Unraveling Connective Tissue Growth Factor as a Therapeutic Target and Assessing Kahweol as a Potential Drug Candidate in Triple-Negative Breast Cancer Treatment
Abstract
:1. Introduction
2. Results
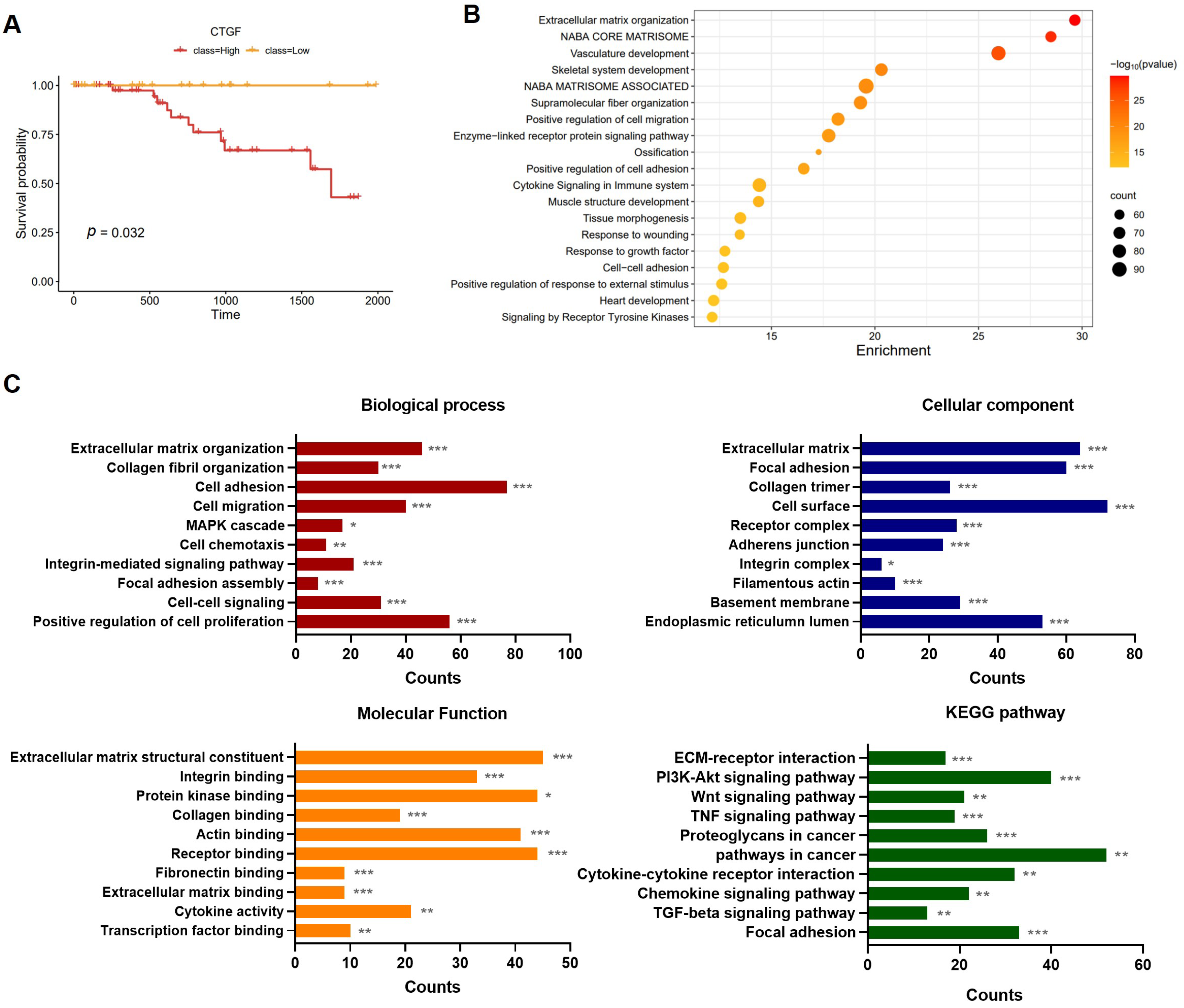
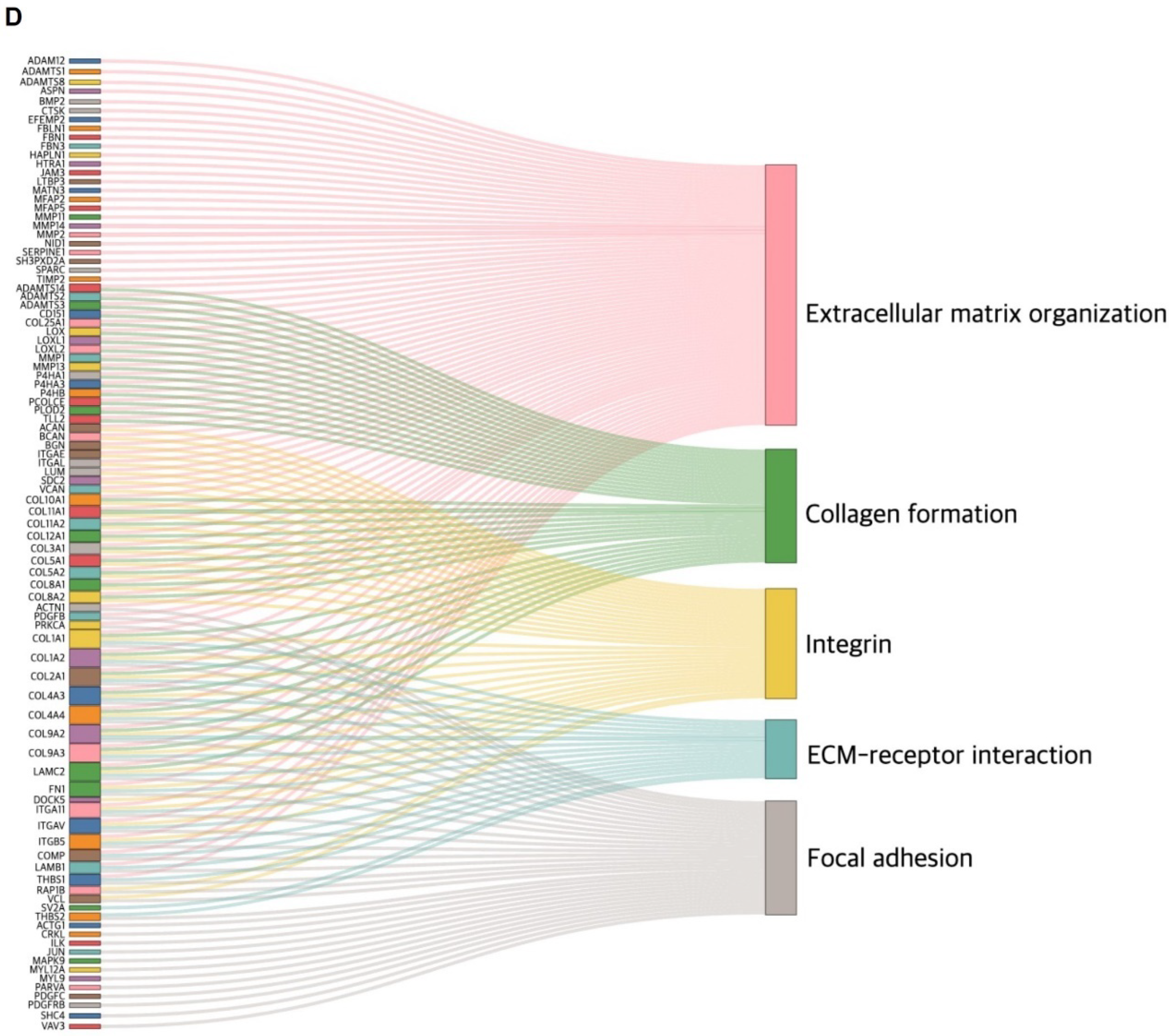
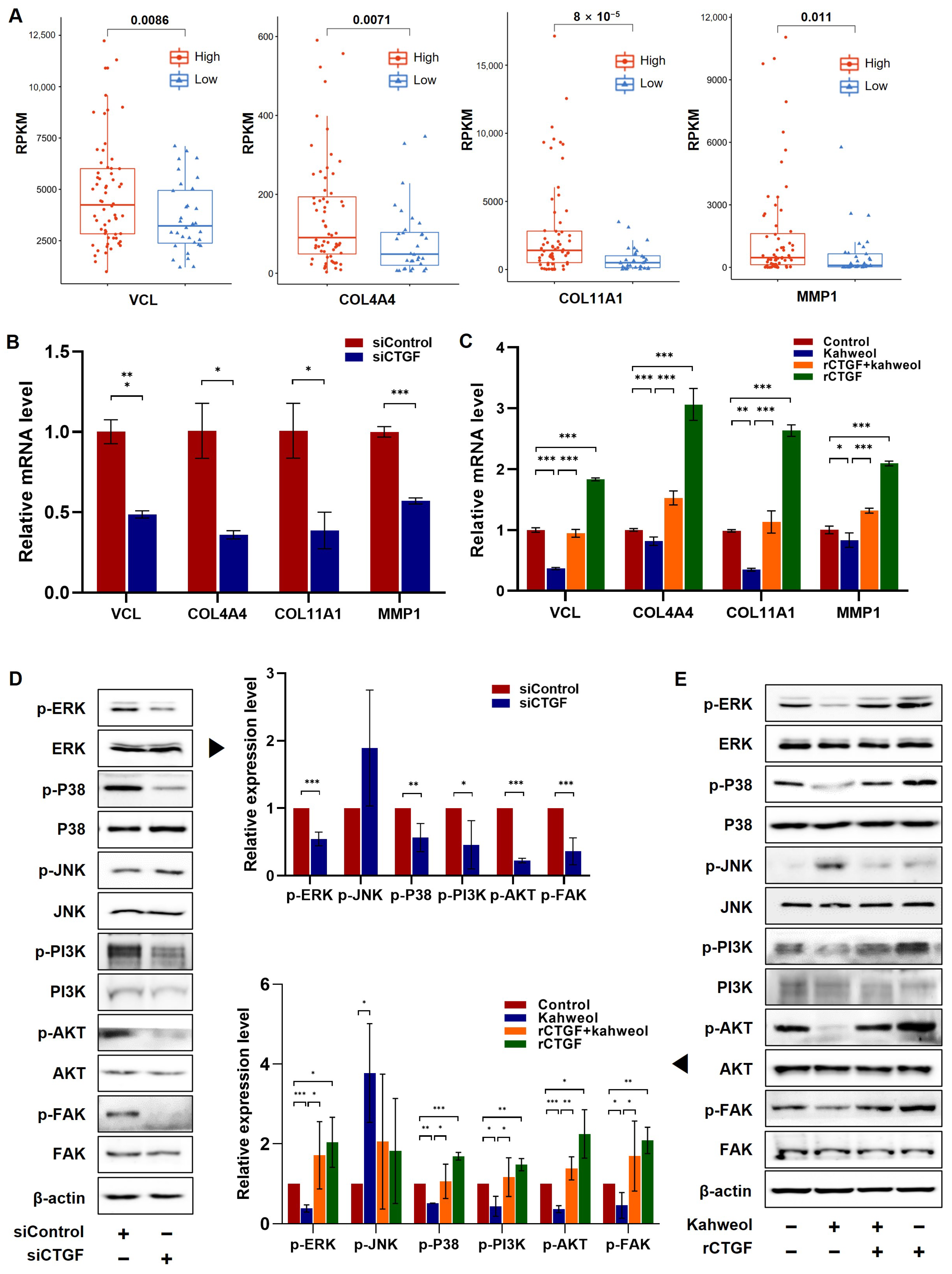
2.1. The Involvement of CTGF Expression in Survival and Cancer Progression in TNBC
2.2. CTGF Contributes to Cell Motility in TNBC
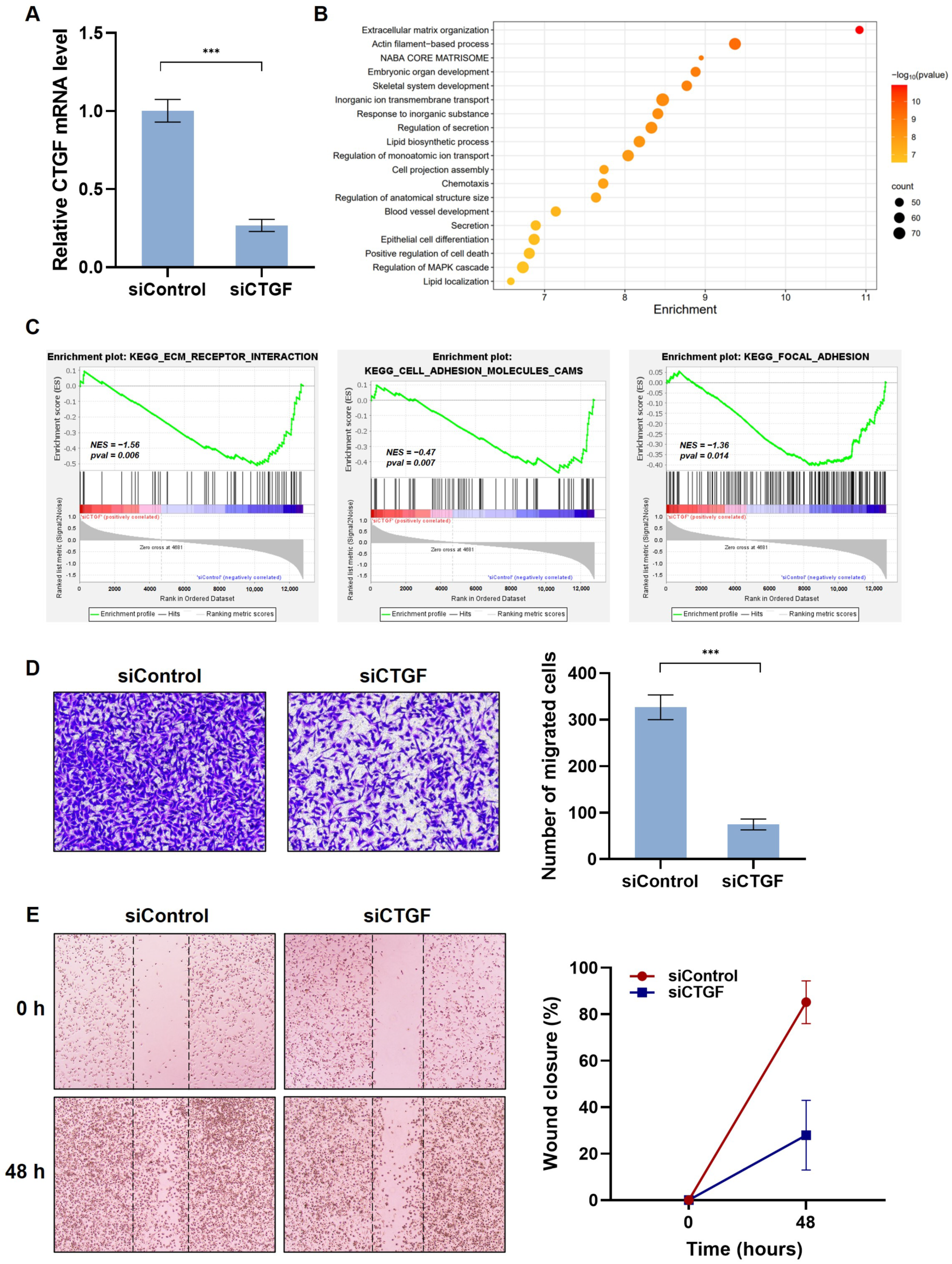
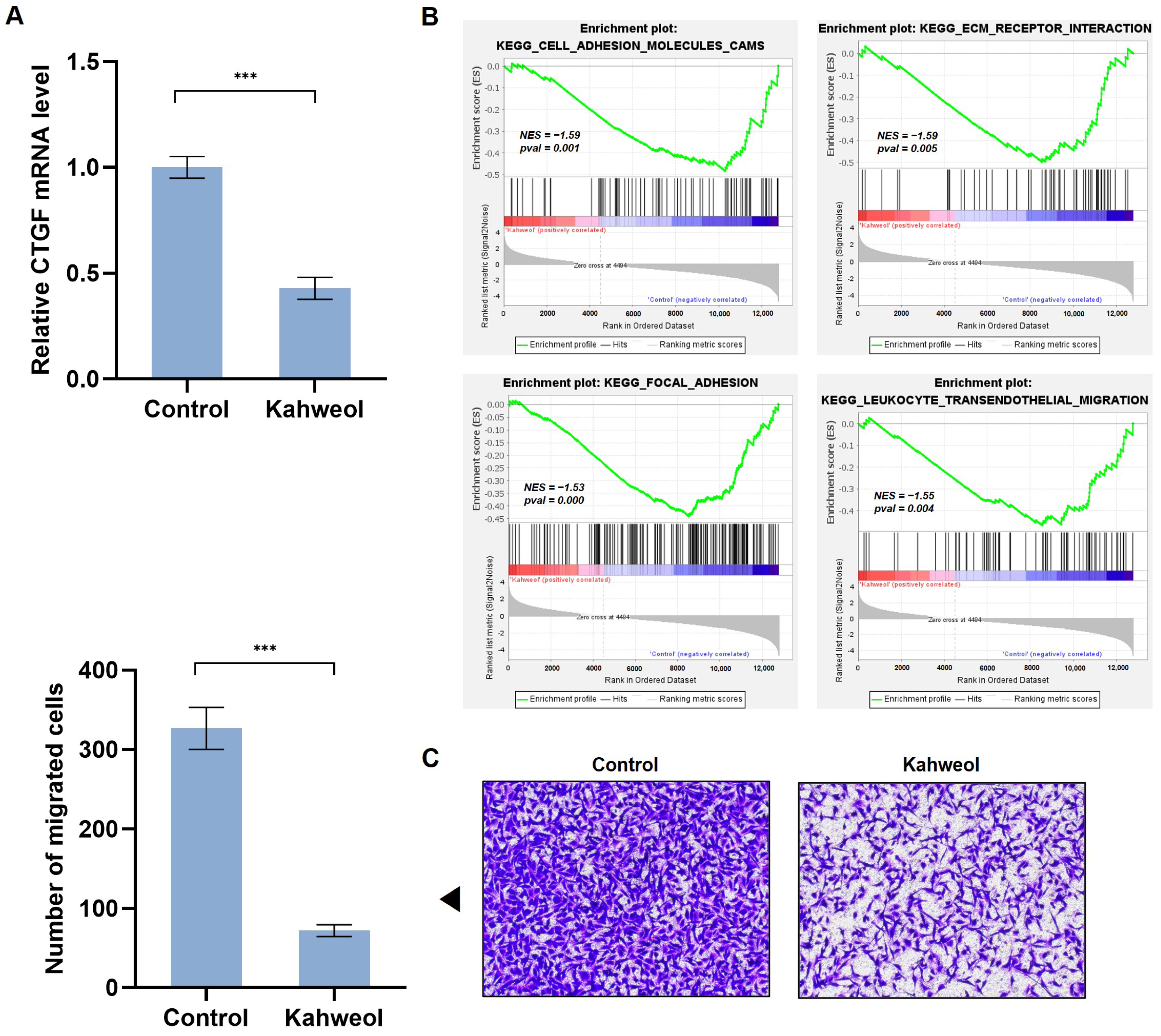
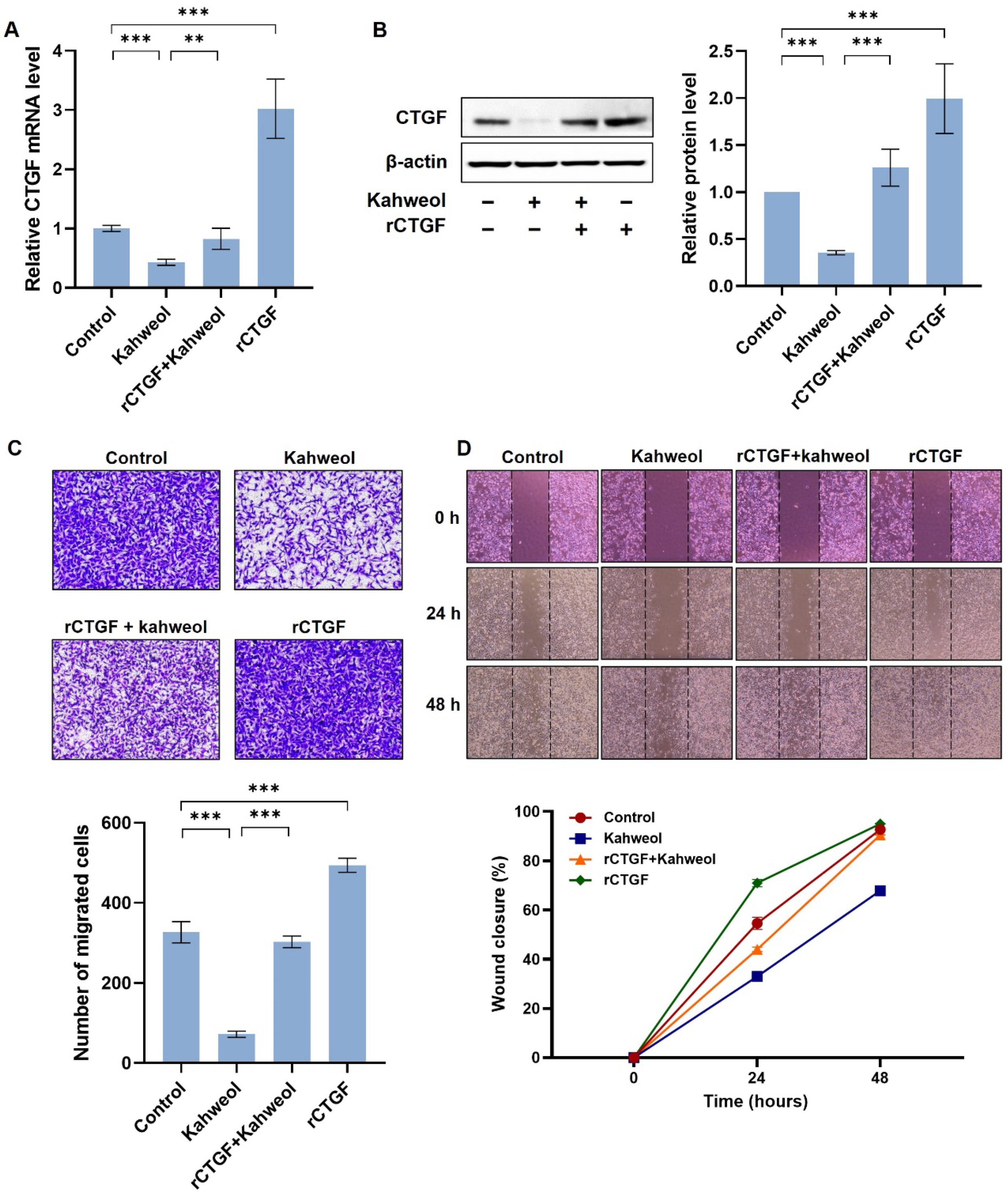
2.3. Kahweol Suppresses CTGF Expression and the Ability of Cell Migration in TNBC
2.4. Kahweol Inhibits Cell Motility by Regulating CTGF Expression in TNBC
2.5. Kahweol Influences the Expression of Genes Associated with Migration by Modulating CTGF via ERK, P38, PI3K, AKT, and FAK Phosphorylation
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Culture and Kahweol Preparation
4.3. Total RNA Sequencing
4.4. Bioinformatics Analysis
4.5. TCGA Data Acquisition and Gene Selection
4.6. Pathway Analysis and Clinical Data Visualization
4.7. Knockdown of CTGF Expression Using Small Interfering RNA (siRNA)
4.8. Western Blot Analysis
4.9. Quantitative Real-Time RT-PCR (qRT-PCR)
4.10. Wound Scratch Assay and Transwell Chamber Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Tsang, J.Y.S.; Tse, G.M. Molecular Classification of Breast Cancer. Adv. Anat. Pathol. 2020, 27, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Sig. Transduct. Target Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Van Zijl, F.; Krupitza, G.; Mikulits, W. Initial Steps of Metastasis: Cell Invasion and Endothelial Transmigration. Mutat. Res. 2011, 728, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Bosch, A.; Eroles, P.; Zaragoza, R.; Viña, J.R.; Lluch, A. Triple-Negative Breast Cancer: Molecular Features, Pathogenesis, Treatment and Current Lines of Research. Cancer Treat. Rev. 2010, 36, 206–215. [Google Scholar] [CrossRef]
- Steward, L.; Conant, L.; Gao, F.; Margenthaler, J.A. Predictive Factors and Patterns of Recurrence in Patients with Triple Negative Breast Cancer. Ann. Surg. Oncol. 2014, 21, 2165–2171. [Google Scholar] [CrossRef]
- Dent, R.; Hanna, W.M.; Trudeau, M.; Rawlinson, E.; Sun, P.; Narod, S.A. Pattern of Metastatic Spread in Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2009, 115, 423–428. [Google Scholar] [CrossRef]
- Chen, P.-S.; Wang, M.-Y.; Wu, S.-N.; Su, J.-L.; Hong, C.-C.; Chuang, S.-E.; Chen, M.-W.; Hua, K.-T.; Wu, Y.-L.; Cha, S.-T.; et al. CTGF Enhances the Motility of Breast Cancer Cells via an Integrin-Avβ3–ERK1/2-Dependent S100A4-Upregulated Pathway. J. Cell Sci. 2007, 120, 2053–2065. [Google Scholar] [CrossRef]
- Chien, W. Expression of Connective Tissue Growth Factor (CTGF/CCN2) in Breast Cancer Cells Is Associated with Increased Migration and Angiogenesis. Int. J. Oncol. 2011, 38, 1741–1747. [Google Scholar] [CrossRef]
- Moran-Jones, K.; Gloss, B.S.; Murali, R.; Chang, D.K.; Colvin, E.K.; Jones, M.D.; Yuen, S.; Howell, V.M.; Brown, L.M.; Wong, C.W.; et al. Connective Tissue Growth Factor as a Novel Therapeutic Target in High Grade Serous Ovarian Cancer. Oncotarget 2015, 6, 44551–44562. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Shih, J.-Y.; Jeng, Y.-M.; Su, J.-L.; Lin, B.-Z.; Chen, S.-T.; Chau, Y.-P.; Yang, P.-C.; Kuo, M.-L. Connective Tissue Growth Factor and Its Role in Lung Adenocarcinoma Invasion and Metastasis. JNCI J. Natl. Cancer Inst. 2004, 96, djh059. [Google Scholar] [CrossRef] [PubMed]
- Urtasun, R.; Latasa, M.U.; Demartis, M.I.; Balzani, S.; Goñi, S.; Garcia-Irigoyen, O.; Elizalde, M.; Azcona, M.; Pascale, R.M.; Feo, F.; et al. Connective Tissue Growth Factor Autocriny in Human Hepatocellular Carcinoma: Oncogenic Role and Regulation by Epidermal Growth Factor Receptor/Yes-Associated Protein-Mediated Activation. Hepatology 2011, 54, 2149–2158. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-Y.; Chang, C.-C.; Prakash, E.; Kuo, M.-L. Connective Tissue Growth Factor (CTGF) and Cancer Progression. J. Biomed. Sci. 2008, 15, 675–685. [Google Scholar] [CrossRef]
- Jia, Q.; Xu, B.; Zhang, Y.; Ali, A.; Liao, X. CCN Family Proteins in Cancer: Insight into Their Structures and Coordination Role in Tumor Microenvironment. Front. Genet. 2021, 12, 649387. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, C.; Xu, J.; Wang, S. Cafestol and Kahweol: A Review on Their Bioactivities and Pharmacological Properties. Int. J. Mol. Sci. 2019, 20, 4238. [Google Scholar] [CrossRef]
- Wu, K.C.; McDonald, P.R.; Liu, J.; Klaassen, C.D. Screening of Natural Compounds as Activators of the Keap1-Nrf2 Pathway. Planta Med. 2014, 80, 97–104. [Google Scholar] [CrossRef]
- Jeon, Y.-J.; Bang, W.; Cho, J.H.; Lee, R.H.; Kim, S.-H.; Kim, M.S.; Park, S.-M.; Shin, J.-C.; Chung, H.-J.; Oh, K.B.; et al. Kahweol Induces Apoptosis by Suppressing BTF3 Expression through the ERK Signaling Pathway in Non-Small Cell Lung Cancer Cells. Int. J. Oncol. 2016, 49, 2294–2302. [Google Scholar] [CrossRef]
- Oh, S.H.; Hwang, Y.P.; Choi, J.H.; Jin, S.W.; Lee, G.H.; Han, E.H.; Chung, Y.H.; Chung, Y.C.; Jeong, H.G. Kahweol Inhibits Proliferation and Induces Apoptosis by Suppressing Fatty Acid Synthase in HER2-Overexpressing Cancer Cells. Food Chem. Toxicol. 2018, 121, 326–335. [Google Scholar] [CrossRef]
- Park, G.H.; Song, H.M.; Jeong, J.B. Kahweol from Coffee Induces Apoptosis by Upregulating Activating Transcription Factor 3 in Human Colorectal Cancer Cells. Biomol. Ther. 2017, 25, 337–343. [Google Scholar] [CrossRef]
- Iwamoto, H.; Izumi, K.; Natsagdorj, A.; Naito, R.; Makino, T.; Kadomoto, S.; Hiratsuka, K.; Shigehara, K.; Kadono, Y.; Narimoto, K.; et al. Coffee Diterpenes Kahweol Acetate and Cafestol Synergistically Inhibit the Proliferation and Migration of Prostate Cancer Cells. Prostate 2019, 79, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Makino, T.; Izumi, K.; Hiratsuka, K.; Kano, H.; Shimada, T.; Nakano, T.; Kadomoto, S.; Naito, R.; Iwamoto, H.; Yaegashi, H.; et al. Anti-Proliferative and Anti-Migratory Properties of Coffee Diterpenes Kahweol Acetate and Cafestol in Human Renal Cancer Cells. Sci. Rep. 2021, 11, 675. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Choi, S.T.; Kang, Y.J. Kahweol, a Diterpenoid Molecule, Inhibits CTGF-Dependent Synthetic Phenotype Switching and Migration in Vascular Smooth Muscle Cells. Molecules 2021, 26, 640. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.-Y.; Jung, Y.-A.; Lee, S.-H.; Hwang, J.S.; Park, K.-G.; Kim, M.-K.; Jang, B.K. Kahweol Decreases Hepatic Fibrosis by Inhibiting the Expression of Connective Tissue Growth Factor via the Transforming Growth Factor-Beta Signaling Pathway. Oncotarget 2017, 8, 87086–87094. [Google Scholar] [CrossRef] [PubMed]
- Arauz, J.; Moreno, M.G.-; Cortés-Reynosa, P.; Salazar, E.P.; Muriel, P. Coffee Attenuates Fibrosis by Decreasing the Expression of TGF-β and CTGF in a Murine Model of Liver Damage. J. Appl. Toxicol. 2013, 33, 970–979. [Google Scholar] [CrossRef]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and Interpreting Genome Data at the Network Level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef]
- Huang, C.; Jacobson, K.; Schaller, M.D. MAP Kinases and Cell Migration. J. Cell Sci. 2004, 117, 4619–4628. [Google Scholar] [CrossRef]
- Takeda, T.; Yamamoto, Y.; Tsubaki, M.; Matsuda, T.; Kimura, A.; Shimo, N.; Nishida, S. PI3K/Akt/YAP Signaling Promotes Migration and Invasion of DLD-1 Colorectal Cancer Cells. Oncol. Lett. 2022, 23, 106. [Google Scholar] [CrossRef]
- Yu, H.; Gao, M.; Ma, Y.; Wang, L.; Shen, Y.; Liu, X. Inhibition of Cell Migration by Focal Adhesion Kinase: Time-Dependent Difference in Integrin-Induced Signaling between Endothelial and Hepatoblastoma Cells. Int. J. Mol. Med. 2018, 41, 2573–2588. [Google Scholar] [CrossRef]
- Braig, S.; Wallner, S.; Junglas, B.; Fuchshofer, R.; Bosserhoff, A.-K. CTGF Is Overexpressed in Malignant Melanoma and Promotes Cell Invasion and Migration. Br. J. Cancer 2011, 105, 231–238. [Google Scholar] [CrossRef]
- Zhang, S.; Li, B.; Tang, W.; Ni, L.; Ma, H.; Lu, M.; Meng, Q. Effects of Connective Tissue Growth Factor on Prostate Cancer Bone Metastasis and Osteoblast Differentiation. Oncol. Lett. 2018, 16, 2305–2311. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Gao, K.; Hu, G.; Wen, Y.; Lin, C.; Li, X. CTGF Enhances Resistance to 5-FU-Mediating Cell Apoptosis through FAK/MEK/ERK Signal Pathway in Colorectal Cancer. Onco Targets Ther. 2016, 9, 7285–7295. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Son, S.; Ko, Y.; Shin, I. CTGF Regulates Cell Proliferation, Migration, and Glucose Metabolism through Activation of FAK Signaling in Triple-Negative Breast Cancer. Oncogene 2021, 40, 2667–2681. [Google Scholar] [CrossRef] [PubMed]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.U.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic Behavior of Breast Cancer Subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef]
- Jin, J.; Gao, Y.; Zhang, J.; Wang, L.; Wang, B.; Cao, J.; Shao, Z.; Wang, Z. Incidence, Pattern and Prognosis of Brain Metastases in Patients with Metastatic Triple Negative Breast Cancer. BMC Cancer 2018, 18, 446. [Google Scholar] [CrossRef]
- Kuksis, M.; Gao, Y.; Tran, W.; Hoey, C.; Kiss, A.; Komorowski, A.S.; Dhaliwal, A.J.; Sahgal, A.; Das, S.; Chan, K.K.; et al. The Incidence of Brain Metastases among Patients with Metastatic Breast Cancer: A Systematic Review and Meta-Analysis. Neuro Oncol. 2020, 23, 894–904. [Google Scholar] [CrossRef]
- Heng, E.C.K.; Huang, Y.; Black, S.A.; Trackman, P.C. CCN2, Connective tissue growth factor, stimulates collagen deposition by gingival fibroblasts via module 3 and A6- and Β1 integrins. J. Cell Biochem. 2006, 98, 409–420. [Google Scholar] [CrossRef]
- Eldesouki, S.; Qadri, R.; Abu Helwa, R.; Barqawi, H.; Bustanji, Y.; Abu-Gharbieh, E.; El-Huneidi, W. Recent Updates on the Functional Impact of Kahweol and Cafestol on Cancer. Molecules 2022, 27, 7332. [Google Scholar] [CrossRef]
- Cárdenas, C.; Quesada, A.R.; Medina, M.Á. Insights on the Antitumor Effects of Kahweol on Human Breast Cancer: Decreased Survival and Increased Production of Reactive Oxygen Species and Cytotoxicity. Biochem. Biophys. Res. Commun. 2014, 447, 452–458. [Google Scholar] [CrossRef]
- Kwon, M.J.; Kim, J.N.; Park, J.; Kim, Y.T.; Lee, M.J.; Kim, B.J. Alisma Canaliculatum Extract Affects AGS Gastric Cancer Cells by Inducing Apoptosis. Int. J. Med. Sci. 2021, 18, 2155–2161. [Google Scholar] [CrossRef]
- Chung, Y.C.; Hyun, C.-G. Inhibitory Effects of Pinostilbene Hydrate on Melanogenesis in B16F10 Melanoma Cells via ERK and P38 Signaling Pathways. Int. J. Mol. Sci. 2020, 21, 4732. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Hwang, Y.P.; Jin, S.W.; Lee, G.H.; Kim, H.G.; Han, E.H.; Kim, S.K.; Kang, K.W.; Chung, Y.C.; Jeong, H.G. Suppression of PMA-Induced Human Fibrosarcoma HT-1080 Invasion and Metastasis by Kahweol via Inhibiting Akt/JNK1/2/P38 MAPK Signal Pathway and NF-κB Dependent Transcriptional Activities. Food Chem. Toxicol. 2019, 125, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.G.; Kim, J.Y.; Hwang, Y.P.; Lee, K.J.; Lee, K.Y.; Kim, D.H.; Kim, D.H.; Jeong, H.G. The Coffee Diterpene Kahweol Inhibits Tumor Necrosis Factor-α-Induced Expression of Cell Adhesion Molecules in Human Endothelial Cells. Toxicol. Appl. Pharmacol. 2006, 217, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhou, C.; Yuan, Q.; Zhang, D.; Xie, J.; Zou, S. CTGF Facilitates Cell-cell Communication in Chondrocytes via PI3K/Akt Signalling Pathway. Cell Prolif. 2021, 54, e13001. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.T.; Martin, K.H.; Slack, J.K.; Taylor, J.M.; Weed, S.A. Focal Adhesion Kinase: A Regulator of Focal Adhesion Dynamics and Cell Movement. Oncogene 2000, 19, 5606–5613. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Cao, B.; Wang, Y.; Ma, C.; Zeng, Z.; Liu, L.; Li, X.; Tao, D.; Gong, J.; Xie, D. Hippo Component YAP Promotes Focal Adhesion and Tumour Aggressiveness via Transcriptionally Activating THBS1/FAK Signalling in Breast Cancer. J. Exp. Clin. Cancer Res. 2018, 37, 175. [Google Scholar] [CrossRef]
- Yu, B.; Jin, G.; Ma, M.; Liang, H.; Zhang, B.; Chen, X.; Ding, Z. Taurocholate Induces Connective Tissue Growth Factor Expression in Hepatocytes Through ERK-YAP Signaling. Cell Physiol. Biochem. 2018, 50, 1711–1725. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Wilson, R.G.; Smith, A.N.; Bird, C.C. Immunohistochemical Detection of Abnormal Cell Proliferation in Colonic Mucosa of Subjects with Polyps. J. Clin. Pathol. 1990, 43, 744–747. [Google Scholar] [CrossRef]
- Kutmon, M.; Riutta, A.; Nunes, N.; Hanspers, K.; Willighagen, E.L.; Bohler, A.; Mélius, J.; Waagmeester, A.; Sinha, S.R.; Miller, R.; et al. WikiPathways: Capturing the Full Diversity of Pathway Knowledge. Nucleic Acids Res. 2016, 44, D488–D494. [Google Scholar] [CrossRef]
- Jewison, T.; Su, Y.; Disfany, F.M.; Liang, Y.; Knox, C.; Maciejewski, A.; Poelzer, J.; Huynh, J.; Zhou, Y.; Arndt, D.; et al. SMPDB 2.0: Big Improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014, 42, D478–D484. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef] [PubMed]
- Hewett, M.; Oliver, D.E.; Rubin, D.L.; Easton, K.L.; Stuart, J.M.; Altman, R.B.; Klein, T.E. PharmGKB: The Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002, 30, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef]
- Hao, T.; Ma, H.-W.; Zhao, X.-M.; Goryanin, I. Compartmentalization of the Edinburgh Human Metabolic Network. BMC Bioinform. 2010, 11, 393. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc Database of Metabolic Pathways and Enzymes and the BioCyc Collection of Pathway/Genome Databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef]
- Kandasamy, K.; Mohan, S.S.; Raju, R.; Keerthikumar, S.; Kumar, G.S.S.; Venugopal, A.K.; Telikicherla, D.; Navarro, J.D.; Mathivanan, S.; Pecquet, C.; et al. NetPath: A Public Resource of Curated Signal Transduction Pathways. Genome Biol. 2010, 11, R3. [Google Scholar] [CrossRef]
- Fazekas, D.; Koltai, M.; Türei, D.; Módos, D.; Pálfy, M.; Dúl, Z.; Zsákai, L.; Szalay-Bekő, M.; Lenti, K.; Farkas, I.J.; et al. SignaLink 2—A Signaling Pathway Resource with Multi-Layered Regulatory Networks. BMC Syst. Biol. 2013, 7, 7. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.H.; Kim, J.; Kim, H.S.; Kang, Y.J. Unraveling Connective Tissue Growth Factor as a Therapeutic Target and Assessing Kahweol as a Potential Drug Candidate in Triple-Negative Breast Cancer Treatment. Int. J. Mol. Sci. 2023, 24, 16307. https://doi.org/10.3390/ijms242216307
Lee JH, Kim J, Kim HS, Kang YJ. Unraveling Connective Tissue Growth Factor as a Therapeutic Target and Assessing Kahweol as a Potential Drug Candidate in Triple-Negative Breast Cancer Treatment. International Journal of Molecular Sciences. 2023; 24(22):16307. https://doi.org/10.3390/ijms242216307
Chicago/Turabian StyleLee, Jeong Hee, Jongsu Kim, Hong Sook Kim, and Young Jin Kang. 2023. "Unraveling Connective Tissue Growth Factor as a Therapeutic Target and Assessing Kahweol as a Potential Drug Candidate in Triple-Negative Breast Cancer Treatment" International Journal of Molecular Sciences 24, no. 22: 16307. https://doi.org/10.3390/ijms242216307
APA StyleLee, J. H., Kim, J., Kim, H. S., & Kang, Y. J. (2023). Unraveling Connective Tissue Growth Factor as a Therapeutic Target and Assessing Kahweol as a Potential Drug Candidate in Triple-Negative Breast Cancer Treatment. International Journal of Molecular Sciences, 24(22), 16307. https://doi.org/10.3390/ijms242216307





