Emerging Role of Circular RNAs in Hepatocellular Carcinoma Immunotherapy
Abstract
:1. Introduction
2. Circular RNAs (CircRNAs)
2.1. What Are CircRNAs?
2.2. Biogenesis of CircRNAs
2.2.1. Intron Pairing-Driven Circularization
2.2.2. RBP-Induced Circularization
2.2.3. Lariat-Induced Circularization Driven by Spliceosomes
2.2.4. Self-Circularization of Introns
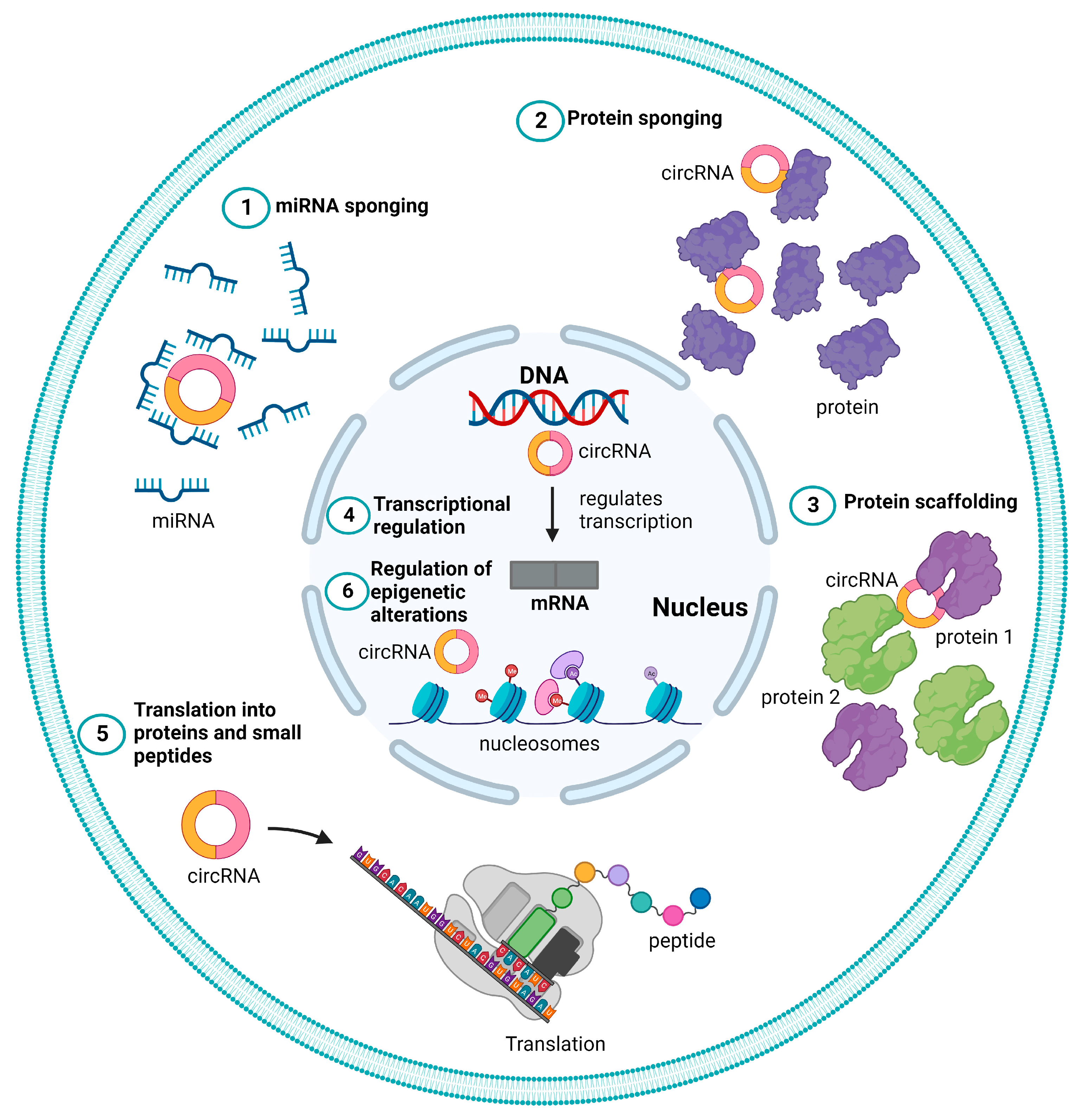
2.3. Functional Roles of CircRNAs
2.3.1. miRNA Sponge
2.3.2. Protein Sponge or Decoy
2.3.3. Protein Scaffolding
2.3.4. Transcriptional Regulation
2.3.5. Translation to Proteins and Peptides
2.3.6. Regulation of Epigenetic Alterations
2.4. Involvement of circRNAs in HCC Tumor Development and Progression
3. Immunotherapy
3.1. HCC Immunotherapy
3.2. CircRNAs in HCC Immunotherapy
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Ringelhan, M.; Pfister, D.; O’Connor, T.; Pikarsky, E.; Heikenwalder, M. The immunology of hepatocellular carcinoma. Nat. Immunol. 2018, 19, 222–232. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Youssef, S.S.; Abbas, E.; Youness, R.A.; Elemeery, M.N.; Nasr, A.S.; Seif, S. PNPLA3 and IL 28B signature for predicting susceptibility to chronic hepatitis C infection and fibrosis progression. Arch. Physiol. Biochem. 2022, 128, 483–489. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M.; American Association for the Study of Liver, D. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Bustamante, J.; Castells, A.; Vilana, R.; Ayuso Mdel, C.; Sala, M.; Bru, C.; Rodes, J.; Bruix, J. Natural history of untreated nonsurgical hepatocellular carcinoma: Rationale for the design and evaluation of therapeutic trials. Hepatology 1999, 29, 62–67. [Google Scholar] [CrossRef]
- Kumada, T.; Nakano, S.; Takeda, I.; Sugiyama, K.; Osada, T.; Kiriyama, S.; Sone, Y.; Toyoda, H.; Shimada, S.; Takahashi, M.; et al. Patterns of recurrence after initial treatment in patients with small hepatocellular carcinoma. Hepatology 1997, 25, 87–92. [Google Scholar] [CrossRef]
- Youssef, S.S.; Youness, R.A.; Abbas, E.A.E.; Osman, N.M.; ELFiky, A.; El-Kassas, M. miR-516a-3P, a potential circulating biomarker in hepatocellular carcinoma, correlated with rs738409 polymorphism in PNPLA3. Per Med. 2022, 19, 483–493. [Google Scholar] [CrossRef]
- Lohitesh, K.; Chowdhury, R.; Mukherjee, S. Resistance a major hindrance to chemotherapy in hepatocellular carcinoma: An insight. Cancer Cell Int. 2018, 18, 44. [Google Scholar] [CrossRef] [PubMed]
- Miyahara, K.; Nouso, K.; Yamamoto, K. Chemotherapy for advanced hepatocellular carcinoma in the sorafenib age. World J. Gastroenterol. 2014, 20, 4151–4159. [Google Scholar] [CrossRef] [PubMed]
- Ferrin, G.; Guerrero, M.; Amado, V.; Rodriguez-Peralvarez, M.; De la Mata, M. Activation of mTOR Signaling Pathway in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 1266ra. [Google Scholar] [CrossRef]
- Youness, R.A.; El-Tayebi, H.M.; Assal, R.A.; Hosny, K.; Esmat, G.; Abdelaziz, A.I. MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncol. Lett. 2016, 12, 2567–2573. [Google Scholar] [CrossRef]
- Abdel-Latif, M.; Youness, R.A. Why natural killer cells in triple negative breast cancer? World J. Clin. Oncol. 2020, 11, 464–476. [Google Scholar] [CrossRef] [PubMed]
- El Din, G.S.; Youness, R.A.; Assal, R.A.; Gad, M.Z. miRNA-506-3p Directly Regulates rs10754339 (A/G) in the Immune Checkpoint Protein B7-H4 in Breast Cancer. Microrna 2020, 9, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Ramzy, A.; ElSafy, S.; Elshoky, H.A.; Soliman, A.; Youness, R.; Mansour, S.; Sebak, A. Drugless nanoparticles tune-up an array of intertwined pathways contributing to immune checkpoint signaling and metabolic reprogramming in triple-negative breast cancer. Biomed. Mater. 2022, 18, 015023. [Google Scholar] [CrossRef] [PubMed]
- Selem, N.A.; Nafae, H.; Manie, T.; Youness, R.A.; Gad, M.Z. Let-7a/cMyc/CCAT1/miR-17-5p Circuit Re-sensitizes Atezolizumab Resistance in Triple Negative Breast Cancer through Modulating PD-L1. Pathol. Res. Pract. 2023, 248, 154579. [Google Scholar] [CrossRef]
- Mekky, R.Y.; Ragab, M.F.; Manie, T.; Attia, A.A.; Youness, R.A. MALAT-1: Immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Transl. Oncol. 2023, 31, 101653. [Google Scholar] [CrossRef]
- Youness, R.A.; Rahmoon, M.A.; Assal, R.A.; Gomaa, A.I.; Hamza, M.T.; Waked, I.; El Tayebi, H.M.; Abdelaziz, A.I. Contradicting interplay between insulin-like growth factor-1 and miR-486-5p in primary NK cells and hepatoma cell lines with a contemporary inhibitory impact on HCC tumor progression. Growth Factors 2016, 34, 128–140. [Google Scholar] [CrossRef]
- Soliman, A.H.; Youness, R.A.; Sebak, A.A.; Handoussa, H. Phytochemical-derived tumor-associated macrophage remodeling strategy using Phoenix dactylifera L. boosted photodynamic therapy in melanoma via H19/iNOS/PD-L1 axis. Photodiagnosis Photodyn. Ther. 2023, 44, 103792. [Google Scholar] [CrossRef]
- Pinato, D.J.; Guerra, N.; Fessas, P.; Murphy, R.; Mineo, T.; Mauri, F.A.; Mukherjee, S.K.; Thursz, M.; Wong, C.N.; Sharma, R.; et al. Immune-based therapies for hepatocellular carcinoma. Oncogene 2020, 39, 3620–3637. [Google Scholar] [CrossRef]
- Zhu, A.X.; Kang, Y.K.; Yen, C.J.; Finn, R.S.; Galle, P.R.; Llovet, J.M.; Assenat, E.; Brandi, G.; Pracht, M.; Lim, H.Y.; et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased alpha-fetoprotein concentrations (REACH-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019, 20, 282–296. [Google Scholar] [CrossRef]
- Saung, M.T.; Pelosof, L.; Casak, S.; Donoghue, M.; Lemery, S.; Yuan, M.; Rodriguez, L.; Schotland, P.; Chuk, M.; Davis, G.; et al. FDA Approval Summary: Nivolumab Plus Ipilimumab for the Treatment of Patients with Hepatocellular Carcinoma Previously Treated with Sorafenib. Oncologist 2021, 26, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Casak, S.J.; Donoghue, M.; Fashoyin-Aje, L.; Jiang, X.; Rodriguez, L.; Shen, Y.L.; Xu, Y.; Jiang, X.; Liu, J.; Zhao, H.; et al. FDA Approval Summary: Atezolizumab Plus Bevacizumab for the Treatment of Patients with Advanced Unresectable or Metastatic Hepatocellular Carcinoma. Clin. Cancer Res. 2021, 27, 1836–1841. [Google Scholar] [CrossRef]
- Psilopatis, I.; Damaskos, C.; Garmpi, A.; Sarantis, P.; Koustas, E.; Antoniou, E.A.; Dimitroulis, D.; Kouraklis, G.; Karamouzis, M.V.; Vrettou, K.; et al. FDA-Approved Monoclonal Antibodies for Unresectable Hepatocellular Carcinoma: What Do We Know So Far? Int. J. Mol. Sci. 2023, 24, 2685. [Google Scholar] [CrossRef]
- Vogel, A.; Cervantes, A.; Chau, I.; Daniele, B.; Llovet, J.M.; Meyer, T.; Nault, J.C.; Neumann, U.; Ricke, J.; Sangro, B.; et al. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv238–iv255. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Vogrig, A.; Muniz-Castrillo, S.; Farina, A.; Honnorat, J.; Joubert, B. How to diagnose and manage neurological toxicities of immune checkpoint inhibitors: An update. J. Neurol. 2022, 269, 1701–1714. [Google Scholar] [CrossRef]
- Remash, D.; Prince, D.S.; McKenzie, C.; Strasser, S.I.; Kao, S.; Liu, K. Immune checkpoint inhibitor-related hepatotoxicity: A review. World J. Gastroenterol. 2021, 27, 5376–5391. [Google Scholar] [CrossRef]
- Chen, R.; Zhou, M.; Zhu, F. Immune Checkpoint Inhibitors Related to Cardiotoxicity. J. Cardiovasc. Dev. Dis. 2022, 9, 378. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- El-Daly, S.M.; Talaat, R.M.; Braoudaki, M.; Youness, R.A.; Cho, W.C. Editorial: Recent breakthroughs in the decoding of circulating nucleic acids and their applications to human diseases. Front. Mol. Biosci. 2023, 10, 1203495. [Google Scholar] [CrossRef] [PubMed]
- Youness, R.A.; Hafez, H.M.; Khallaf, E.; Assal, R.A.; Abdel Motaal, A.; Gad, M.Z. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J. Cell Physiol. 2019, 234, 20286–20297. [Google Scholar] [CrossRef] [PubMed]
- Selem, N.A.; Youness, R.A.; Gad, M.Z. What is beyond LncRNAs in breast cancer: A special focus on colon cancer-associated Transcript-1 (CCAT-1). Noncoding RNA Res. 2021, 6, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Youness, R.A.; Assal, R.A.; Abdel Motaal, A.; Gad, M.Z. A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric Oxide 2018, 80, 12–23. [Google Scholar] [CrossRef]
- Mekky, R.Y.; El-Ekiaby, N.; El Sobky, S.A.; Elemam, N.M.; Youness, R.A.; El-Sayed, M.; Hamza, M.T.; Esmat, G.; Abdelaziz, A.I. Epigallocatechin gallate (EGCG) and miR-548m reduce HCV entry through repression of CD81 receptor in HCV cell models. Arch. Virol. 2019, 164, 1587–1595. [Google Scholar] [CrossRef]
- Nafea, H.; Youness, R.A.; Abou-Aisha, K.; Gad, M.Z. LncRNA HEIH/miR-939-5p interplay modulates triple-negative breast cancer progression through NOS2-induced nitric oxide production. J. Cell Physiol. 2021, 236, 5362–5372. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Dawoud, A.; Zeinelabdeen, Y.A.; Kiriacos, C.J.; Daniel, K.A.; Eltahtawy, O.; Abdelhalim, M.M.; Braoudaki, M.; Youness, R.A. Molecular Engines, Therapeutic Targets, and Challenges in Pediatric Brain Tumors: A Special Emphasis on Hydrogen Sulfide and RNA-Based Nano-Delivery. Cancers 2022, 14, 5244. [Google Scholar] [CrossRef]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef]
- Xu, Z.; Li, P.; Fan, L.; Wu, M. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front. Immunol. 2018, 9, 9. [Google Scholar] [CrossRef]
- Dawoud, A.; Ihab Zakaria, Z.; Hisham Rashwan, H.; Braoudaki, M.; Youness, R.A. Circular RNAs: New layer of complexity evading breast cancer heterogeneity. Noncoding RNA Res. 2023, 8, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Fang, S.; Jiang, M.; Zhao, X.; Zhou, C.; Gong, Z. Circular RNAs: Regulatory functions in respiratory tract cancers. Clin. Chim. Acta 2020, 510, 264–271. [Google Scholar] [CrossRef]
- Wang, F.; Li, X.; Li, Z.; Wang, S.; Fan, J. Functions of Circular RNAs in Regulating Adipogenesis of Mesenchymal Stem Cells. Stem Cells Int. 2020, 2020, 3763069. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Bala, S. Emerging role of non-coding RNA in oral cancer. Cell Signal 2018, 42, 134–143. [Google Scholar] [CrossRef]
- Chen, L.; Shan, G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. 2021, 505, 49–57. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Q.; Zhang, K.; Wang, F.; Qiao, X.; Cui, J. Circ SMARCA5 Inhibited Tumor Metastasis by Interacting with SND1 and Downregulating the YWHAB Gene in Cervical Cancer. Cell Transpl. 2021, 30, 963689720983786. [Google Scholar] [CrossRef]
- El-Aziz, M.K.A.; Dawoud, A.; Kiriacos, C.J.; Fahmy, S.A.; Hamdy, N.M.; Youness, R.A. Decoding hepatocarcinogenesis from a noncoding RNAs perspective. J. Cell Physiol. 2023, 238, 1982–2009. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, X.; Su, X. Noncoding RNAs in cancer immunity: Functions, regulatory mechanisms, and clinical application. Mol. Cancer 2020, 19, 48. [Google Scholar] [CrossRef] [PubMed]
- Youness, R.A.; Gad, A.Z.; Sanber, K.; Ahn, Y.J.; Lee, G.J.; Khallaf, E.; Hafez, H.M.; Motaal, A.A.; Ahmed, N.; Gad, M.Z. Targeting hydrogen sulphide signaling in breast cancer. J. Adv. Res. 2021, 27, 177–190. [Google Scholar] [CrossRef]
- Papatsirou, M.; Artemaki, P.I.; Scorilas, A.; Kontos, C.K. The role of circular RNAs in therapy resistance of patients with solid tumors. Per Med. 2020, 17, 469–490. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Dragomir, M.; Calin, G.A. Circular RNAs in Cancer—Lessons Learned From microRNAs. Front. Oncol. 2018, 8, 179. [Google Scholar] [CrossRef]
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Geng, X.; Jia, Y.; Zhang, Y.; Shi, L.; Li, Q.; Zang, A.; Wang, H. Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 2020, 12, 267–283. [Google Scholar] [CrossRef]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef]
- Wang, M.; Yu, F.; Li, P. Circular RNAs: Characteristics, Function and Clinical Significance in Hepatocellular Carcinoma. Cancers 2018, 10, 258. [Google Scholar] [CrossRef]
- Bolha, L.; Ravnik-Glavac, M.; Glavac, D. Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. Int. J. Genom. 2017, 2017, 6218353. [Google Scholar] [CrossRef] [PubMed]
- Papatsirou, M.; Artemaki, P.I.; Karousi, P.; Scorilas, A.; Kontos, C.K. Circular RNAs: Emerging Regulators of the Major Signaling Pathways Involved in Cancer Progression. Cancers 2021, 13, 2744. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Floris, G.; Zhang, L.; Follesa, P.; Sun, T. Regulatory Role of Circular RNAs and Neurological Disorders. Mol. Neurobiol. 2017, 54, 5156–5165. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Shen, T.; Han, M.; Wei, G.; Ni, T. An intriguing RNA species--perspectives of circularized RNA. Protein Cell 2015, 6, 871–880. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Errichelli, L.; Dini Modigliani, S.; Laneve, P.; Colantoni, A.; Legnini, I.; Capauto, D.; Rosa, A.; De Santis, R.; Scarfo, R.; Peruzzi, G.; et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017, 8, 14741. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.L.; Cherry, S.; Wilusz, J.E. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol. Cell 2017, 68, 940–954.e3. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Yang, Y.; Niu, G.; Tang, Z.; Li, K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 2017, 24, 523–535. [Google Scholar] [CrossRef]
- Li, P.; Chen, H.; Chen, S.; Mo, X.; Li, T.; Xiao, B.; Yu, R.; Guo, J. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. Cancer 2017, 116, 626–633. [Google Scholar] [CrossRef]
- Huang, S.; Yang, B.; Chen, B.J.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Gong, X.J.; Sun, L.; Zhou, Q.Y.; Lu, B.L.; Zhu, L.Y. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS ONE 2016, 11, e0158347. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, M.; Zheng, X.; Yi, P.; Lan, C.; Xu, M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 17–27. [Google Scholar] [CrossRef]
- Ren, S.; Xin, Z.; Xu, Y.; Xu, J.; Wang, G. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle 2017, 16, 2204–2211. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yang, Z.; Zheng, B.; Zhang, X.H.; Zhang, M.L.; Zhao, X.S.; Zhao, H.Y.; Suzuki, T.; Wen, J.K. A Novel Regulatory Mechanism of Smooth Muscle alpha-Actin Expression by NRG-1/circACTA2/miR-548f-5p Axis. Circ. Res. 2017, 121, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yao, M.D.; Li, C.P.; Shan, K.; Yang, H.; Wang, J.J.; Liu, B.; Li, X.M.; Yao, J.; Jiang, Q.; et al. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics 2017, 7, 2863–2877. [Google Scholar] [CrossRef]
- Rahmoon, M.A.; Youness, R.A.; Gomaa, A.I.; Hamza, M.T.; Waked, I.; El Tayebi, H.M.; Abdelaziz, A.I. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors 2017, 35, 76–87. [Google Scholar] [CrossRef]
- Zhong, Z.; Huang, M.; Lv, M.; He, Y.; Duan, C.; Zhang, L.; Chen, J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017, 403, 305–317. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, G.C.; Ding, C.; Liu, P.; Wang, R.K.; Ding, W.B.; Tong, D.K.; Wu, D.J.; Li, C.; Wei, Q.; et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 2017, 8, 61687–61697. [Google Scholar] [CrossRef]
- Liang, H.F.; Zhang, X.Z.; Liu, B.G.; Jia, G.T.; Li, W.L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017, 7, 1566–1576. [Google Scholar]
- Wang, X.H.; Fang, L. Advances in circular RNAs and their roles in breast Cancer. J. Exp. Clin. Cancer Res. 2018, 37, 206. [Google Scholar] [CrossRef]
- Yu, T.; Ran, L.; Zhao, H.; Yin, P.; Li, W.; Lin, J.; Mao, H.; Cai, D.; Ma, Q.; Pan, X.; et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol. Ther. Nucleic Acids 2021, 26, 649–664. [Google Scholar] [CrossRef] [PubMed]
- Shaalan, Y.M.; Handoussa, H.; Youness, R.A.; Assal, R.A.; El-Khatib, A.H.; Linscheid, M.W.; El Tayebi, H.M.; Abdelaziz, A.I. Destabilizing the interplay between miR-1275 and IGF2BPs by Tamarix articulata and quercetin in hepatocellular carcinoma. Nat. Prod. Res. 2018, 32, 2217–2220. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Xu, B.; Liu, Y.; Kong, P.; Li, C.; Li, B. circ-SIRT1 Promotes Colorectal Cancer Proliferation and EMT by Recruiting and Binding to eIF4A3. Anal. Cell Pathol. 2021, 2021, 5739769. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, M.; Zhang, Y. Circ_0000079 Decoys the RNA-Binding Protein FXR1 to Interrupt Formation of the FXR1/PRCKI Complex and Decline Their Mediated Cell Invasion and Drug Resistance in NSCLC. Cell Transplant. 2020, 29, 0963689720961070. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Wu, S.; Zhou, Z.; Ding, X.; Shi, R.; Thorne, R.F.; Zhang, X.D.; Hu, W.; Wu, M. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab. 2019, 30, 157–173.e7. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, J.; Tian, Y.; Gao, Y.; Dong, X.; Chen, W.; Yuan, X.; Yin, W.; Xu, J.; Chen, K.; et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 2020, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, N.; Yang, X.; Luo, J.; Yan, S.; Xiao, F.; Chen, W.; Gao, X.; Zhao, K.; Zhou, H.; et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 2018, 37, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, L.; Zhou, Y.; Wang, Q.; Zheng, Z.; Xu, B.; Wu, C.; Zhou, Q.; Hu, W.; Wu, C.; et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 2019, 18, 47. [Google Scholar] [CrossRef]
- Liang, W.C.; Wong, C.W.; Liang, P.P.; Shi, M.; Cao, Y.; Rao, S.T.; Tsui, S.K.; Waye, M.M.; Zhang, Q.; Fu, W.M.; et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019, 20, 84. [Google Scholar] [CrossRef]
- Di Timoteo, G.; Dattilo, D.; Centron-Broco, A.; Colantoni, A.; Guarnacci, M.; Rossi, F.; Incarnato, D.; Oliviero, S.; Fatica, A.; Morlando, M.; et al. Modulation of circRNA Metabolism by m(6)A Modification. Cell Rep. 2020, 31, 107641. [Google Scholar] [CrossRef]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes. Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.F.; Hu, L.; Zhuo, W.; Zhang, C.M.; Zhou, H.H.; Fan, L. Epigenetic alternations and cancer chemotherapy response. Cancer Chemother. Pharmacol. 2016, 77, 673–684. [Google Scholar] [CrossRef]
- Chen, N.; Zhao, G.; Yan, X.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z.; et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018, 19, 218. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Su, M.; Xiao, Y.; Tang, J.; Wu, J.; Ma, J.; Tian, B.; Zhou, Y.; Wang, H.; Yang, D.; Liao, Q.J.; et al. Role of lncRNA and EZH2 Interaction/Regulatory Network in Lung Cancer. J. Cancer 2018, 9, 4156–4165. [Google Scholar] [CrossRef]
- Li, B.; Xie, F.; Zheng, F.X.; Jiang, G.S.; Zeng, F.Q.; Xiao, X.Y. Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. Curr. Med. Sci. 2017, 37, 886–890. [Google Scholar] [CrossRef]
- Qu, D.; Yan, B.; Xin, R.; Ma, T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am. J. Cancer Res. 2018, 8, 1387–1402. [Google Scholar] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Zhang, J.; Chang, Y.; Xu, L.; Qin, L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J. Cell Biochem. 2019, 120, 9487–9494. [Google Scholar] [CrossRef]
- Yu, J.; Yang, M.; Zhou, B.; Luo, J.; Zhang, Z.; Zhang, W.; Yan, Z. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin. Sci. 2019, 133, 1487–1503. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Zhao, Z.; Liu, Q.; Wang, X.; Ma, Z.; Li, H. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene 2019, 683, 253–261. [Google Scholar] [CrossRef]
- Gong, Y.; Mao, J.; Wu, D.; Wang, X.; Li, L.; Zhu, L.; Song, R. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018, 18, 116. [Google Scholar] [CrossRef]
- Guan, Z.; Tan, J.; Gao, W.; Li, X.; Yang, Y.; Li, X.; Li, Y.; Wang, Q. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J. Cell Physiol. 2018, 234, 500–508. [Google Scholar] [CrossRef]
- Wei, X.; Zheng, W.; Tian, P.; He, Y.; Liu, H.; Peng, M.; Li, X.; Liu, X. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle 2020, 19, 817–824. [Google Scholar] [CrossRef]
- Liu, B.; Yang, G.; Wang, X.; Liu, J.; Lu, Z.; Wang, Q.; Xu, B.; Liu, Z.; Li, J. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J. Cell Physiol. 2020, 235, 6929–6941. [Google Scholar] [CrossRef]
- Pu, J.; Wang, J.; Li, W.; Lu, Y.; Wu, X.; Long, X.; Luo, C.; Wei, H. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J. Cell Mol. Med. 2020. [Google Scholar] [CrossRef]
- Yang, G.; Wang, X.; Liu, B.; Lu, Z.; Xu, Z.; Xiu, P.; Liu, Z.; Li, J. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle 2019, 18, 976–989. [Google Scholar] [CrossRef]
- Chen, W.; Quan, Y.; Fan, S.; Wang, H.; Liang, J.; Huang, L.; Chen, L.; Liu, Q.; He, P.; Ye, Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020, 475, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Li, D.; Liu, H.; Sun, H.; Liu, Z.; Zhang, L.; Hu, Y. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. J. Cell Physiol. 2019, 234, 2460–2470. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yu, Y.; Huang, Z.; Kong, Y.; Hu, X.; Xiao, W.; Quan, J.; Fan, X. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019, 10, 900. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Dai, Z.H.; Liu, F.C.; Guo, X.G.; Ge, C.M.; Ding, J.; Liu, H.; Yang, F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 2019, 45, 155–167. [Google Scholar] [CrossRef]
- Wang, L.; Long, H.; Zheng, Q.; Bo, X.; Xiao, X.; Li, B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol. Cancer 2019, 18, 119. [Google Scholar] [CrossRef]
- Huang, X.Y.; Huang, Z.L.; Huang, J.; Xu, B.; Huang, X.Y.; Xu, Y.H.; Zhou, J.; Tang, Z.Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 20. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Guo, L.; Deng, Z.; Li, P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann. Hepatol. 2020, 19, 269–279. [Google Scholar] [CrossRef]
- Li, Q.; Pan, X.; Zhu, D.; Deng, Z.; Jiang, R.; Wang, X. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. Hepatology 2019, 70, 1298–1316. [Google Scholar] [CrossRef]
- Hu, Z.Q.; Zhou, S.L.; Li, J.; Zhou, Z.J.; Wang, P.C.; Xin, H.Y.; Mao, L.; Luo, C.B.; Yu, S.Y.; Huang, X.W.; et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology 2020, 72, 906–922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.F.; Gao, C.; Huang, X.Y.; Lu, J.C.; Guo, X.J.; Shi, G.M.; Cai, J.B.; Ke, A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; He, R.; Dang, Y.; Wu, H.; Feng, Z.; Chen, G. The Latest Overview of circRNA in the Progression, Diagnosis, Prognosis, Treatment, and Drug Resistance of Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 608257. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, X.; Liang, C.; Ling, Y.; Yang, X.; Ye, X.; Zhang, H.; Yang, P.; Cui, X.; Ren, Y.; et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology 2020, 71, 130–147. [Google Scholar] [CrossRef]
- Zhai, Z.; Fu, Q.; Liu, C.; Zhang, X.; Jia, P.; Xia, P.; Liu, P.; Liao, S.; Qin, T.; Zhang, H. Emerging Roles Of hsa-circ-0046600 Targeting The miR-640/HIF-1alpha Signalling Pathway In The Progression Of HCC. Onco Targets Ther. 2019, 12, 9291–9302. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Xu, X.; Luo, L.; Zhang, Y.; Luo, L.; Yao, Y.; Xiang, G.; Huang, X.; Wang, G. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle 2019, 18, 2398–2413. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, J.; He, X.; Yan, X.; Wei, J.; Huang, M.; Liu, Y.; Lin, J.; Hu, H.; Liu, L. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J. Cancer 2020, 11, 3318–3326. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Liu, W.; Zou, Y.; Wang, G.; Deng, Y.; Luo, J.; Zhang, Y.; Li, H.; Zhang, Q.; Yang, Y.; et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 2019, 40, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, T.; Ge, S.; Liu, Y.; Bai, M.; Zhu, K.; Fan, Q.; Li, J.; Ning, T.; Tian, F.; et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 2019, 38, 2844–2859. [Google Scholar] [CrossRef]
- Su, Y.; Lv, X.; Yin, W.; Zhou, L.; Hu, Y.; Zhou, A.; Qi, F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging 2019, 11, 8183–8203. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Y.; Xiao, B.; Cai, H.; Liu, M.; Ma, L.; Yin, H.; Wang, F. The circular RNA PVT1/miR-203/HOXD3 pathway promotes the progression of human hepatocellular carcinoma. Biol. Open 2019, 8, bio043687. [Google Scholar] [CrossRef] [PubMed]
- Bu, N.; Dong, Z.; Zhang, L.; Zhu, W.; Wei, F.; Zheng, S. CircPVT1 Regulates Cell Proliferation, Apoptosis and Glycolysis in Hepatocellular Carcinoma via miR-377/TRIM23 Axis. Cancer Manag. Res. 2020, 12, 12945–12956. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Dai, Y.; Guo, X.; Chen, W.; Zhao, J.; Cao, L.; Wu, Z. Silencing Of hsa_circ_0008450 Represses Hepatocellular Carcinoma Progression Through Regulation Of microRNA-214-3p/EZH2 Axis. Cancer Manag. Res. 2019, 11, 9133–9143. [Google Scholar] [CrossRef]
- Zhan, W.; Liao, X.; Chen, Z.; Li, L.; Tian, T.; Yu, L.; Wang, W.; Hu, Q. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J. Cell Physiol. 2020, 235, 1733–1745. [Google Scholar] [CrossRef]
- Li, Z.; Hu, Y.; Zeng, Q.; Wang, H.; Yan, J.; Li, H.; Yu, Z. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. 2019, 19, 211. [Google Scholar] [CrossRef]
- Li, Y.; Zang, H.; Zhang, X.; Huang, G. Exosomal Circ-ZNF652 Promotes Cell Proliferation, Migration, Invasion and Glycolysis in Hepatocellular Carcinoma via miR-29a-3p/GUCD1 Axis. Cancer Manag. Res. 2020, 12, 7739–7751. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Tang, L.; Jiang, H.; Li, X.; Wang, R.; Gao, J.; Li, Q. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J. Cell Biochem. 2019, 120, 11350–11357. [Google Scholar] [CrossRef]
- Wang, W.; Li, Y.; Li, X.; Liu, B.; Han, S.; Li, X.; Zhang, B.; Li, J.; Sun, S. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed. Pharmacother. 2020, 121, 109517. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Qian, Z.; Zheng, W.; Wu, Q.; Chen, Y.; Zhu, G.; Liu, Y.; Bian, Z.; Xu, W.; et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 1091. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Wang, G.; Wang, J.; Li, C.; Zhang, L. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol. Cell Biol. 2019, 97, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gu, H.; Huang, Y.; Peng, Q.; Zhou, R.; Yi, P.; Chen, R.; Huang, Z.; Hu, X.; Huang, Y.; et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle 2018, 17, 2349–2359. [Google Scholar] [CrossRef]
- Bai, N.; Peng, E.; Qiu, X.; Lyu, N.; Zhang, Z.; Tao, Y.; Li, X.; Wang, Z. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J. Exp. Clin. Cancer Res. 2018, 37, 172. [Google Scholar] [CrossRef]
- Cai, H.; Hu, B.; Ji, L.; Ruan, X.; Zheng, Z. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am. J. Transl. Res. 2018, 10, 1690–1702. [Google Scholar]
- Wang, B.; Chen, H.; Zhang, C.; Yang, T.; Zhao, Q.; Yan, Y.; Zhang, Y.; Xu, F. Effects of hsa_circRBM23 on Hepatocellular Carcinoma Cell Viability and Migration as Produced by Regulating miR-138 Expression. Cancer Biother. Radiopharm. 2018, 33, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Li, M.F.; Li, Y.H.; He, Y.H.; Wang, Q.; Zhang, Y.; Li, X.F.; Meng, X.M.; Huang, C.; Li, J. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed. Pharmacother. 2018, 99, 848–858. [Google Scholar] [CrossRef]
- Bai, N.; Peng, E.; Xia, F.; Wang, D.; Li, X.; Li, X. CircABCC2 Regulates Hepatocellular Cancer Progression by Decoying MiR-665. J. Cancer 2019, 10, 3893–3898. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Huang, Z.L.; Zhang, P.B.; Huang, X.Y.; Huang, J.; Wang, H.C.; Xu, B.; Zhou, J.; Tang, Z.Y. CircRNA-100338 Is Associated With mTOR Signaling Pathway and Poor Prognosis in Hepatocellular Carcinoma. Front. Oncol. 2019, 9, 392. [Google Scholar] [CrossRef]
- Ji, C.; Hong, X.; Lan, B.; Lin, Y.; He, Y.; Chen, J.; Liu, X.; Ye, W.; Mo, Z.; She, Z.; et al. Circ_0091581 Promotes the Progression of Hepatocellular Carcinoma Through Targeting miR-591/FOSL2 Axis. Dig. Dis. Sci. 2021, 66, 3074–3085. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Fan, X.; Hu, X.; Fu, X.; Wei, Q.; Zang, Y. circPCNX and Pecanex Promote Hepatocellular Carcinoma Cell Viability by Inhibiting miR-506. Cancer Manag. Res. 2019, 11, 10957–10967. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Y.; Yan, J.; Zeng, Q.; Hu, Y.; Wang, H.; Li, H.; Li, J.; Yu, Z. Circular RNA hsa_circ_0056836 functions an oncogenic gene in hepatocellular carcinoma through modulating miR-766-3p/FOSL2 axis. Aging 2020, 12, 2485–2497. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Dong, G.; Meng, Q.; Lin, S.; Li, X. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J. Cell Biochem. 2020, 121, 4440–4449. [Google Scholar] [CrossRef]
- Liu, M.; Guo, B.; Zhang, G.; Qi, H. Circ_0091579 Knockdown Inhibited HCC Proliferation and Glutamine Metabolism Through miR-1270/YAP1 Axis. Biochem. Genet. 2023. [Google Scholar] [CrossRef]
- Mao, Y.; Ding, Z.; Jiang, M.; Yuan, B.; Zhang, Y.; Zhang, X. Circ_0091579 exerts an oncogenic role in hepatocellular carcinoma via mediating miR-136-5p/TRIM27. Biomed. J. 2022, 45, 883–895. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Y.; Zhang, X.; Zhai, H.; Sun, X.; Li, Y. Circ_0091579 Serves as a Tumor-Promoting Factor in Hepatocellular Carcinoma Through miR-1225-5p/PLCB1 Axis. Dig. Dis. Sci. 2022, 67, 585–597. [Google Scholar] [CrossRef]
- Ding, B.; Fan, W.; Lou, W. hsa_circ_0001955 Enhances In Vitro Proliferation, Migration, and Invasion of HCC Cells through miR-145-5p/NRAS Axis. Mol. Ther. Nucleic Acids 2020, 22, 445–455. [Google Scholar] [CrossRef]
- Yao, Z.; Xu, R.; Yuan, L.; Xu, M.; Zhuang, H.; Li, Y.; Zhang, Y.; Lin, N. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019, 10, 945. [Google Scholar] [CrossRef]
- Li, X.; Lv, J.; Hou, L.; Guo, X. Circ_0001955 Acts as a miR-646 Sponge to Promote the Proliferation, Metastasis and Angiogenesis of Hepatocellular Carcinoma. Dig. Dis. Sci. 2022, 67, 2257–2268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.F.; Wei, C.Y.; Huang, X.Y.; Peng, R.; Yang, X.; Lu, J.C.; Zhang, C.; Gao, C.; Cai, J.B.; Gao, P.T.; et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol. Cancer 2019, 18, 105. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Y.; Wang, Y.; Jin, C. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed. Pharmacother. 2019, 116, 108932. [Google Scholar] [CrossRef]
- Chen, Z.; Zuo, X.; Pu, L.; Zhang, Y.; Han, G.; Zhang, L.; Wu, J.; Wang, X. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 2019, 110, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, Q.; Wang, W.; Yu, J. Mechanism of the Regulatory Effect of Overexpression of circMTO1 on Proliferation and Apoptosis of Hepatoma Cells via miR-9-5p/NOX4 Axis. Cancer Manag. Res. 2020, 12, 3915–3925. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, Z.; Deng, X.; Lu, Y.; Huang, X.; Lin, J.; Lan, X.; Su, Q.; Wang, C. Circular RNA CircITCH (has-circ-0001141) suppresses hepatocellular carcinoma (HCC) progression by sponging miR-184. Cell Cycle 2022, 21, 1557–1577. [Google Scholar] [CrossRef]
- Chen, X.; Li, H.D.; Bu, F.T.; Li, X.F.; Chen, Y.; Zhu, S.; Wang, J.N.; Chen, S.Y.; Sun, Y.Y.; Pan, X.Y.; et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics 2020, 10, 4851–4870. [Google Scholar] [CrossRef]
- Matboli, M.; Hassan, M.K.; Ali, M.A.; Mansour, M.T.; Elsayed, W.; Atteya, R.; Aly, H.S.; Meteini, M.E.; Elghazaly, H.; El-Khamisy, S.; et al. Impact of circ-0000221 in the Pathogenesis of Hepatocellular via Modulation of miR-661-PTPN11 mRNA Axis. Pharmaceutics 2022, 14, 138. [Google Scholar] [CrossRef]
- Shen, H.; Li, H.; Zhou, J. Circular RNA hsa_circ_0032683 inhibits the progression of hepatocellular carcinoma by sponging microRNA-338-5p. Bioengineered 2022, 13, 2321–2335. [Google Scholar] [CrossRef]
- Xu, Z.X.; Li, J.Z.; Li, Q.; Xu, M.Y.; Li, H.Y. CircRNA608-microRNA222-PINK1 axis regulates the mitophagy of hepatic stellate cells in NASH related fibrosis. Biochem. Biophys. Res. Commun. 2022, 610, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Chen, G.F.; Wang, J.C.; Ji, S.H.; Wu, X.W.; Lu, X.J.; Chen, J.L.; Li, J.T. Hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging 2020, 12, 1643–1655. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Liang, H.; Huang, X.; Zeng, Q.; Liu, Y.; Lv, J.; Ming, L. Circular RNA Hsa_circ_0004018 Inhibits Wnt/beta-Catenin Signaling Pathway by Targeting microRNA-626/DKK3 in Hepatocellular Carcinoma. Onco Targets Ther. 2020, 13, 9351–9364. [Google Scholar] [CrossRef]
- van den Bulk, J.; Verdegaal, E.M.; de Miranda, N.F. Cancer immunotherapy: Broadening the scope of targetable tumours. Open Biol. 2018, 8, 180037. [Google Scholar] [CrossRef]
- Feins, S.; Kong, W.; Williams, E.F.; Milone, M.C.; Fraietta, J.A. An introduction to chimeric antigen receptor (CAR) T-cell immunotherapy for human cancer. Am. J. Hematol. 2019, 94, S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fu, M.Y.; Wang, M.N.; Wan, D.D.; Wei, Y.Q.; Wei, X.W. Cancer vaccines as promising immuno-therapeutics: Platforms and current progress. J. Hematol. Oncol. 2022, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, H.L.; Kohlhapp, F.J.; Zloza, A. Oncolytic viruses: A new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662. [Google Scholar] [CrossRef] [PubMed]
- Carlino, M.S.; Larkin, J.; Long, G.V. Immune checkpoint inhibitors in melanoma. Lancet 2021, 398, 1002–1014. [Google Scholar] [CrossRef] [PubMed]
- Koerner, J.; Horvath, D.; Herrmann, V.L.; MacKerracher, A.; Gander, B.; Yagita, H.; Rohayem, J.; Groettrup, M. PLGA-particle vaccine carrying TLR3/RIG-I ligand Riboxxim synergizes with immune checkpoint blockade for effective anti-cancer immunotherapy. Nat. Commun. 2021, 12, 2935. [Google Scholar] [CrossRef]
- Ahmed Youness, R.; Amr Assal, R.; Mohamed Ezzat, S.; Zakaria Gad, M.; Abdel Motaal, A. A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Nat. Prod. Res. 2020, 34, 1475–1480. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, Z.B.; Lai, W.F.; Cui, L.; Zhu, X. How to overcome the side effects of tumor immunotherapy. Biomed. Pharmacother. 2020, 130, 110639. [Google Scholar] [CrossRef]
- Galli, F.; Aguilera, J.V.; Palermo, B.; Markovic, S.N.; Nistico, P.; Signore, A. Relevance of immune cell and tumor microenvironment imaging in the new era of immunotherapy. J. Exp. Clin. Cancer Res. 2020, 39, 89. [Google Scholar] [CrossRef] [PubMed]
- Mandlik, D.S.; Mandlik, S.K.; Choudhary, H.B. Immunotherapy for hepatocellular carcinoma: Current status and future perspectives. World J. Gastroenterol. 2023, 29, 1054–1075. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Neves, A.F.; Dias-Oliveira, J.D.; Araujo, T.G.; Marangoni, K.; Goulart, L.R. Prostate cancer antigen 3 (PCA3) RNA detection in blood and tissue samples for prostate cancer diagnosis. Clin. Chem. Lab. Med. 2013, 51, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, R.M.; Elkhouly, A.M.; Soliman, R.A.; El Mechawy, N.; El Sebaei, A.; Motaal, A.A.; El-Askary, H.; Youness, R.A.; Assal, R.A. Hindering the Synchronization Between miR-486-5p and H19 lncRNA by Hesperetin Halts Breast Cancer Aggressiveness Through Tuning ICAM-1. Anticancer. Agents Med. Chem. 2022, 22, 586–595. [Google Scholar] [CrossRef]
- Abdel-Latif, M.; Riad, A.; Soliman, R.A.; Elkhouly, A.M.; Nafae, H.; Gad, M.Z.; Motaal, A.A.; Youness, R.A. MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitin glycoside alters immunogenic and oncogenic profiles of breast cancer. Mol. Cell Biochem. 2022, 477, 1281–1293. [Google Scholar] [CrossRef]
- El Kilany, F.H.; Youness, R.A.; Assal, R.A.; Gad, M.Z. miR-744/eNOS/NO axis: A novel target to halt triple negative breast cancer progression. Breast Dis. 2021, 40, 161–169. [Google Scholar] [CrossRef]
- Dawoud, A.; Youness, R.A.; Nafea, H.; Manie, T.; Abdel-Kader, R.M.; Gad, M. 26P 3MST: A potential workhorse in H2S signaling trimmed by microRNA-548 in breast cancer. ESMO Open 2023, 8, 100992. [Google Scholar] [CrossRef]
- Soliman, R.; Youness, R.A.; El-Shazly, M.; Handoussa, H.; Gad, M. Regulatory interacting network between the immunomodulatory non-coding RNAs: miR-17-5p, MALAT1 and H19 lncRNAs in modulating the tumour microenvironment in TNBC. Ann. Oncol. 2019, 30, xi57. [Google Scholar] [CrossRef]
- Awad, A.R.; Youness, R.A.; Ibrahim, M.; Motaal, A.A.; El-Askary, H.I.; Assal, R.A.; Gad, M.Z. An acetylated derivative of vitexin halts MDA-MB-231 cellular progression and improves its immunogenic profile through tuning miR- 20a-MICA/B axis. Nat. Prod. Res. 2021, 35, 3126–3130. [Google Scholar] [CrossRef]
- Arita, T.; Ichikawa, D.; Konishi, H.; Komatsu, S.; Shiozaki, A.; Shoda, K.; Kawaguchi, T.; Hirajima, S.; Nagata, H.; Kubota, T.; et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer. Res. 2013, 33, 3185–3193. [Google Scholar] [PubMed]
- Yu, L.L.; Xiao, Q.; Yu, B.; Lv, Q.L.; Liu, Z.Q.; Yin, J.Y. CircRNAs in tumor immunity and immunotherapy: Perspectives from innate and adaptive immunity. Cancer Lett. 2023, 564, 216219. [Google Scholar] [CrossRef]
- Cheng, H.; Sun, G.; Chen, H.; Li, Y.; Han, Z.; Li, Y.; Zhang, P.; Yang, L.; Li, Y. Trends in the treatment of advanced hepatocellular carcinoma: Immune checkpoint blockade immunotherapy and related combination therapies. Am. J. Cancer Res. 2019, 9, 1536–1545. [Google Scholar] [PubMed]
- Huang, X.Y.; Zhang, P.F.; Wei, C.Y.; Peng, R.; Lu, J.C.; Gao, C.; Cai, J.B.; Yang, X.; Fan, J.; Ke, A.W.; et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol. Cancer 2020, 19, 92. [Google Scholar] [CrossRef]
- Chen, Z.Q.; Zuo, X.L.; Cai, J.; Zhang, Y.; Han, G.Y.; Zhang, L.; Ding, W.Z.; Wu, J.D.; Wang, X.H. Hypoxia-associated circPRDM4 promotes immune escape via HIF-1alpha regulation of PD-L1 in hepatocellular carcinoma. Exp. Hematol. Oncol. 2023, 12, 17. [Google Scholar] [CrossRef]
- Cai, J.; Chen, Z.; Zhang, Y.; Wang, J.; Zhang, Z.; Wu, J.; Mao, J.; Zuo, X. CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol. Ther. Oncolytics 2022, 24, 755–771. [Google Scholar] [CrossRef]
- Lu, J.C.; Zhang, P.F.; Huang, X.Y.; Guo, X.J.; Gao, C.; Zeng, H.Y.; Zheng, Y.M.; Wang, S.W.; Cai, J.B.; Sun, Q.M.; et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J. Hematol. Oncol. 2021, 14, 200. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, G.; Zhao, Y.; Gao, H.; Li, L.; Yin, Y.; Jiang, J.; Wang, L.; Mang, Y.; Gao, Y.; et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol. Cancer 2023, 22, 55. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017, 27, 109–118. [Google Scholar] [CrossRef]
- Zheng, C.; Zheng, L.; Yoo, J.K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356.e16. [Google Scholar] [CrossRef]
- Huang, M.; Huang, X.; Huang, N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022, 113, 1968–1983. [Google Scholar] [CrossRef] [PubMed]
- Huntington, N.D.; Cursons, J.; Rautela, J. The cancer-natural killer cell immunity cycle. Nat. Rev. Cancer 2020, 20, 437–454. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, C.; Zhang, B.; Yu, H.; Yu, Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol. Lett. 2019, 17, 388–397. [Google Scholar] [CrossRef] [PubMed]


| Circular RNA | Class | Molecular Targets | In Vitro/In Vivo/Ex Vivo Model | References |
|---|---|---|---|---|
| SCD-circRNA2 | Oncogenic | MAPK1, RBM3 | Huh7 HepG2 HCT-15 NCI-N87 | [121] |
| circRHOT1 | Oncogenic | NR2F6 | HCC Tissues | [122] |
| circ-100338 | Oncogenic | MMP2, MMP9 | Hep3B HLE Huh7 BEL7402 SMCC7721 MHCC97L MHCC97H HCCLM3 HCCLM6 | [123] |
| circ-0000092 | Oncogenic | miR-338-3p | Hep3B LM3 MHCC97L SK-hep1 HepG2 | [116] |
| circPRMT5 | Oncogenic | miR-188-5p/HK2 axis | HCC tissues HCCLM3 SNU-387 | [124] |
| circMAT2B | Oncogenic | PKM2 | HepG2 Huh7 SMMC-772 MHCC-97L MHCC-97H | [125] |
| circASAP1 | Oncogenic | MAPK1 | MHCC97L MHCC97H HCCLM3 | [126] |
| circβ-catenin | Oncogenic | β-catenin | Huh7 | [97] |
| circUHRF1 | Oncogenic | UHRF1 | HepG2 HCCLM3 SMMC-7721 Huh 7 PLC/PRF/5 Hep3B | [127] |
| circ-CDYL | Oncogenic | PI3K-AKT-MTORC1/β-catenin and NOTCH2 | HCCLM SMMC7721 | [128,129] |
| circ-0046600 | Oncogenic | HIF-1α | HepG2 SK-HEP-1 | [130] |
| hsa_circ_0101432 | Oncogenic | MAPK1 | Huh-7 SK-HEP-1 HepG2 HLE | [131] |
| circMAN2B2 | Oncogenic | MAPK1 | HL-7702 | [132] |
| circPTGR1 | Oncogenic | MET | HepG2 97L LM3 | [133] |
| circ-DB | Oncogenic | miR-34a, and USP7 | HepG2 Hepa 1-6 3T3L1 | [134] |
| circRNA Cdr1as | Oncogenic | AFP | SMMC-7721 Bel-7402 HepG2 Hep3B Huh-7 HB611 | [135] |
| circRNA PVT1 | Oncogenic | miR-203/HOXD3 pathway | SMMC-7721 Huh-7 | [136] |
| circPVT1 | Oncogenic | TRIM23/miR-377 axis | SNU-387 Huh-7 | [137] |
| hsa_circ_0008450 | Oncogenic | EZH2 | SMMC7721 Sk-Hep-1 HepG2 Huh-7 HCCLM3 | [138] |
| circ_0008450 | Oncogenic | miR-548 | HepG2 Huh-7, SMMC7721 Sk-Hep-1 HCCLM3 | [109] |
| hsa_circRNA_103809 | Oncogenic | miR-377-3p/FGFR1/MAPK1 axis | MHCC97L Huh7 SK-HEP-1 Hep3B HCCLM3 | [139] |
| circRNA-104718 | Oncogenic | miR-218-5p/TXNDC5 | HCC nude mice model | [110] |
| circMYLK | Oncogenic | miR-362-3p/Rab23 | Huh7 Hep3B | [140] |
| circ-ZNF652 | Oncogenic | miR-29a-3p/GUCD1 Axis | SNU-387 Huh-7 | [141] |
| circ_0000267 | Oncogenic | miR-646 | HepG2 Huh-7 SMMC7721 Sk-Hep-1 HCCLM3 | [142] |
| circ-FOXP1 | Oncogenic | miR-875-3p, miR-421, SOX9 factor | SNU-387 HepG2 Hep3B Huh7 SMMC-7721 HCCLM3 | [143] |
| circRNA_104075 | Oncogenic | YAP-dependent tumorigenesis through regulating HNF4a | Bel-7402 SMMC-7721 Huh7 HepG2 Hep1 Bel-7404 THLE-3 HL-7702 | [144] |
| hsa_circ_101280 | Oncogenic | miR-375/JAK2 | HepG2 SNU-398 | [145] |
| circRNA-101368 | Oncogenic | HMGB1/RAGE | HCCLM3 HepG2 | [146] |
| circ-ZEB1.33 | Oncogenic | miR-200a-3p-CDK6 | 97H Huh7 HepG2 SNU423 SNU475 L02 | [112] |
| circFBLIM1 | Oncogenic | miR-346 | HCC tissues HCC mouse model | [147] |
| hsa_circ_0103809 | Oncogenic | miR-490-5p/SOX2 signaling pathway | MHCC97H HepG2 Huh7 SMMC7721 SK-Hep1 | [148] |
| hsa_circ_0016788 | Oncogenic | miR-486/CDK4 | HepG2 Hep3B Huh7 HCCLM3 MHCC97L | [113] |
| hsa_circRBM23 | Oncogenic | miR-138 | HCC tissues HepG2 Huh7 Bel-7402 | [149] |
| hsa_circ_0005075 | Oncogenic | miR-431 | SMMC-7721 | [150] |
| circABCC2 | Oncogenic | miR-665 | HepG2 Bel-7402 MHCC97H | [151] |
| hsa_circ_100338 | Oncogenic | MTOR signaling pathway | SMMC7721 Bel-7402 Hep3B | [152] |
| circ_0091581 | Oncogenic | miR-591/FOSL2 axis | THLE-2 | [153] |
| circPCNX | Oncogenic | miR-506 | HL-7702 SMMC-7721 HuH-7 Hep3B HepG2 | [154] |
| hsa_circ_0056836 | Oncogenic | miR-766-3p/FOSL2 axis | Huh7 HepG2 SNU449 SK-HEP-1 | [155] |
| circ- HOMER1 | Oncogenic | miR-1322 on CXCL6 | Sk-Hep-1 SMMC7721 HCCLM3 Huh-7 HepG2 | [156] |
| circ_0091579 | Oncogenic | miR-136-5p/TRIM27 miR-1270/YAP1 miR-1225/PLCB1 | HCCLM3 MHCC97H Huh-7 | [157,158,159] |
| circ_0001955 | Oncogenic | miR-516a-5p miR-646 miR-145-5p/NRAS | Huh-7 HepG2 SMMC-7721 Bel-7402 Hep-3B | [160,161,162] |
| circTRIM33-12 | Tumor suppressor | miR-191 | HCC tissues MHCC97-L MHCC97-H LM3 | [163] |
| circHIAT1 | Tumor suppressor | PTEN | Hep3B SMMC-7721 HepG2 LM3 | [164] |
| circLARP4 | Tumor suppressor | miR-761/RUNX3/p53/CDKN1A pathway | Huh7 Hep3B SMMC7721 HepG2 | [165] |
| circMTO1 | Tumor suppressor | miR-9-5p/NOX4 axis | HepG2 Hep3B | [166] |
| circITCH | Tumor suppressor | miR-184 | Huh7 HCCLM3 SMMC-7721 MHCC97H HepG2 | [167] |
| circFBXW4 | Tumor suppressor | miR-18b-3p/FBXW7 axis | LX-2 | [168] |
| mmu_circ_34116 | Tumor suppressor | miR-661/PTPN11 | HepG2, SNU449 | [169] |
| hsa_circ_0007874/cMTO1 | Tumor suppressor | miR-338-5p | HCCLM3 MHCC97-L Hep3B SMMC-7721 Huh7 Bel-7402 MHCC97-H | [170] |
| circ608 | Tumor suppressor | miR-222/PINK1 | Primary hepatic stellate cells (PHSCs) from C57BL/6 mice | [171] |
| hsa_circ_0070963 | Tumor suppressor | miR-223-3p LEMD3 | LX2 | [172] |
| hsa_circ_0004018 | Tumor suppressor | miR-626/DKK3 | Huh7 Bel7402 SNU182 Hep3B SNU449 | [173] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abaza, T.; El-Aziz, M.K.A.; Daniel, K.A.; Karousi, P.; Papatsirou, M.; Fahmy, S.A.; Hamdy, N.M.; Kontos, C.K.; Youness, R.A. Emerging Role of Circular RNAs in Hepatocellular Carcinoma Immunotherapy. Int. J. Mol. Sci. 2023, 24, 16484. https://doi.org/10.3390/ijms242216484
Abaza T, El-Aziz MKA, Daniel KA, Karousi P, Papatsirou M, Fahmy SA, Hamdy NM, Kontos CK, Youness RA. Emerging Role of Circular RNAs in Hepatocellular Carcinoma Immunotherapy. International Journal of Molecular Sciences. 2023; 24(22):16484. https://doi.org/10.3390/ijms242216484
Chicago/Turabian StyleAbaza, Tasneem, Mostafa K. Abd El-Aziz, Kerolos Ashraf Daniel, Paraskevi Karousi, Maria Papatsirou, Sherif Ashraf Fahmy, Nadia M. Hamdy, Christos K. Kontos, and Rana A. Youness. 2023. "Emerging Role of Circular RNAs in Hepatocellular Carcinoma Immunotherapy" International Journal of Molecular Sciences 24, no. 22: 16484. https://doi.org/10.3390/ijms242216484
APA StyleAbaza, T., El-Aziz, M. K. A., Daniel, K. A., Karousi, P., Papatsirou, M., Fahmy, S. A., Hamdy, N. M., Kontos, C. K., & Youness, R. A. (2023). Emerging Role of Circular RNAs in Hepatocellular Carcinoma Immunotherapy. International Journal of Molecular Sciences, 24(22), 16484. https://doi.org/10.3390/ijms242216484













