A Regulator Role for the ATP-Binding Cassette Subfamily C Member 6 Transporter in HepG2 Cells: Effect on the Dynamics of Cell–Cell and Cell–Matrix Interactions
Abstract
:1. Introduction
2. Results
2.1. Whole Transcriptome of the ABCC6-Silenced HepG2 Cell Line
2.2. Functional Enrichment Analysis of Differentially Expressed Genes
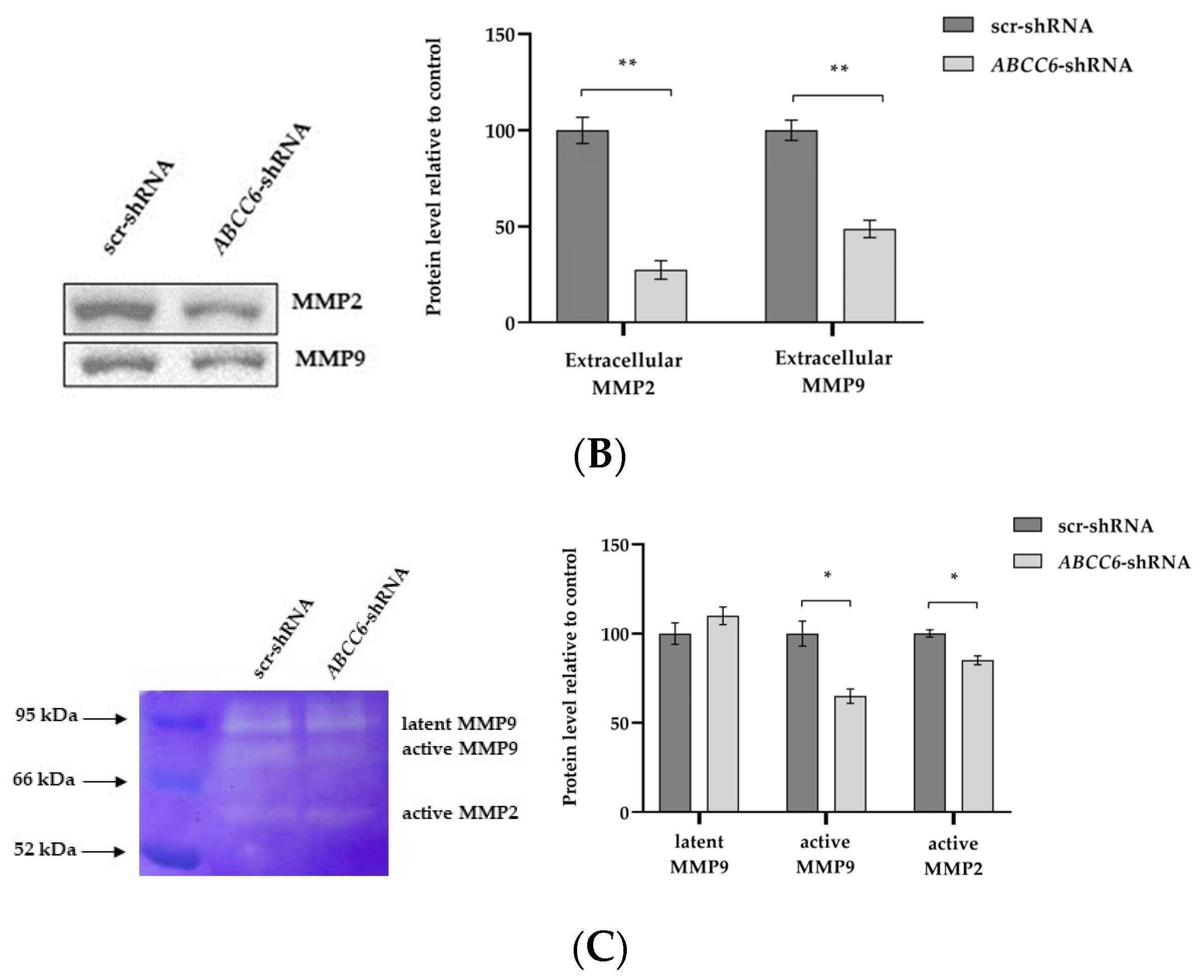
2.3. The Silencing of ABCC6 Changes the Expression of Some Proteins Involved in Cell–Cell and Cell–Matrix Interactions
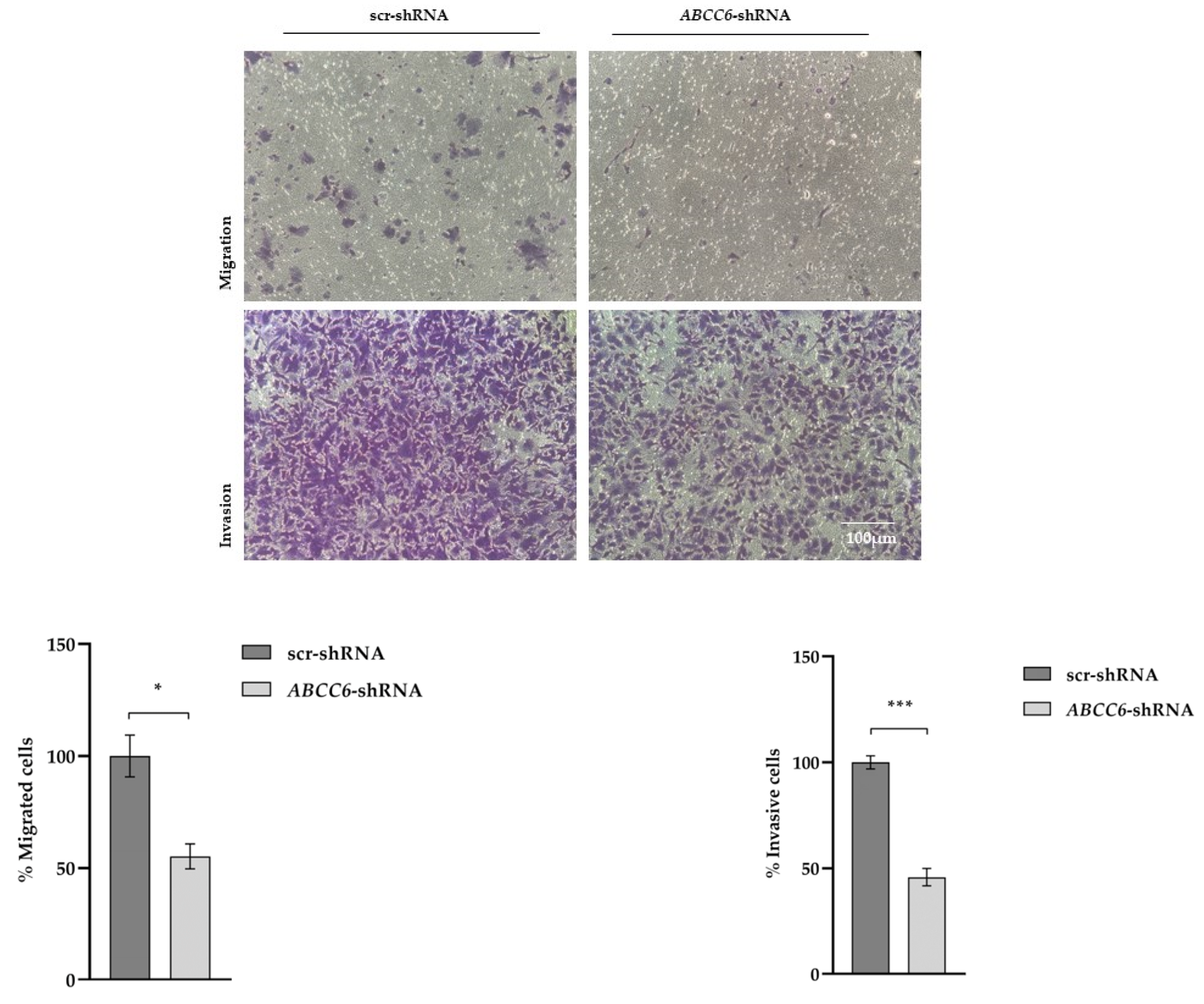
2.4. ABCC6-Silenced Cells Modify Their Clonogenic Potential and Ability to Invade the Extracellular Matrix
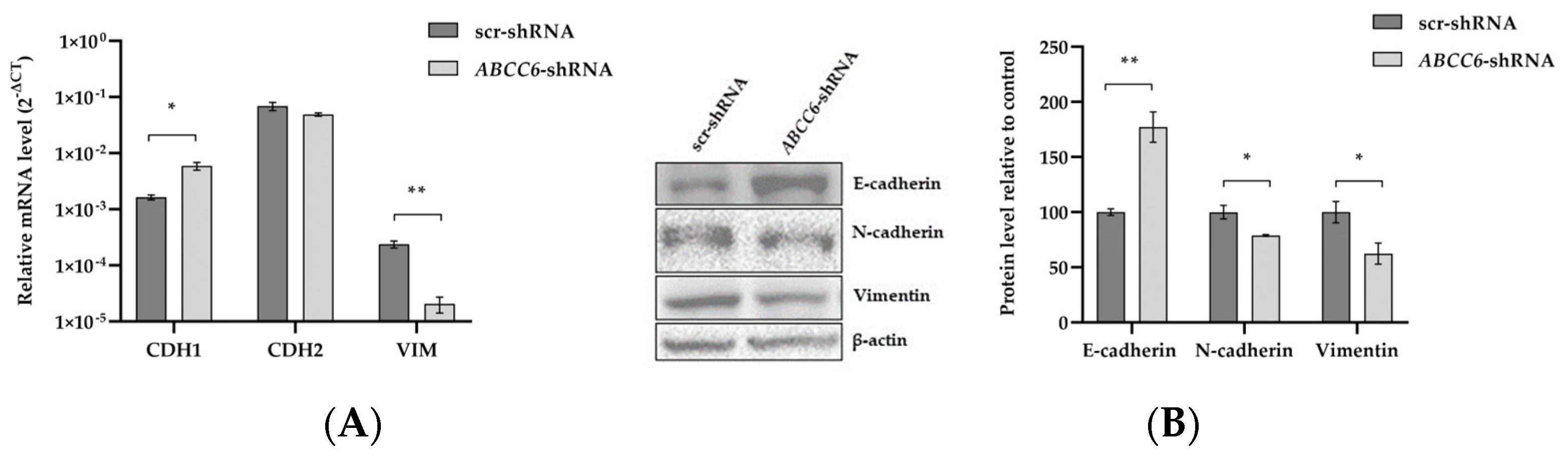
2.5. ABCC6 Silencing Dysregulates Genes Involved in the Epithelial–Mesenchymal Transition
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Maintenance of Cell Culture
4.2. Gene Knockdown of Stable Cell Lines
4.3. RNA Sequencing
4.4. Functional and Pathway Analysis of Differentially Regulated Genes
4.5. Real-Time Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
4.6. Western Blot Analysis
4.7. Cell Adhesion Assay
4.8. Migration and Invasion Assays
4.9. SDS–Gelatin Gel Zymogram
4.10. Colony Formation Assay
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borst, P.; Váradi, A.; van de Wetering, K. PXE, a Mysterious Inborn Error Clarified. Trends Biochem. Sci. 2019, 44, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Favre, G.; Laurain, A.; Aranyi, T.; Szeri, F.; Fulop, K.; Le Saux, O.; Duranton, C.; Kauffenstein, G.; Martin, L.; Lefthériotis, G. The ABCC6 Transporter: A New Player in Biomineralization. Int. J. Mol. Sci. 2017, 18, 1941. [Google Scholar] [CrossRef] [PubMed]
- Terry, S.F. The Human Face of ABCC6. FEBS Lett. 2020, 594, 4151–4157. [Google Scholar] [CrossRef] [PubMed]
- Pomozi, V.; Le Saux, O.; Brampton, C.; Apana, A.; Iliás, A.; Szeri, F.; Martin, L.; Monostory, K.; Paku, S.; Sarkadi, B.; et al. ABCC6 Is a Basolateral Plasma Membrane Protein. Circ. Res. 2013, 112, e148–e151. [Google Scholar] [CrossRef]
- Jansen, R.S.; Duijst, S.; Mahakena, S.; Sommer, D.; Szeri, F.; Váradi, A.; Plomp, A.; Bergen, A.A.; Oude Elferink, R.P.J.; Borst, P.; et al. ABCC6-Mediated ATP Secretion by the Liver Is the Main Source of the Mineralization Inhibitor Inorganic Pyrophosphate in the Systemic Circulation—Brief Report. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1985–1989. [Google Scholar] [CrossRef]
- Jansen, R.S.; Küçükosmanoǧlu, A.; De Haas, M.; Sapthu, S.; Otero, J.A.; Hegman, I.E.M.; Bergen, A.A.B.; Gorgels, T.G.M.F.; Borst, P.; Van De Wetering, K. ABCC6 Prevents Ectopic Mineralization Seen in Pseudoxanthoma Elasticum by Inducing Cellular Nucleotide Release. Proc. Natl. Acad. Sci. USA 2013, 110, 20206–20211. [Google Scholar] [CrossRef]
- Uitto, J.; Li, Q.; van de Wetering, K.; Váradi, A.; Terry, S.F. Insights into Pathomechanisms and Treatment Development in Heritable Ectopic Mineralization Disorders: Summary of the PXE Inte\rnational Biennial Research Symposium—2016. J. Investig. Dermatol. 2017, 137, 790–795. [Google Scholar] [CrossRef]
- Ferrari, D.; Malavasi, F.; Antonioli, L. A Purinergic Trail for Metastases. Trends Pharmacol. Sci. 2017, 38, 277–290. [Google Scholar] [CrossRef]
- Belinsky, M.G.; Chen, Z.-S.; Shchaveleva, I.; Zeng, H.; Kruh, G.D. Characterization of the Drug Resistance and Transport Properties of Multidrug Resistance Protein 6 (MRP6, ABCC6). Cancer Res. 2002, 62, 6172–6177. [Google Scholar]
- Chen, Z.-S.; Tiwari, A.K. Multidrug Resistance Proteins (MRPs/ABCCs) in Cancer Chemotherapy and Genetic Diseases. FEBS J. 2011, 278, 3226–3245. [Google Scholar] [CrossRef]
- Ikeda, R.; Vermeulen, L.C.; Lau, E.; Jiang, Z.; Sachidanandam, K.; Yamada, K.; Kolesar, J.M. Isolation and Characterization of Gemcitabine-Resistant Human Non-Small Cell Lung Cancer A549 Cells. Int. J. Oncol. 2011, 38, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Eadie, L.N.; Dang, P.; Goyne, J.M.; Hughes, T.P.; White, D.L. ABCC6 Plays a Significant Role in the Transport of Nilotinib and Dasatinib, and Contributes to TKI Resistance in Vitro, in Both Cell Lines and Primary Patient Mononuclear Cells. PLoS ONE 2018, 13, e0192180. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Nam, H.J.; Kang, H.J.; Samuels, T.L.; Johnston, N.; Lim, Y.C. Valproic Acid Suppresses the Self-Renewal and Proliferation of Head and Neck Cancer Stem Cells. Oncol. Rep. 2015, 34, 2065–2071. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-Paolillo, A.; Tesser-Gamba, F.; Seixas Alves, M.T.; Filho, R.J.G.; Oliveira, R.; Petrilli, A.S.; Toledo, S.R.C. Pharmacogenetics of the Primary and Metastatic Osteosarcoma: Gene Expression Profile Associated with Outcome. Int. J. Mol. Sci. 2023, 24, 5607. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; He, Z.; Zhou, F.; Huang, Y.; Zhu, G. Prognostic Significance and Molecular Mechanisms of Adenosine Triphosphate-Binding Cassette Subfamily C Members in Gastric Cancer. Medicine 2019, 98, e18347. [Google Scholar] [CrossRef]
- Elsnerova, K.; Mohelnikova-Duchonova, B.; Cerovska, E.; Ehrlichova, M.; Gut, I.; Rob, L.; Skapa, P.; Hruda, M.; Bartakova, A.; Bouda, J.; et al. Gene Expression of Membrane Transporters: Importance for Prognosis and Progression of Ovarian Carcinoma. Oncol. Rep. 2016, 35, 2159–2170. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, P.; Huang, C.; Jiang, L.; Lu, Z.; Wang, P. Varied Clinical Significance of ATP-Binding Cassette C Sub-Family Members for Lung Adenocarcinoma. Medicine 2021, 100, e25246. [Google Scholar] [CrossRef]
- Januchowski, R.; Sterzyńska, K.; Zawierucha, P.; Ruciński, M.; Świerczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brazert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-Based Detection and Expression Analysis of New Genes Associated with Drug Resistance in Ovarian Cancer Cell Lines. Oncotarget 2017, 8, 49944–49958. [Google Scholar] [CrossRef]
- Hlavata, I.; Mohelnikova-Duchonova, B.; Vaclavikova, R.; Liska, V.; Pitule, P.; Novak, P.; Bruha, J.; Vycital, O.; Holubec, L.; Treska, V.; et al. The Role of ABC Transporters in Progression and Clinical Outcome of Colorectal Cancer. Mutagenesis 2012, 27, 187–196. [Google Scholar] [CrossRef]
- Mohelnikova-Duchonova, B.; Brynychova, V.; Oliverius, M.; Honsova, E.; Kala, Z.; Muckova, K.; Soucek, P. Differences in Transcript Levels of ABC Transporters between Pancreatic Adenocarcinoma and Nonneoplastic Tissues. Pancreas 2013, 42, 707–716. [Google Scholar] [CrossRef]
- Meng, X.; Dong, S.; Yangyang, L.; Wang, S.; Xu, X.; Liu, T.; Zhuang, X. Adenosine Triphosphate-Binding Cassette Subfamily C Members in Liver Hepatocellular Carcinoma. Medicine 2022, 101, e28869. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhao, Z.; Wang, J.; Zhang, H.; Xi, Z.; Xia, Q. ABCC6 Knockdown Fuels Cell Proliferation by Regulating PPARα in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 840287. [Google Scholar] [CrossRef] [PubMed]
- Bisaccia, F.; Koshal, P.; Abruzzese, V.; Morelli, M.A.C.; Ostuni, A. Structural and Functional Characterization of the ABCC6 Transporter in Hepatic Cells: Role on PXE, Cancer Therapy and Drug Resistance. Int. J. Mol. Sci. 2021, 22, 2858. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, F.; Cuviello, F.; Pace, M.C.; Armentano, M.F.; Miglionico, R.; Ostuni, A.; Bisaccia, F. Extracellular ATP Regulates CD73 and ABCC6 Expression in HepG2 Cells. Front. Mol. Biosci. 2018, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Ostuni, A.; Carmosino, M.; Miglionico, R.; Abruzzese, V.; Martinelli, F.; Russo, D.; Laurenzana, I.; Petillo, A.; Bisaccia, F. Inhibition of ABCC6 Transporter Modifies Cytoskeleton and Reduces Motility of HepG2 Cells via Purinergic Pathway. Cells 2020, 9, 1410. [Google Scholar] [CrossRef]
- Abruzzese, V.; Matera, I.; Martinelli, F.; Carmosino, M.; Koshal, P.; Milella, L.; Bisaccia, F.; Ostuni, A. Effect of Quercetin on ABCC6 Transporter: Implication in HepG2 Migration. Int. J. Mol. Sci. 2021, 22, 3871. [Google Scholar] [CrossRef]
- Paskeh, M.D.A.; Ghadyani, F.; Hashemi, M.; Abbaspour, A.; Zabolian, A.; Javanshir, S.; Razzazan, M.; Mirzaei, S.; Entezari, M.; Goharrizi, M.A.S.B.; et al. Biological Impact and Therapeutic Perspective of Targeting PI3K/Akt Signaling in Hepatocellular Carcinoma: Promises and Challenges. Pharmacol. Res. 2023, 187, 106553. [Google Scholar] [CrossRef]
- Abruzzese, V.; Sukowati, C.H.C.; Tiribelli, C.; Matera, I.; Ostuni, A.; Bisaccia, F. The Expression Level of ABCC6 Transporter in Colon Cancer Cells Correlates with the Activation of Different Intracellular Signaling Pathways. Pathophysiology 2022, 29, 173–186. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Welch, D.R. Dormancy in Cancer Metastasis: Keys to Moving Forward. Cancer Metastasis Rev. 2023, 42, 5–7. [Google Scholar] [CrossRef]
- Janiszewska, M.; Primi, M.C.; Izard, T. Cell Adhesion in Cancer: Beyond the Migration of Single Cells. J. Biol. Chem. 2020, 295, 2495–2505. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [PubMed]
- Niland, S.; Riscanevo, A.X.; Eble, J.A. Matrix Metalloproteinases Shape the Tumor Microenvironment in Cancer Progression. Int. J. Mol. Sci. 2022, 23, 146. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.H.; Yang, J. Epithelial–Mesenchymal Plasticity in Carcinoma Metastasis. Genes Dev. 2013, 27, 2192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Jiao, S.; Han, G.; Zhu, J.; Liu, T. Functional and Clinical Characteristics of Focal Adhesion Kinases in Cancer Progression. Front. Cell Dev. Biol. 2022, 10, 1040311. [Google Scholar] [CrossRef] [PubMed]
- Geiger, B.; Boujemaa-Paterski, R.; Winograd-Katz, S.E.; Balan Venghateri, J.; Chung, W.L.; Medalia, O. The Actin Network Interfacing Diverse Integrin-Mediated Adhesions. Biomolecules 2023, 13, 294. [Google Scholar] [CrossRef]
- Zhang, P.F.; Li, K.S.; Shen, Y.H.; Gao, P.T.; Dong, Z.R.; Cai, J.B.; Zhang, C.; Huang, X.Y.; Tian, M.X.; Hu, Z.Q.; et al. Galectin-1 Induces Hepatocellular Carcinoma EMT and Sorafenib Resistance by Activating FAK/PI3K/AKT Signaling. Cell Death Dis. 2016, 7, e2201. [Google Scholar] [CrossRef]
- Ren, D.; Zhao, J.; Sun, Y.; Li, D.; Meng, Z.; Wang, B.; Fan, P.; Liu, Z.; Jin, X.; Wu, H. Overexpressed ITGA2 Promotes Malignant Tumor Aggression by Up-Regulating PD-L1 Expression through the Activation of the STAT3 Signaling Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 485. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Wu, H.Y.; Lin, Y.L.; Tzou, S.C.; Chuang, C.H.; Jian, T.Y.; Chen, P.R.; Chang, Y.C.; Lin, C.H.; Huang, T.H.; et al. Blockade of ITGA2 Induces Apoptosis and Inhibits Cell Migration in Gastric Cancer. Biol. Proced. Online 2018, 20, 10. [Google Scholar] [CrossRef]
- Deichmann, S.; Schindel, L.; Braun, R.; Bolm, L.; Taylor, M.; Deshpande, V.; Schilling, O.; Bronsert, P.; Keck, T.; Ferrone, C.; et al. Overexpression of Integrin Alpha 2 (ITGA2) Correlates with Poor Survival in Patients with Pancreatic Ductal Adenocarcinoma. J. Clin. Pathol. 2022, 76, 541–547. [Google Scholar] [CrossRef]
- Jiang, H.; Li, H. Prognostic Values of Tumoral MMP2 and MMP9 Overexpression in Breast Cancer: A Systematic Review and Meta-Analysis. BMC Cancer 2021, 21, 149. [Google Scholar] [CrossRef]
- Kaszak, I.; Witkowska-Piłaszewicz, O.; Niewiadomska, Z.; Dworecka-Kaszak, B.; Toka, F.N.; Jurka, P. Role of Cadherins in Cancer—A Review. Int. J. Mol. Sci. 2020, 21, 7624. [Google Scholar] [CrossRef]
- Yu, W.; Yang, L.; Li, T.; Zhang, Y. Cadherin Signaling in Cancer: Its Functions and Role as a Therapeutic Target. Front. Oncol. 2019, 9, 989. [Google Scholar] [CrossRef]
- Aiello, N.M.; Kang, Y. Context-Dependent EMT Programs in Cancer Metastasis. J. Exp. Med. 2019, 216, 1016. [Google Scholar] [CrossRef]
- Usman, S.; Waseem, N.H.; Nguyen, T.K.N.; Mohsin, S.; Jamal, A.; Teh, M.T.; Waseem, A. Vimentin Is at the Heart of Epithelial Mesenchymal Transition (EMT) Mediated Metastasis. Cancers 2021, 13, 4985. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the Extracellular Matrix in Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef] [PubMed]
- Miglionico, R.; Ostuni, A.; Armentano, M.F.; Milella, L.; Crescenzi, E.; Carmosino, M.; Bisaccia, F. ABCC6 Knockdown in HepG2 Cells Induces a Senescent-like Cell Phenotype. Cell. Mol. Biol. Lett. 2017, 22, 7. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Xu, F.; Chen, L.; Li, Y.; Xu, Z.; Zhang, Y.; Wei, W.; Su, N.; Zhang, T.; Fan, F.; et al. Quantitative Proteomics Reveals FLNC as a Potential Progression Marker for the Development of Hepatocellular Carcinoma. Oncotarget 2016, 7, 68242. [Google Scholar] [CrossRef] [PubMed]
- Kamil, M.; Shinsato, Y.; Higa, N.; Hirano, T.; Idogawa, M.; Takajo, T.; Minami, K.; Shimokawa, M.; Yamamoto, M.; Kawahara, K.; et al. High Filamin-C Expression Predicts Enhanced Invasiveness and Poor Outcome in Glioblastoma Multiforme. Br. J. Cancer 2019, 120, 819–826. [Google Scholar] [CrossRef]
- Klatt, A.R.; Becker, A.K.A.; Neacsu, C.D.; Paulsson, M.; Wagener, R. The Matrilins: Modulators of Extracellular Matrix Assembly. Int. J. Biochem. Cell Biol. 2011, 43, 320–330. [Google Scholar] [CrossRef]
- Szabó, E.; Korpos, É.; Batmunkh, E.; Lotz, G.; Holczbauer, Á.; Kovalszky, I.; Deák, F.; Kiss, I.; Schaff, Z.; Kiss, A. Expression of Matrilin-2 in Liver Cirrhosis and Hepatocellular Carcinoma. Pathol. Oncol. Res. POR 2008, 14, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Jia, J.; Zhang, D. Purinergic Signalling in Liver Diseases: Pathological Functions and Therapeutic Opportunities. JHEP Rep. 2020, 2, 100165. [Google Scholar] [CrossRef] [PubMed]
- Dimri, M.; Satyanarayana, A. Molecular Signaling Pathways and Therapeutic Targets in Hepatocellular Carcinoma. Cancers 2020, 12, 491. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, G.-D.; Shi, Z.; Qi, L.-L.; Zhou, L.-Y.; Fu, Z.-X. The Ras/Raf/MEK/ERK Signaling Pathway and Its Role in the Occurrence and Development of HCC. Oncol. Lett. 2016, 12, 3045–3050. [Google Scholar] [CrossRef]
- Krchniakova, M.; Skoda, J.; Neradil, J.; Chlapek, P.; Veselska, R. Repurposing Tyrosine Kinase Inhibitors to Overcome Multidrug Resistance in Cancer: A Focus on Transporters and Lysosomal Sequestration. Int. J. Mol. Sci. 2020, 21, 3157. [Google Scholar] [CrossRef]
- Granito, A.; Marinelli, S.; Terzi, E.; Piscaglia, F.; Renzulli, M.; Venerandi, L.; Benevento, F.; Bolondi, L. Metronomic capecitabine as second-line treatment in hepatocellular carcinoma after sorafenib failure. Dig. Liver Dis. 2015, 47, 518–522. [Google Scholar] [CrossRef]
- Pelizzaro, F.; Sammarco, A.; Dadduzio, V.; Pastorelli, D.; Giovanis, P.; Soldà, C.; Rizzato, M.D.; Lombardi, G.; Lonardi, S.; Peserico, G.; et al. Capecitabine in advanced hepatocellular carcinoma: A multicenter experience. Dig. Liver Dis. 2019, 51, 1713–1719. [Google Scholar] [CrossRef]
- Simon, A. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Babraham, UK, 2010. [Google Scholar]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Ggplot2. WIREs Comp. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Ulgen, E.; Ozisik, O.; Sezerman, O.U. pathfindR: An R Package for Comprehensive Identification of Enriched Pathways in Omics Data Through Active Subnetworks. Front. Genet. 2019, 10, 858. [Google Scholar] [CrossRef] [PubMed]
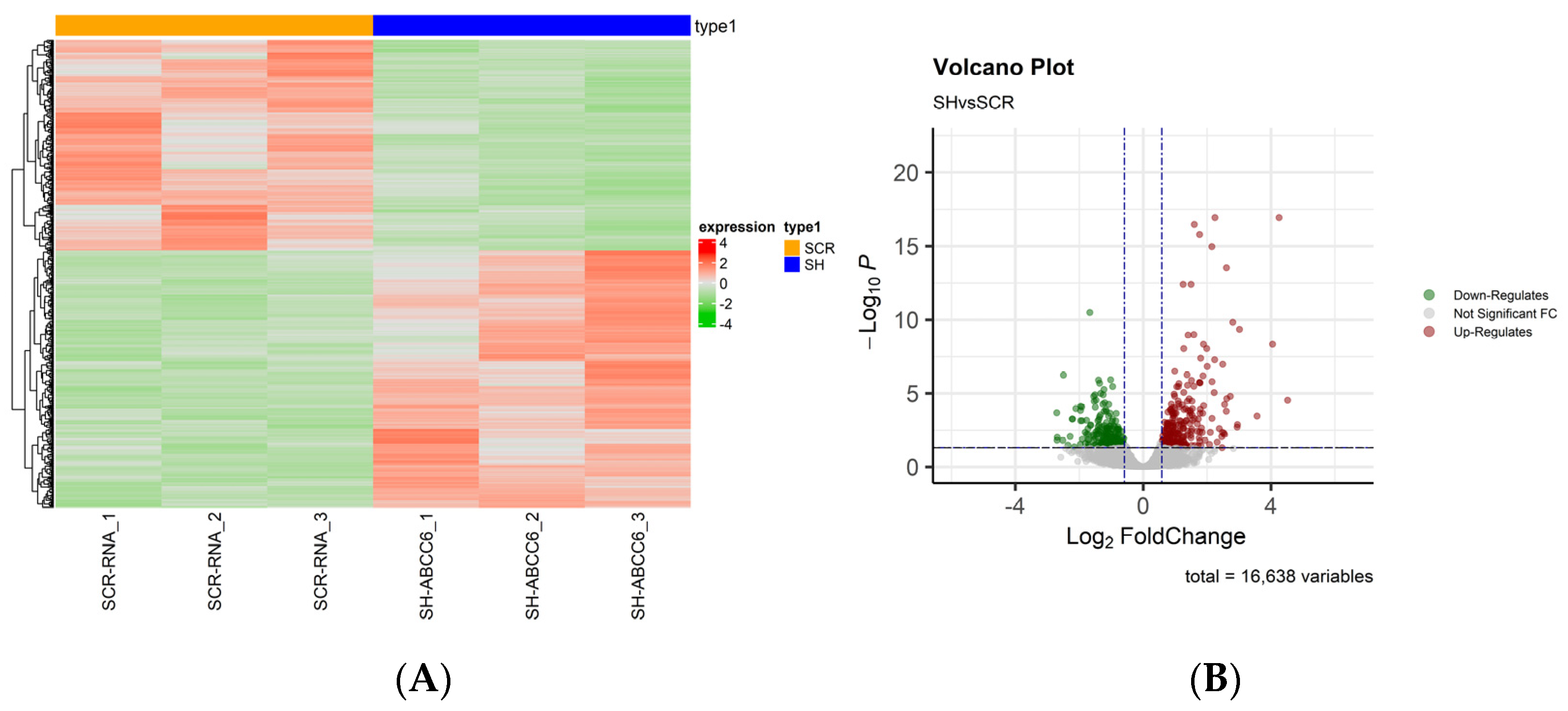
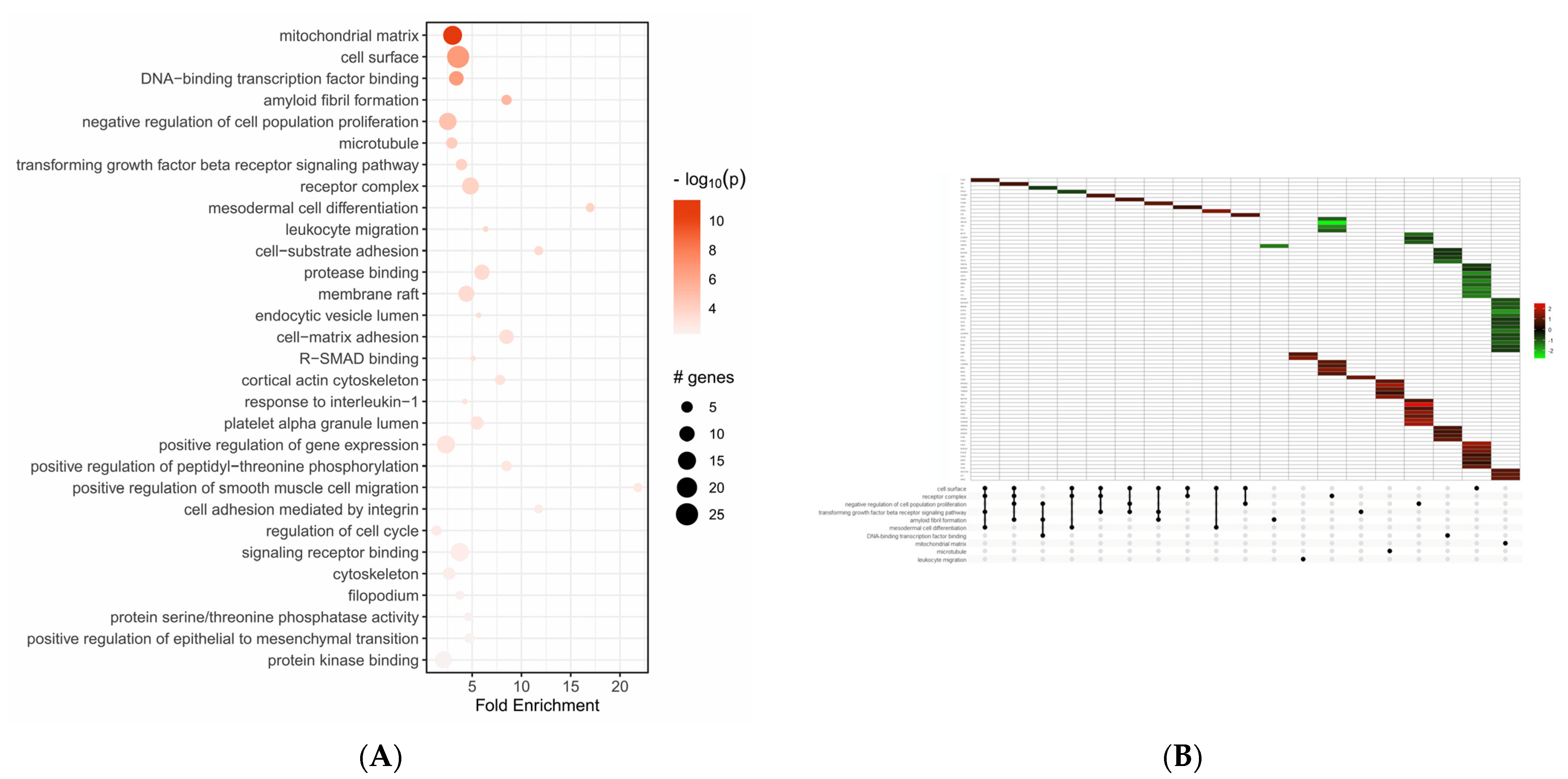



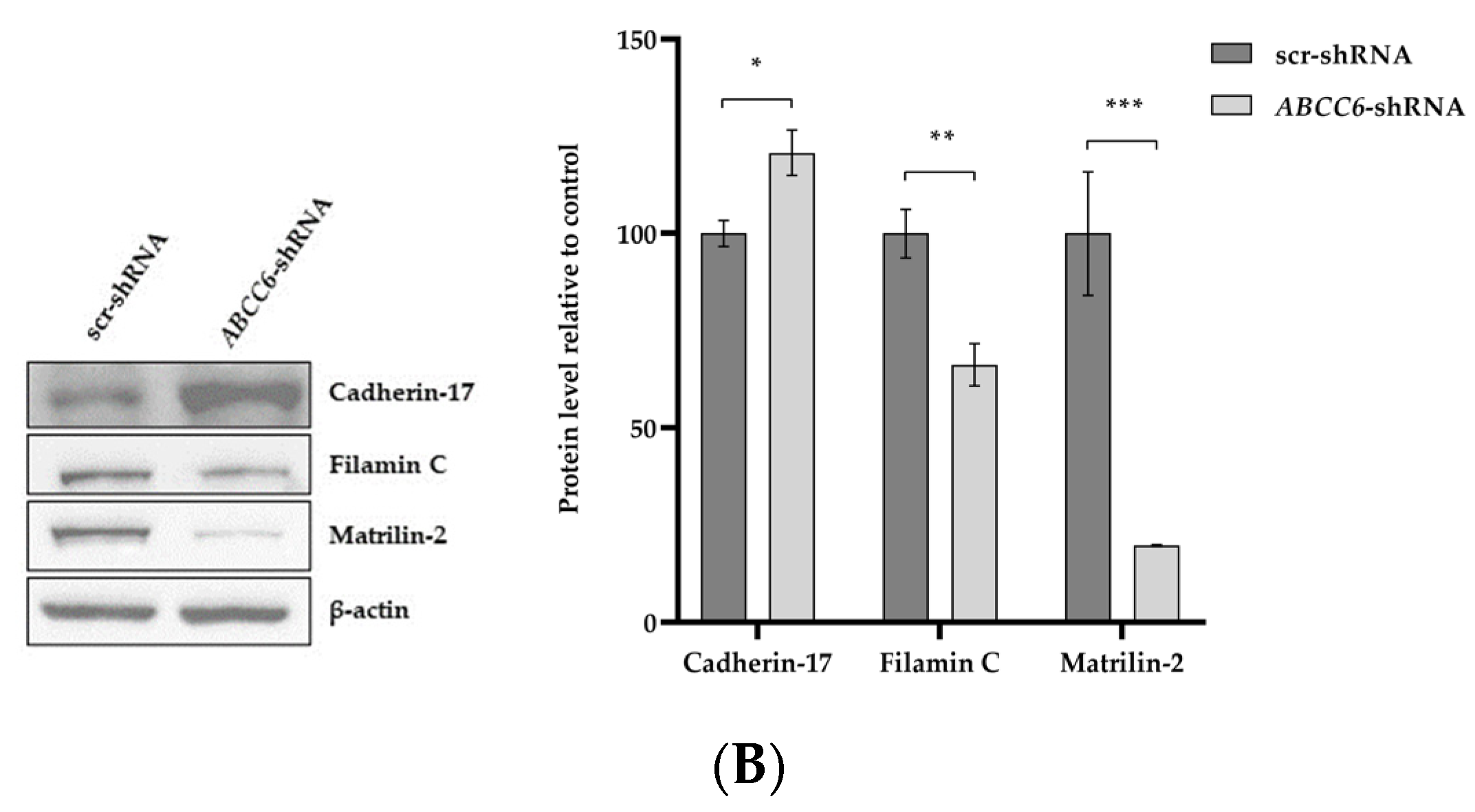
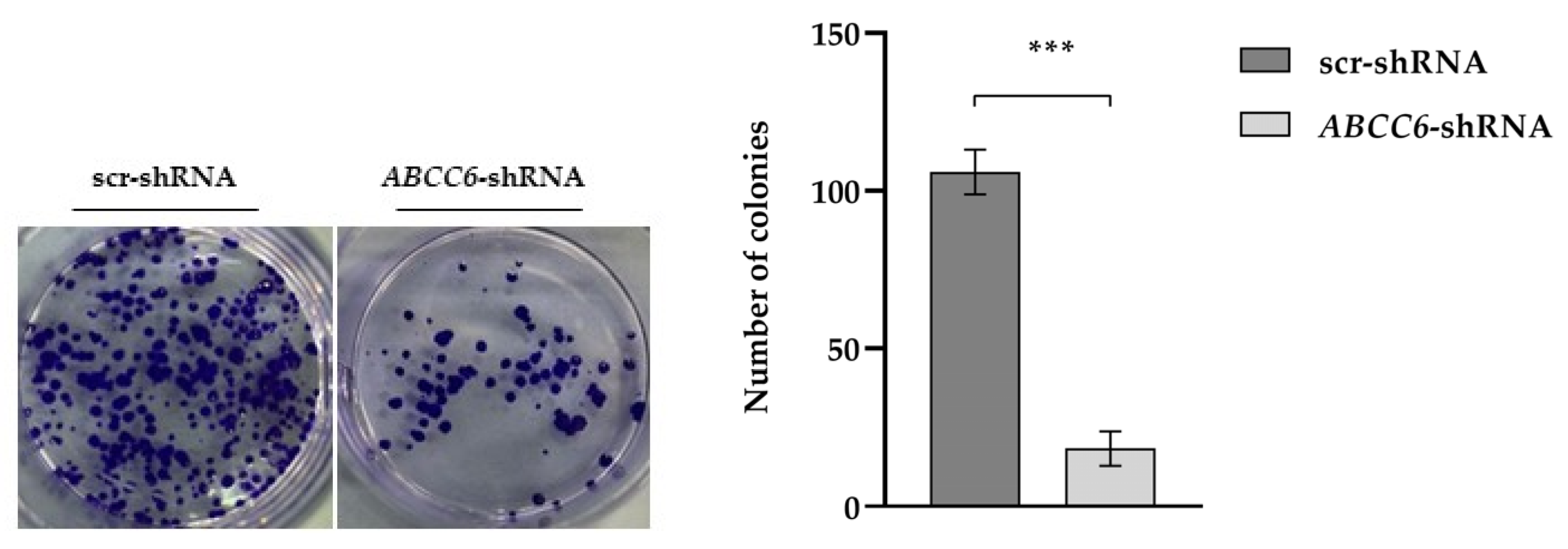


| Gene Symbol | Fold Change | padj | Regulation |
|---|---|---|---|
| MATN2 | −6.52 | 2.08 × 10−4 | |
| ABCG8 | −6.48 | 1.55 × 10−2 | Down |
| PITPNM3 | −6.46 | 9.42 × 10−3 | |
| FADS6 | −5.71 | 1.60 × 10−2 | |
| FLNC | −5.64 | 5.94 × 10−7 | |
| JAM3 | −5.12 | 3.35 × 10−2 | |
| ICAM2 | −4.88 | 8.39 × 10−3 | |
| UNC13C | −4.66 | 5.54 × 10−4 | |
| PLCB2 | −4.63 | 5.55 × 10−4 | |
| SLC22A7 | −4.46 | 4.40 × 10−2 | |
| GLIPR1 | 6.14 | 2.30 × 10−5 | |
| NXPH3 | 6.63 | 1.62 × 10−5 | Up |
| ISM1 | 6.99 | 1.50 × 10−10 | |
| CDH17 | 7.68 | 1.94 × 10−3 | |
| RAB27B | 7.72 | 1.32 × 10−3 | |
| RIPPLY3 | 8.08 | 4.54 × 10−10 | |
| PAUPAR | 11.81 | 3.53 × 10−4 | |
| LYVE1 | 16.55 | 4.57 × 10−9 | |
| MMP7 | 19.08 | 1.17 × 10−17 | |
| DHRS9 | 22.97 | 2.90 × 10−5 |
| ID | Term Description | Fold- Enrichment | Occurrence | Support | Lowest_p | Highest_p | Up_Regulated | Down_Regulated |
|---|---|---|---|---|---|---|---|---|
| hsa05414 | Dilated cardiomyopathy | 5.36 | 10 | 0.12 | 5.41 × 10−9 | 3.54 × 10−7 | ITGA2, ITGA6, ITGB1, ACTG1, TPM4, ADCY7, TGFB1 | ITGA3, TTN, ADCY9 |
| hsa04510 | Focal adhesion | 3.36 | 10 | 0.14 | 1.68 × 10−6 | 8.16 × 10−6 | LAMB3, SPP1, ITGA2, ITGA6, ITGB1, ARHGAP5, ACTG1, CAPN2, FLNA | VTN, ITGA3, PPP1CC, FLNC |
| hsa05410 | Hypertrophic cardiomyopathy | 4.58 | 10 | 0.09 | 3.04 × 10−7 | 9.07 × 10−6 | ITGA2, ITGA6, ITGB1, ACTG1, TPM4, TGFB1 | ITGA3, TTN |
| hsa05205 | Proteoglycans in cancer | 3.26 | 10 | 0.14 | 3.28 × 10−6 | 1.23 × 10−5 | ACTG1, FLNA, ITPR3, CD63, TGFB1, PLAUR, ITGA2, ITGB1, RDX | FLNC, PPP1CC, TIMP3, VTN |
| hsa05132 | Salmonella infection | 2.06 | 10 | 0.03 | 3.86 × 10−5 | 3.86 × 10−5 | DYNLL2, DYNLRB1, TUBA1A, TUBB2A, FLNA, ACTG1, SNX9 | FLNC, CYFIP2, CD14 |
| hsa04610 | Complement and coagulation cascades | 5.99 | 10 | 0.04 | 4.46 × 10−5 | 4.46 × 10−5 | F2R, SERPINE1, PLAUR | FGG, KNG1, MBL2, MASP2, C3, CLU, VTN |
| hsa05412 | Arrhythmogenic right ventricular cardiomyopathy | 4.02 | 10 | 0.07 | 2.07 × 10−5 | 4.53 × 10−5 | ITGA2, ITGA6, ITGB1, ACTG1, DSP | ITGA3 |
| hsa04512 | ECM–receptor interaction | 4.63 | 10 | 0.05 | 2.09 × 10−5 | 8.19 × 10−5 | LAMB3, SPP1, NPNT, ITGA2, ITGA6, ITGB1 | VTN, ITGA3 |
| hsa05145 | Toxoplasmosis | 2.27 | 10 | 0.03 | 8.49 × 10−5 | 1.02 × 10−4 | LAMB3, ITGA6, ITGB1, LDLR, TGFB1 | |
| hsa05222 | Small-cell lung cancer | 3.32 | 10 | 0.05 | 2.61 × 10−5 | 1.02 × 10−4 | CDKN2B, LAMB3, ITGA2, ITGA6, ITGB1 | ITGA3 |
| Gene | Accession Number | Forward Primer | Reverse Primer |
|---|---|---|---|
| β-actin | NM_001101.3 | 5′-CCTGGCACCCAGCACAAT-3′ | 5′-GCCGATCCACACGGAGTACT-3′ |
| ABCC6 | NM_001171.5 | 5′-AAGGAACCACCATCAGGAGGAG-3′ | 5′-ACCAGCGACACAGAGAAGAGG-3′ |
| CDH1 | NM_001317184.2 | 5′-CTCCCTTCACAGCAGAACTAACAC-3′ | 5′-GTCCTCTTCTCCGCCTCCTTC-3′ |
| CDH2 | NM_001308176.2 | 5′-GGATCAAAGCCTGGAACATAT-3′ | 5′-TTGGAGCCTGAGACACGATT-3′ |
| CDH17 | NM_004063.4 | 5′-TCAAAATCACTCAGGTGCGG-3′ | 5′-GAAAAATGGGAATCTTGGGAGC-3′ |
| FLNC | NM_001458.5 | 5′-CTGTCCATGTGTCGGAAGCC-3′ | 5′-ACACCTTGAAGTCAGCCACC -3′ |
| ITGA2 | NM_002203.4 | 5′-AAATGATATTCTGATGCTGGG-3′ | 5′- CCA GCC TTT TCT AGT AGA GC-3′ |
| ITGA6 | NM_000210.4 | 5′-CTT AGG TTT TTC TTT GGA CTC A-3′ | 5′-TCTCTTCAGCAAAACCACGG-3′ |
| MATN2 | NM_002380.5 | 5′-AAACGCTGCCGAAGGAAGG -3′ | 5′-TCCTCAGCTAGAACAAATCCG -3′ |
| VIM | NM_003380.5 | 5′-ATGGACAGGTTATCAACGAAA-3′ | 5′-AAGTTTGGAAGAGGCAGAGA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matera, I.; Miglionico, R.; Abruzzese, V.; Marchese, G.; Ventola, G.M.; Castiglione Morelli, M.A.; Bisaccia, F.; Ostuni, A. A Regulator Role for the ATP-Binding Cassette Subfamily C Member 6 Transporter in HepG2 Cells: Effect on the Dynamics of Cell–Cell and Cell–Matrix Interactions. Int. J. Mol. Sci. 2023, 24, 16391. https://doi.org/10.3390/ijms242216391
Matera I, Miglionico R, Abruzzese V, Marchese G, Ventola GM, Castiglione Morelli MA, Bisaccia F, Ostuni A. A Regulator Role for the ATP-Binding Cassette Subfamily C Member 6 Transporter in HepG2 Cells: Effect on the Dynamics of Cell–Cell and Cell–Matrix Interactions. International Journal of Molecular Sciences. 2023; 24(22):16391. https://doi.org/10.3390/ijms242216391
Chicago/Turabian StyleMatera, Ilenia, Rocchina Miglionico, Vittorio Abruzzese, Giovanna Marchese, Giovanna Maria Ventola, Maria Antonietta Castiglione Morelli, Faustino Bisaccia, and Angela Ostuni. 2023. "A Regulator Role for the ATP-Binding Cassette Subfamily C Member 6 Transporter in HepG2 Cells: Effect on the Dynamics of Cell–Cell and Cell–Matrix Interactions" International Journal of Molecular Sciences 24, no. 22: 16391. https://doi.org/10.3390/ijms242216391
APA StyleMatera, I., Miglionico, R., Abruzzese, V., Marchese, G., Ventola, G. M., Castiglione Morelli, M. A., Bisaccia, F., & Ostuni, A. (2023). A Regulator Role for the ATP-Binding Cassette Subfamily C Member 6 Transporter in HepG2 Cells: Effect on the Dynamics of Cell–Cell and Cell–Matrix Interactions. International Journal of Molecular Sciences, 24(22), 16391. https://doi.org/10.3390/ijms242216391







