Anti-Proteolytic Peptide R7I Protects the Intestinal Barrier and Alleviates Fatty Acid Malabsorption in Salmonella typhimurium-Infected Mice
Abstract
:1. Introduction
2. Results
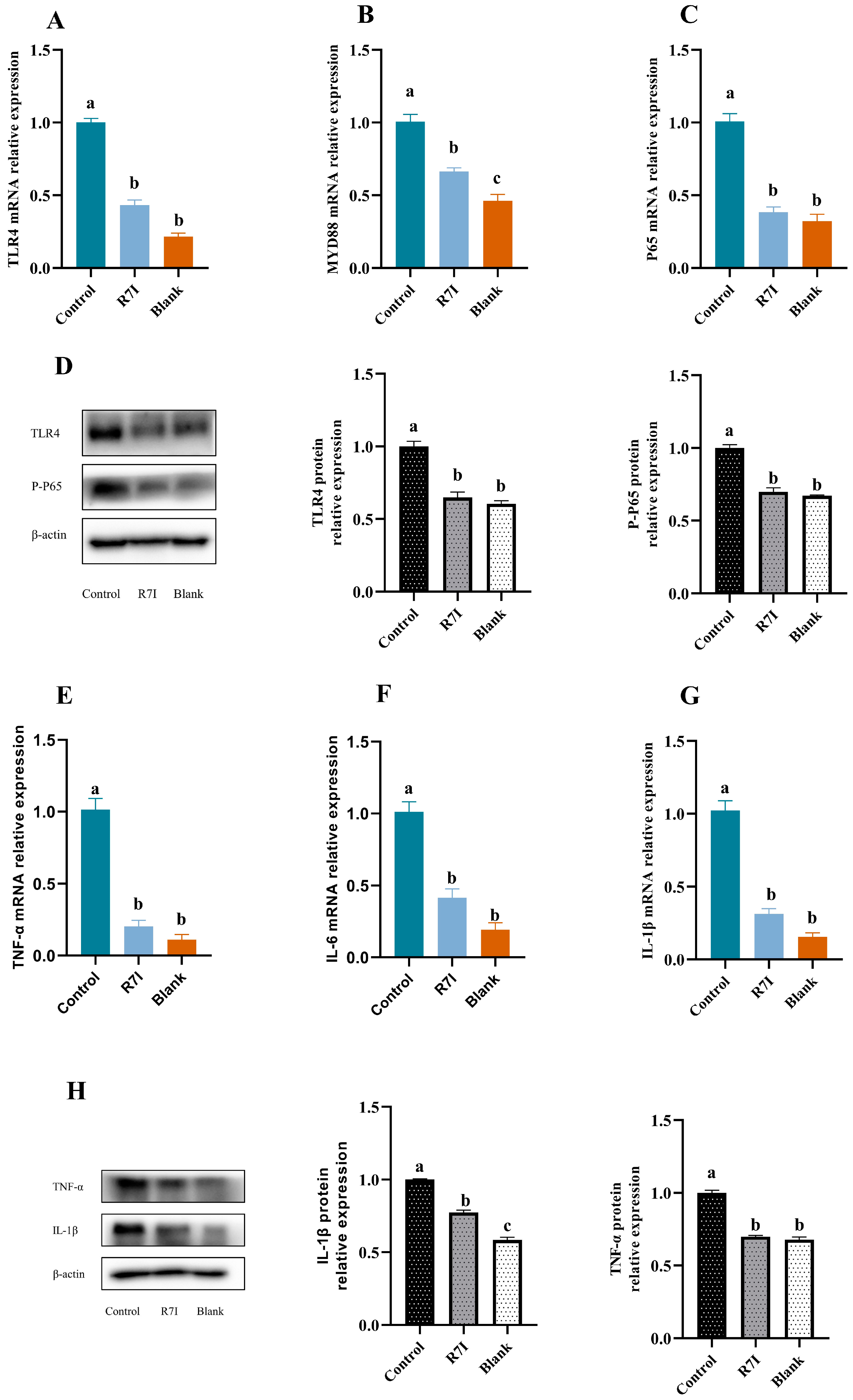
2.1. R7I Could Inhibit the Development of Inflammatory Factors in Jejunal Tissue
2.2. R7I Mitigates Oxidative Damage to the Gut Brought on by Salmonella typhimurium
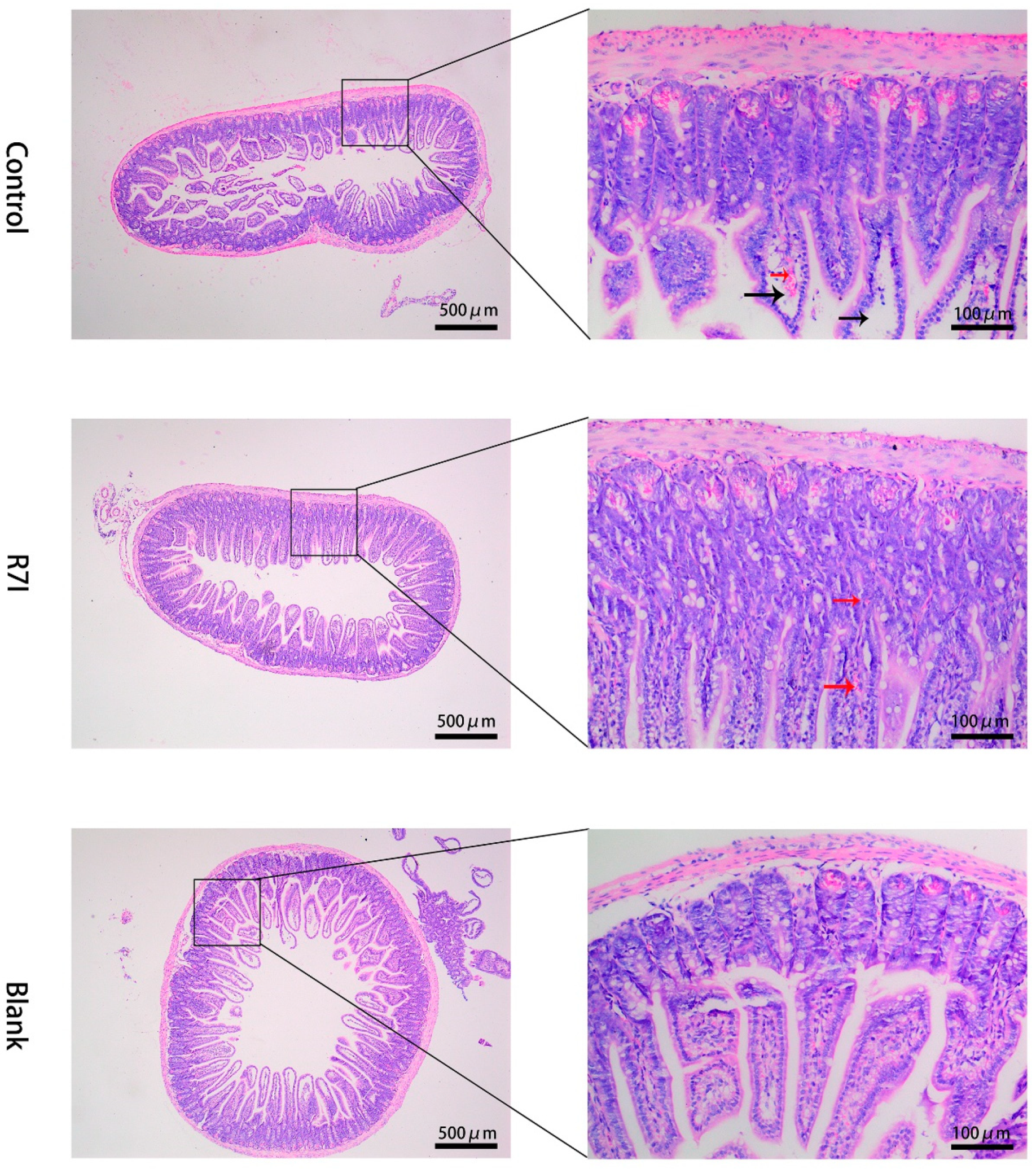
2.3. Effects of R7I on Salmonella typhimurium-Induced Intestinal Barrier Damage
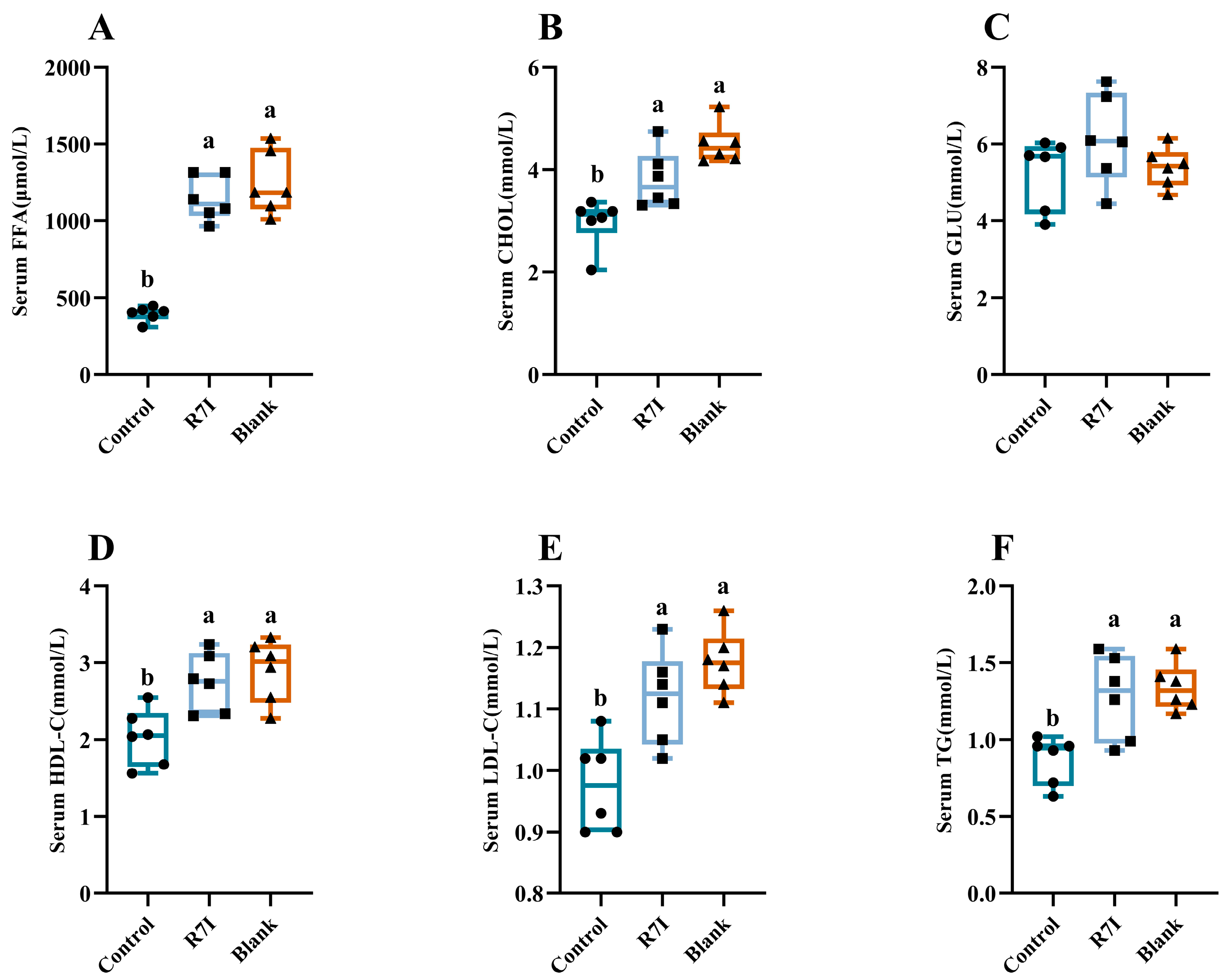
2.4. Alleviating Effect of R7I on Impaired Fatty Acid Absorption Caused by Salmonella typhimurium
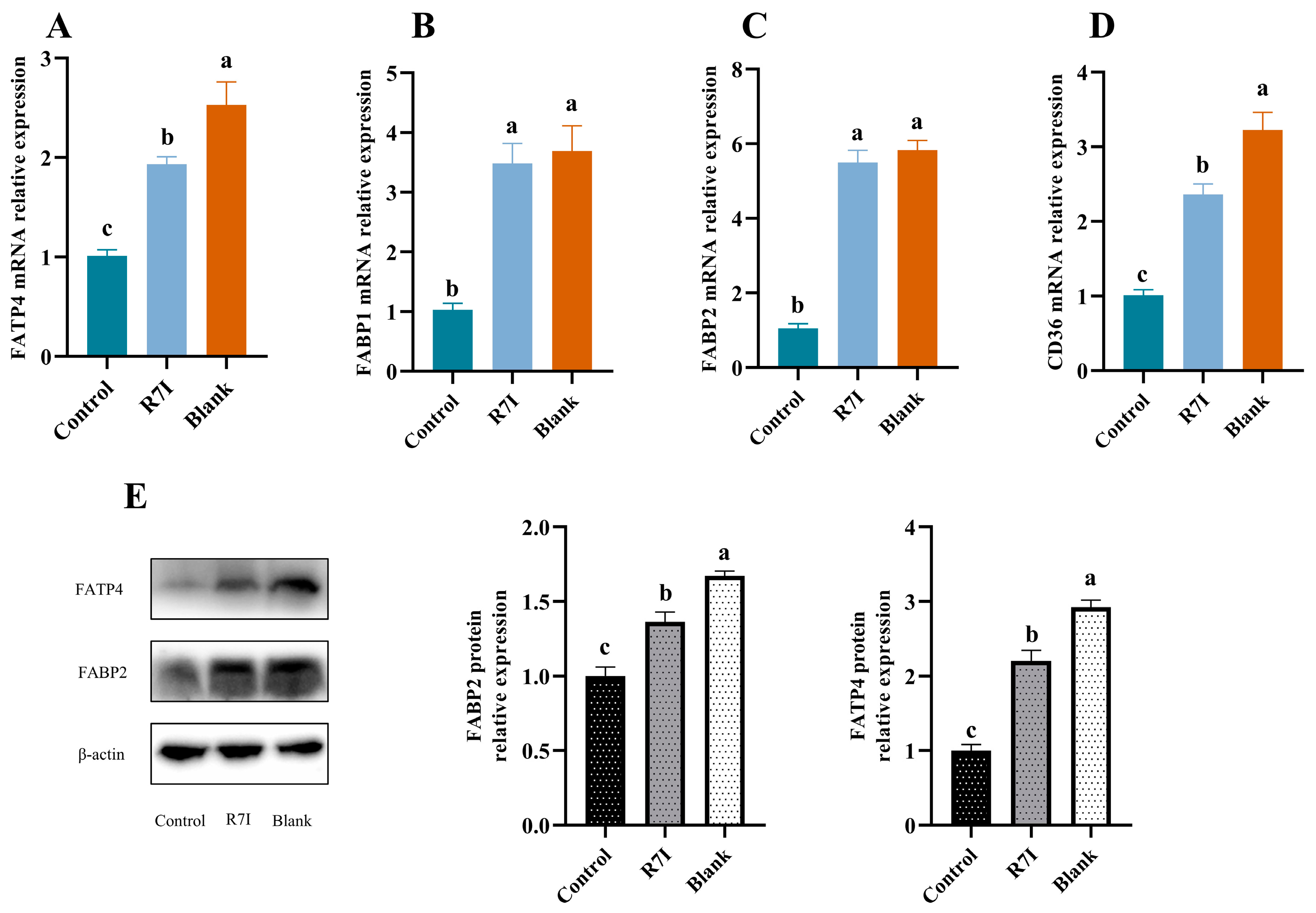
2.5. R7I Enhances the Expression of Fatty Acid Uptake-Related Genes and Proteins in Salmonella typhimurium-Treated Mice
3. Discussion
4. Materials and Methods
4.1. Ethics Approval and Consent to Participate
4.2. Synthesis and Characterization of Peptides
4.3. Construction of an Enteritis Model and Experimental Design
4.4. Collection of Blood, Tissues, and Organs Samples
4.5. Blood Biochemistry Analysis
4.6. Determination of Free Fatty Acid Content
4.7. The Antioxidant Function of the Empty Intestine
4.8. Histological Analysis
4.9. RNA Extraction and Real-Time Quantitative PCR
4.10. Assessment of Protein Expression by Western Blot Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aviv, G.; Cornelius, A.; Davidovich, M.; Cohen, H.; Suwandi, A.; Galeev, A.; Steck, N.; Azriel, S.; Rokney, A.; Valinsky, L.; et al. Differences in the expression of SPI-1 genes pathogenicity and epidemiology between the emerging Salmonella enterica serovar Infantis and the model Salmonella enterica serovar Typhimurium. J. Infect. Dis. 2019, 220, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; International Collaboration on Enteric Disease “Burden of Illness” Studies. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Brumell, J.H.; Tang, P.; Mills, S.D.; Finlay, B.B. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic 2001, 2, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Knodler, L.A. Salmonella enterica: Living a double life in epithelial cells. Curr. Opin. Microbiol. 2015, 23, 23–31. [Google Scholar] [CrossRef]
- Kim, H.B.; Isaacson, R.E. Salmonella in Swine: Microbiota Interactions. Annu. Rev. Anim. Biosci. 2017, 5, 43–63. [Google Scholar] [CrossRef]
- Lan, Y.B.; Wang, S.Z.; Yin, Y.G.; Hoffmann, W.C.; Zheng, X.-Z. Using a Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella typhimurium in Chicken Carcass. J. Bionic Eng. 2008, 5, 239–246. [Google Scholar] [CrossRef]
- Andino, A.; Hanning, I. Salmonella enterica: Survival, colonization, and virulence differences among serovars. Sci. World J. 2015, 2015, 520179. [Google Scholar] [CrossRef]
- Ghilardi, A.C.R.; Tavechio, A.T.; Fernandes, S.A. Antimicrobial susceptibility, phage types, and pulsetypes of Salmonella typhimurium, in Sao Paulo, Brazil. Mem. Inst. Oswaldo Cruz 2006, 101, 281–286. [Google Scholar] [CrossRef]
- Vazquez-Torres, A.; Fang, F.C. Cellular routes of invasion by enteropathogens. Curr. Opin. Microbiol. 2000, 3, 54–59. [Google Scholar] [CrossRef]
- Vazquez-Torres, A.; Jones-Carson, J.; Baumler, A.J.; Falkow, S.; Valdivia, R.; Brown, W.; Le, M.; Berggren, R.; Parks, W.T.; Fang, F.C. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 1999, 401, 804–808. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Wu, Q.; Gao, N.; Wang, Z.; Yang, Y.; Shan, A. Exopolysaccharides of Lactobacillus rhamnosus GG ameliorate Salmonella typhimurium-induced intestinal inflammation via the TLR4/NF-κB/MAPK pathway. J. Anim. Sci. Biotechnol. 2023, 14, 23. [Google Scholar] [CrossRef]
- Lopez, F.E.; Pescaretti, M.D.; Morero, R.; Delgado, M.A. Salmonella typhimurium general virulence factors: A battle of David against Goliath? Food Res. Int. 2012, 45, 842–851. [Google Scholar] [CrossRef]
- Dorsey, C.W.; Laarakker, M.C.; Humphries, A.D.; Weening, E.H.; Bäumler, A.J. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol. Microbiol. 2005, 57, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Haimovich, B.; Venkatesan, M.M. Shigella and Salmonella: Death as a means of survival. Microbes Infect. 2006, 8, 568–577. [Google Scholar] [CrossRef]
- Zolotarevsky, Y.; Hecht, G.; Koutsouris, A.; Gonzalez, D.E.; Quan, C.; Tom, J.; Mrsny, R.J.; Turner, J.R. A membrane-permeant peptide that inhibits MLC kinase restores barrier function in in vitro models of intestinal disease. Gastroenterology 2002, 123, 163–172. [Google Scholar] [CrossRef]
- Ghareeb, K.; Awad, W.A.; Bohm, J.; Zebeli, Q. Impact of luminal and systemic endotoxin exposure on gut function, immune response and performance of chickens. Worlds Poult. Sci. J. 2016, 72, 367–379. [Google Scholar] [CrossRef]
- Albin, D.M.; Wubben, J.E.; Rowlett, J.M.; Tappenden, K.A.; Nowak, R.A. Changes in small intestinal nutrient transport and barrier function after lipopolysaccharide exposure in two pig breeds. J. Anim. Sci. 2007, 85, 2517–2523. [Google Scholar] [CrossRef]
- Kohler, H.; Sakaguchi, T.; Hurley, B.P.; Kase, B.J.; Reinecker, H.-C.; McCormick, B.A. Salmonella enterica serovar Typhimurium regulates intercellular junction proteins and facilitates transepithelial neutrophil and bacterial passage. Am. J. Physiol.-Gastrointest. Liver Physiol. 2007, 293, G178–G187. [Google Scholar] [CrossRef]
- Jepson, M.A.; Schlecht, H.B.; Collares-Buzato, C.B. Localization of dysfunctional tight junctions in Salmonella enterica serovar typhimurium-infected epithelial layers. Infect. Immun. 2000, 68, 7202–7208. [Google Scholar] [CrossRef]
- Rydstrom, A.; Wick, M.J. Monocyte recruitment, activation, and function in the gut-associated lymphoid tissue during oral Salmonella infection. J. Immunol. 2007, 178, 5789–5801. [Google Scholar] [CrossRef]
- Broz, P.; Ohlson, M.B.; Monack, D.M. Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes 2012, 3, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Awad, W.A.; Mann, E.; Dzieciol, M.; Hess, C.; Schmitz-Esser, S.; Wagner, M.; Hess, M. Age-Related Differences in the Luminal and Mucosa-Associated Gut Microbiome of Broiler Chickens and Shifts Associated with Campylobacter jejuni Infection. Front. Cell. Infect. Microbiol. 2016, 6, 154. [Google Scholar] [CrossRef]
- Adams, D.; Fullerton, K.; Jajosky, R.; Sharp, P.; Onweh, D.; Schley, A.; Anderson, W.; Faulkner, A.; Kugeler, K. Summary of Notifiable Infectious Diseases and Conditions—United States, 2013. MMWR Morb. Mortal. Wkly. Rep. 2015, 62, 1–122. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.K.; Sun, Y.; Li, W.Y.; Guo, X.; Liu, X.; Wu, W.; Yu, W.; Wang, J.; Shan, A. Self-Assembly of Antimicrobial Peptide-Based Micelles Breaks the Limitation of Trypsin. ACS Appl. Mater. Interfaces 2023, 15, 494–510. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Wei, Y.X.; Wu, W.P.; Zhang, L.; Wang, J.; Shan, A. Characterization of simplified nonapeptides with broad-spectrum antimicrobial activities as potential food preservatives, and their antibacterial mechanism. Food Funct. 2023, 14, 3139–3154. [Google Scholar] [CrossRef]
- Lyu, Y.; Tan, M.S.; Xue, M.; Hou, W.; Yang, C.; Shan, A.; Xiang, W.; Cheng, B. Broad-spectrum hybrid antimicrobial peptides derived from PMAP-23 with potential LPS binding ability. Biochem. Pharmacol. 2023, 210, 115500. [Google Scholar] [CrossRef]
- Li, Y.M.; Xiang, Q.; Zhang, Q.H.; Huang, Y.; Su, Z. Overview on the recent study of antimicrobial peptides: Origins, functions, relative mechanisms and application. Peptides 2012, 37, 207–215. [Google Scholar] [CrossRef]
- Li, G.; Lai, Z.; Shan, A. Advances of Antimicrobial Peptide-Based Biomaterials for the Treatment of Bacterial Infections. Adv. Sci. 2023, 10, e2206602. [Google Scholar] [CrossRef]
- Sun, T.T.; Liu, X.S.; Su, Y.Z.; Wang, Z.; Cheng, B.; Dong, N.; Wang, J.; Shan, A. The efficacy of anti-proteolytic peptide R7I in intestinal inflammation, function, microbiota, and metabolites by multi-omics analysis in murine bacterial enteritis. Bioeng. Transl. Med. 2023, 8, 20. [Google Scholar] [CrossRef]
- Crump, J.A.; Medalla, F.M.; Joyce, K.W.; Krueger, A.L.; Hoekstra, R.M.; Whichard, J.M.; Barzilay, E.J. Antimicrobial Resistance among Invasive Nontyphoidal Salmonella enterica Isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob. Agents Chemother. 2011, 55, 1148–1154. [Google Scholar] [CrossRef]
- Mezal, E.H.; Stefanova, R.; Khan, A.A. Isolation and molecular characterization of Salmonella enterica serovar Javiana from food, environmental and clinical samples. Int. J. Food Microbiol. 2013, 164, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Preston-Hurlburt, P.; Janeway, C.A., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997, 388, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Keestra, A.M.; van Putten, J.P.M. Unique properties of the chicken TLR4/MD-2 complex: Selective lipopolysaccharide activation of the MyD88-dependent pathway. J. Immunol. 2008, 181, 4354–4362. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, F.; Yang, K.; Han, S.; Jian, S.; Deng, B. Dihydromyricetin alleviates intestinal inflammation by changing intestinal microbial metabolites and inhibiting the expression of the MyD88/NF-κB signaling pathway. Anim. Res. One Health 2023. [Google Scholar] [CrossRef]
- Xagorari, A.; Chlichlia, K. Toll-like receptors and viruses: Induction of innate antiviral immune responses. Open Microbiol. J. 2008, 2, 49–59. [Google Scholar] [CrossRef]
- Awad, W.A.; Molnar, A.; Aschenbach, J.R.; Ghareeb, K.; Khayal, B.; Hess, C.; Liebhart, D.; Dublecz, K.; Hess, M. Campylobacter infection in chickens modulates the intestinal epithelial barrier function. Innate Immun. 2015, 21, 151–160. [Google Scholar] [CrossRef]
- Luo, K.; Cao, S.S. Endoplasmic Reticulum Stress in Intestinal Epithelial Cell Function and Inflammatory Bowel Disease. Gastroenterol. Res. Pract. 2015, 2015, 328791. [Google Scholar] [CrossRef]
- Chaudhari, N.; Talwar, P.; Parimisetty, A.; Lefebvre d’Hellencourt, C.; Ravanan, P. A molecular web: Endoplasmic reticulum stress, inflammation, and oxidative stress. Front. Cell. Neurosci. 2014, 8, 213. [Google Scholar] [CrossRef]
- Yin, S.S.; Cao, W.S. Toll-Like Receptor Signaling Induces Nrf2 Pathway Activation through p62-Triggered Keap1 Degradation. Mol. Cell. Biol. 2015, 35, 2673–2683. [Google Scholar] [CrossRef]
- Mukojima, K.; Mishima, S.; Oda, J.; Homma, H.; Sasaki, H.; Ohta, S.; Yukioka, T. Protective Effects of Free Radical Scavenger Edaravone Against Xanthine Oxidase-Mediated Permeability Increases in Human Intestinal Epithelial Cell Monolayer. J. Burn. Care Res. 2009, 30, 335–340. [Google Scholar] [CrossRef]
- Wakabayashi, N.; Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Kang, M.-I.; Kobayashi, A.; Yamamoto, M.; Kensler, T.W.; Talalay, P. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 2004, 101, 2040–2045. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Wang, H.; Wang, X.; Zhu, L.; Mao, L. The absence of Nrf2 enhances NF-κB-dependent inflammation following scratch injury in mouse primary cultured astrocytes. Mediat. Inflamm. 2012, 2012, 217580. [Google Scholar] [CrossRef] [PubMed]
- Doege, H.; Baillie, R.A.; Ortegon, A.M.; Tsang, B.; Wu, Q.; Punreddy, S.; Hirsch, D.; Watson, N.; Gimeno, R.E.; Stahl, A. Targeted deletion of FATP5 reveals multiple functions in liver metabolism: Alterations in hepatic lipid Homeostasis. Gastroenterology 2006, 130, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Abumrad, N.A. Cellular fatty acid uptake: A pathway under construction. Trends Endocrinol. Metab. 2009, 20, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Jay, A.G.; Hamilton, J.A. The enigmatic membrane fatty acid transporter CD36: New insights into fatty acid binding and their effects on uptake of oxidized LDL. Prostaglandins Leukot. Essent. Fatty. Acids 2018, 138, 64–70. [Google Scholar] [CrossRef] [PubMed]


| Gene | Sequences (5′→3′) | Fragments Sizes | Gen Bank No. |
|---|---|---|---|
| IL-6 | F: CTCCCAACAGACCTGTCTATAC | 97 bp | NM_031168.2 |
| R: CCATTGCACAACTCTTTTCTCA | |||
| IL-1β | F: GAAATGCCACCTTTTGACAGTG | 116 bp | NM_008361.4 |
| R: TGGATGCTCTCATCAGGACAG | |||
| TNF-α | F: ATGTCTCAGCCTCTTCTCATTC | 179 bp | NM_013693.3 |
| R: GCTTGTCACTCGAATTTTGAGA | |||
| NF-κB-P65 | F: AGACCCAGGAGTGTTCACAGACC | 141 bp | NM_001402548.1 |
| R: GTCACCAGGCGAGTTATAGCTTCAG | |||
| MYD88 | F: TCATGTTCTCCATACCCTTGGT | 175 bp | NM_010851.3 |
| R: AAACTGCGAGTGGGGTCAG | |||
| TLR4 | F: TCCCTGCATAGAGGTAGTTCC | 119 bp | NM_021297.3 |
| R: TCAAGGGGTTGAAGCTCAGA | |||
| NRF2 | F: TAGATGACCATGAGTCGCTTGC | 153 bp | NM_010902.5 |
| R: GCCAAACTTGCTCCATGTCC | |||
| KEAP-1 | F: TGCCCCTGTGGTCAAAGTG | 104 bp | NM_001110307.1 |
| R: GGTTCGGTTACCGTCCTGC | |||
| I-NOS | F: ACATCGACCCGTCCACAGTAT | 177 bp | NM_001313922.1 |
| R: CAGAGGGGTAGGCTTGTCTC | |||
| FATP4 | F: ACCAGGGTGCCAACAACAAGAAG | 121 bp | NM_011989.5 |
| R: GTGCGATCTCGGAAGTACAGGTAAC | |||
| CD36 | F: GCAGGTCTATCTACGCTGTGTTCG | 111 bp | XM_030254088.1 |
| R: TGTCTGGATTCTGGAGGGGTGATG | |||
| FABP1 | F: AAGTGGTCCGCAATGAGTTCACC | 87 bp | NM_017399.5 |
| R: CCAGCTTGACGACTGCCTTGAC | |||
| FABP2 | F: GATTGCTGTCCGAGAGGTTTCTGG | 80 bp | NM_007980.3 |
| R: TAAAGAATCGCTTGGCCTCAACTCC | |||
| Occludin | F: TGCTTCATCGCTTCCTTAGTAA | 155 bp | NM_001360536.1 |
| R: GGGTTCACTCCCATTATGTACA | |||
| ZO-1 | F: CTGGTGAAGTCTCGGAAAAATG | 97 bp | NM_009386.2 |
| R: CATCTCTTGCTGCCAAACTATC | |||
| Claudin 1 | F: AGATACAGTGCAAAGTCTTCGA | 86 bp | NM_016674.4 |
| R: CAGGATGCCAATTACCATCAAG | |||
| β-actin | F: CTACCTCATGAAGATCCTGACC | 100 bp | NM_007393.5 |
| R: CACAGCTTCTCTTTGATGTCAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Y.; Sun, T.; Gao, J.; Zhang, C.; Liu, X.; Bi, C.; Wang, J.; Shan, A. Anti-Proteolytic Peptide R7I Protects the Intestinal Barrier and Alleviates Fatty Acid Malabsorption in Salmonella typhimurium-Infected Mice. Int. J. Mol. Sci. 2023, 24, 16409. https://doi.org/10.3390/ijms242216409
Su Y, Sun T, Gao J, Zhang C, Liu X, Bi C, Wang J, Shan A. Anti-Proteolytic Peptide R7I Protects the Intestinal Barrier and Alleviates Fatty Acid Malabsorption in Salmonella typhimurium-Infected Mice. International Journal of Molecular Sciences. 2023; 24(22):16409. https://doi.org/10.3390/ijms242216409
Chicago/Turabian StyleSu, Yunzhe, Taotao Sun, Junhan Gao, Chenxu Zhang, Xuesheng Liu, Chongpeng Bi, Jiajun Wang, and Anshan Shan. 2023. "Anti-Proteolytic Peptide R7I Protects the Intestinal Barrier and Alleviates Fatty Acid Malabsorption in Salmonella typhimurium-Infected Mice" International Journal of Molecular Sciences 24, no. 22: 16409. https://doi.org/10.3390/ijms242216409
APA StyleSu, Y., Sun, T., Gao, J., Zhang, C., Liu, X., Bi, C., Wang, J., & Shan, A. (2023). Anti-Proteolytic Peptide R7I Protects the Intestinal Barrier and Alleviates Fatty Acid Malabsorption in Salmonella typhimurium-Infected Mice. International Journal of Molecular Sciences, 24(22), 16409. https://doi.org/10.3390/ijms242216409






