A. Prepregnancy Body Mass Index and Congenital Heart Defects among Offspring: A Population-Based Study: Prepregnancy BMI and Congenital Heart Defects. Congenit. Heart Dis. 2013, 8, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Deputy, N.P.; Dub, B.; Sharma, A.J. Prevalence and Trends in Prepregnancy Normal Weight—48 States, New York City, and District of Columbia, 2011–2015. MMWR Morb. Mortal. Wkly. Rep. 2018, 66, 1402–1407. [Google Scholar] [CrossRef]
- Persson, M.; Razaz, N.; Edstedt Bonamy, A.-K.; Villamor, E.; Cnattingius, S. Maternal Overweight and Obesity and Risk of Congenital Heart Defects. J. Am. Coll. Cardiol. 2019, 73, 44–53. [Google Scholar] [CrossRef]
- Wu, X.; Ge, R.; Huang, L.; Tian, F.; Chen, Y.; Wu, L.; Niu, J. Pregestational Diabetes Mediates the Association between Maternal Obesity and the Risk of Congenital Heart Defects. J. Diabetes Invest. 2022, 13, 367–374. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef]
- Preissl, S.; Gaulton, K.J.; Ren, B. Characterizing Cis-Regulatory Elements Using Single-Cell Epigenomics. Nat. Rev. Genet. 2023, 24, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Delpierre, C.; Lepeule, J.; Cordier, S.; Slama, R.; Heude, B.; Charles, M.-A. DOHaD: Les Apports Récents de l’épidémiologie. Med. Sci. 2016, 32, 21–26. [Google Scholar] [CrossRef][Green Version]
- Hammer, C.; Caton, J.; Dahlen, C.; Ward, A.; Borowicz, P.; Reynolds, L. DOHaD: A MENAGERIE OF ADAPTATIONS AND PERSPECTIVES: Large Animal Models of Developmental Programming: Sustenance, Stress, and Sex Matter. Reproduction 2023, 165, F1–F13. [Google Scholar] [CrossRef] [PubMed]
- Dunford, A.R.; Sangster, J.M. Maternal and Paternal Periconceptional Nutrition as an Indicator of Offspring Metabolic Syndrome Risk in Later Life through Epigenetic Imprinting: A Systematic Review. Diabetes Metab. Syndr. 2017, 11 (Suppl. S2), S655–S662. [Google Scholar] [CrossRef] [PubMed]
| Diabetes Mellitus Models | Advantages | Disadvantages | |
|---|---|---|---|
| in vivo | Chemically induced models: STZ-induced | Quick (rapid onset after STZ injection) and cost-effective method. | Cytotoxicity of STZ to liver and kidneys and mild teratogenic effect. |
| Simulate T1D and when combined to HFD can simulate T2D. | Heterogeneous response to STZ (genetic background, and strain). | ||
| Genetic models: spontaneous mutations (NOD, BioBreeding and Cohen rats) or transgenic models (Akita mice) | Simulate T1D. | Use of genetic models in cardiac development is limited. | |
| No chemical toxicity on vital organs. | Time needed to generate transgenic diabetic females (multiple rounds of breeding to get the desired genotype). | ||
| Diabetic models induced by spontaneous mutations can take up to 30 weeks to show diabetic characteristics. | |||
| Diet-induced: HFD | Recapitulate key features of T2D (hyperglycemia, insulin resistance and obesity). | Time consuming: it takes 15 weeks of HFD to get a diabetic state (not recommended for developmental biology studies as old mice have reduced fertility). | |
| No insulin deficiency. | Lipotoxicity making it hard to study the isolated effect of glucotoxicity. | ||
| No chemical toxicity on vital organs. | |||
| in vitro | 2D culture | Permit to study the effect of an isolated environmental factor (ex: hyperglycemia and/or hyperinsulenmia) on a specific cardiac cell type. | Need for in vivo validation. |
| Large scale screening. | Does not reproduce system biology with the complex interactions between different organs and tissues. | ||
| Quick and easy. | |||
| Reduce the use of animal models and less ethical issues. | |||
| 3D culture: organoïds | Permit to study the effect of an isolated environmental factor (ex: hyperglycemia and/or hyperinsulenmia): simultaneously on several subtypes of cardiac cells, and also at different stages of specification and differentiation. | Need for in vivo validation. | |
| More physiological context than 2D culture. | Does not reproduce system biology, with the complex interactions between different organs and tissues. | ||
| Large scale screening. | Lack of standardization and reproducibility in generating cardiac organoids. | ||
| Quick and easy. | |||
| Reduce the use of animal models and less ethical issues. |
| Experimental Animal Model of Pregestational Diabetes | Observed CHD | Dysregulated Genes | Experimental Approach | Reference |
|---|---|---|---|---|
| STZ-induced Sprague Dawley rats | N/A | 68 upregulated genes 271 downregulated genes | RNA-Seq | [103] |
| STZ-induced C57BL/6 mice | VSD | Notch1, Hey2, EfnB2, Nrg1, Bmp10, Jarid2 and Nos3 | RT-PCR | [98] |
| STZ-induced FVB mice | VSD and thin myocardial wall | Hif1α, Nkx2.5, Tbx5, Mef2C, α-SMA Cx43, Nppa | RT-PCR | [104] |
| STZ-induced C57BL/6 mice | ASD, VSD, AVSD, PTA, DORV, pulmonary valve stenosis, aortic valve stenosis, RV hypertrophy, HLHS, narrowing of the aorta | Gata4, Tbx5, Nkx2.5, Gata5, Bmp10, Notch1, Gch1 and Dhfr | RT-PCR | [105] |
| STZ-induced CD1 mice | N/A | Bcl2, Casp3, and Casp9 | RT-PCR | [106] |
| STZ-induced Swiss Albino mice | PTA, VSD, AVSD and defective valve development | Bmp4, Msx1, and Pax3 | RT-PCR | [87] |
| STZ-induced Swiss Albino mice | N/A | eNOS and VEGF | RT-PCR | [107] |
| STZ-induced Swiss Albino mice | N/A | 411 upregulated and 458 downregulated genes at E13.5 | Affymetrix Mouse Genome 430 2.0 microarrays (GSE32078) | [102,108] |
| 802 upregulated and 1295 downregulated genes at E15.5 | ||||
| STZ-induced C57BL/6 mice | N/A | Isl1, Tbx1, Tbx20, Fgf10, Mef2c, Nkx2-5, and Hand2 | scRNA-Seq | [77] |
| STZ-induced C57BL/6 mice | VSD, ASD, AVSD, TGA, DORV and TOF | Gata4, Gata5, and Vegfa | RT-PCR | [109] |
| STZ-induced Sprague Dawley rats | OFT defects | Vegf | In situ hybridization | [110] |
| STZ-induced C57BL/6 mice | ASD, VSD, AVSD, DORV, pulmonary valve stenosis, aortic valve stenosis, HLHS, HRHS | Gata4, Hif1a, Cyclin D1, Notch1 and Snail1 | RT-PCR | [111] |
| Glucose treated fertilized chicken eggs | Glut1, p21 and Cyclin D1 | RT-PCR | [112] | |
| STZ-induced C57BL/6 mice | PTA, VSD, right sided aortic arch | Cyclin D1, Cyclin D3, p21, p27, BMP4, Nkx2.5, and GATA4 | RT-PCR | [113] |
| STZ-induced C57BL/6 mice | VSD, PTA | sFRP1, Dkk1, β-catenin, Islet1, Gja1, Versican, Wnt5a, NFAT2/4, Mrtf-b, Tpm1, and Rcan1 | RT-PCR | [114] |
| STZ-induced C57BL/6 mice | TGFβ1, TGFβ3, TβRII, Smad2/3, Snai2, CTGF, and GDF1 | RT-PCR | [115] | |
| HFD-induced C57BL/6 mice | PTA, VSD | BiP, CHOP, calnexin, PDIA, GRP94, and XBP1 | RT-PCR | [116] |
| STZ-induced C57BL/6 mice | Aortic and pulmonary stenosis, PTA | 24 genes of the WNT pathway were differentially expressed | RT-PCR | [117] |
| STZ-induced C57BL/6 mice | HRHS, heterotaxia, thin myocardial wall, endocardial cushions defect, VSD | 22 genes of the TGFβ signaling were differentially expressed | RT-PCR | [118] |
| STZ-induced FVB mice | N/A | 1024 differentially expressed genes on E8.5, and 2148 genes on E9.5 | SOLiD SAGE mRNA deep sequencing (PRJNA275285) | [119] |
| Ndufa6, Actg1, Glut4, Fgf2, Tpm4, Marcksl1, Myosin1H, Axin1, Mrto4, Psmc4, Ptpn11, Mrps23, Rnaseh2c, Cdk1 and Ubxn8 | RT-PCR | |||
| STZ-induced FVB mice | OFT defects | Pax3 | LacZ reporter assay | [120] |
| STZ-induced Sprague Dawley rats | N/A | Nppa, Nppb, Myh2, Myh3, Atp2a2, Kcnip2, Ucp2/3, Slc2a4, Egln3, and Tnfrsf12a | RT-PCR | [88] |
| STZ-induced Sprague Dawley rats | N/A | CuZnSOD, MnSOD, Gpx-1, AR, p53, PARP, Shh, Ret, Bmp-4, VEGF-A, and TNF-α | RT-PCR | [121] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, S.; Gaborit, B.; Lenoir, M.; Collod-Beroud, G.; Stefanovic, S. Maternal Pre-Existing Diabetes: A Non-Inherited Risk Factor for Congenital Cardiopathies. Int. J. Mol. Sci. 2023, 24, 16258. https://doi.org/10.3390/ijms242216258
Ibrahim S, Gaborit B, Lenoir M, Collod-Beroud G, Stefanovic S. Maternal Pre-Existing Diabetes: A Non-Inherited Risk Factor for Congenital Cardiopathies. International Journal of Molecular Sciences. 2023; 24(22):16258. https://doi.org/10.3390/ijms242216258
Chicago/Turabian StyleIbrahim, Stéphanie, Bénédicte Gaborit, Marien Lenoir, Gwenaelle Collod-Beroud, and Sonia Stefanovic. 2023. "Maternal Pre-Existing Diabetes: A Non-Inherited Risk Factor for Congenital Cardiopathies" International Journal of Molecular Sciences 24, no. 22: 16258. https://doi.org/10.3390/ijms242216258
APA StyleIbrahim, S., Gaborit, B., Lenoir, M., Collod-Beroud, G., & Stefanovic, S. (2023). Maternal Pre-Existing Diabetes: A Non-Inherited Risk Factor for Congenital Cardiopathies. International Journal of Molecular Sciences, 24(22), 16258. https://doi.org/10.3390/ijms242216258








