Genome-Wide Identification and Expression Analysis of Malate Dehydrogenase Gene Family in Sweet Potato and Its Two Diploid Relatives
Abstract
:1. Introduction
2. Results
2.1. Identification of MDHs in Sweet Potato and Its Two Diploid Wild Relatives
2.2. Collinearity Analysis of MDH Genes and Ka/Ks Analysis
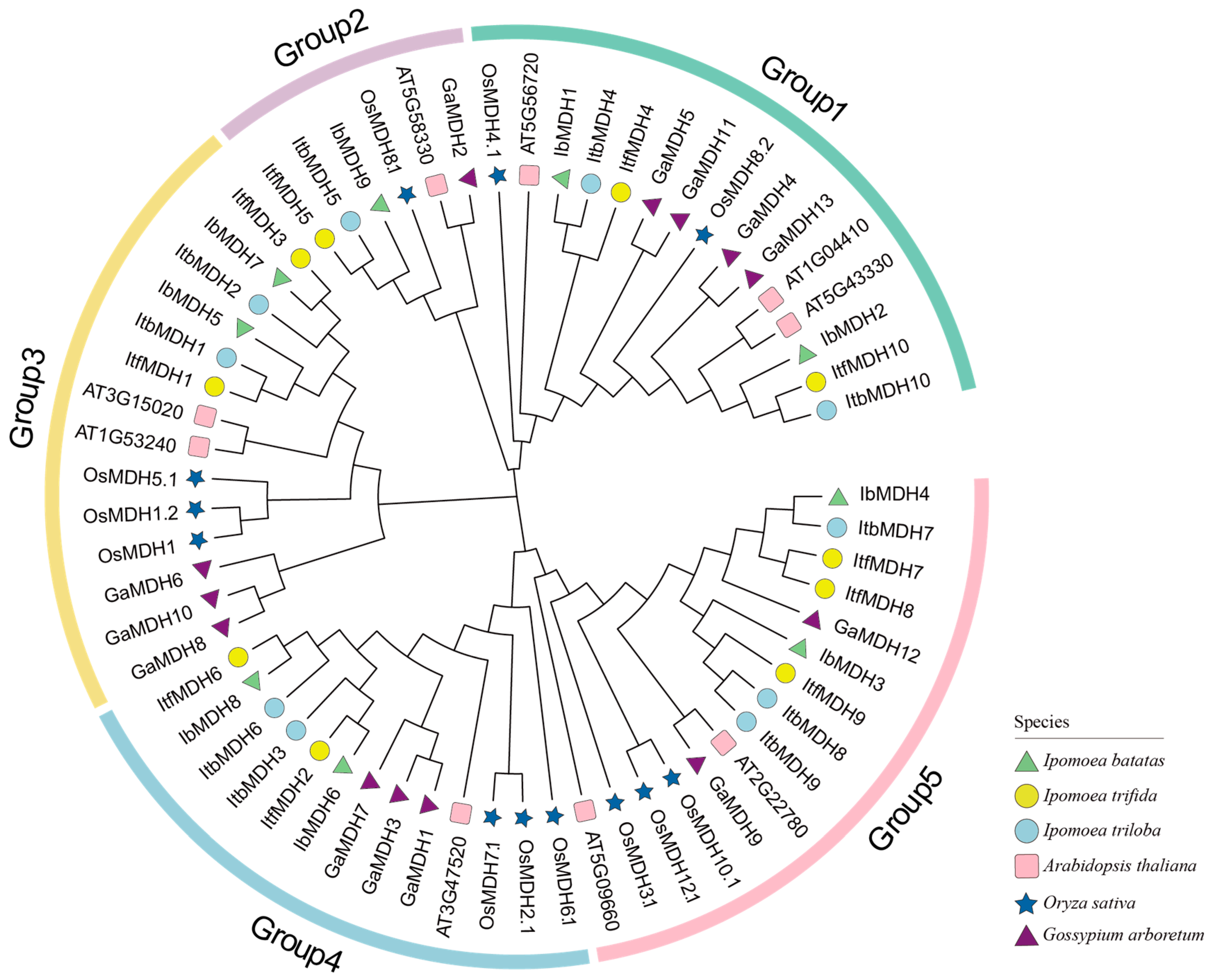
2.3. Phylogenetic Relationship Analysis of MDHs in Sweet Potato and Its Two Diploid Wild Relatives
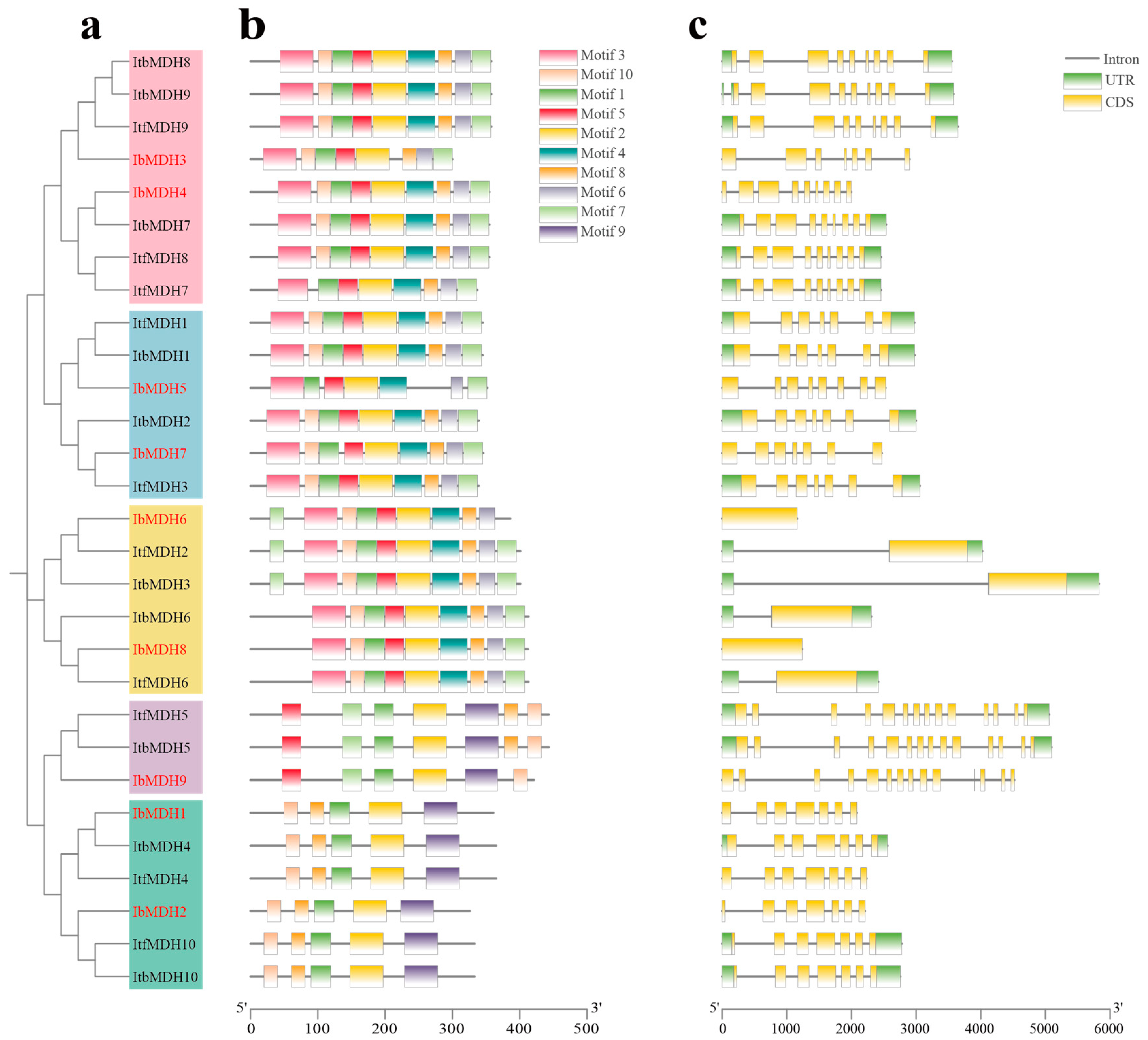
2.4. Conserved Motif and Exon-Intron Structure Analysis of MDHs in Sweet Potato and Its Two Diploid Wild Relatives
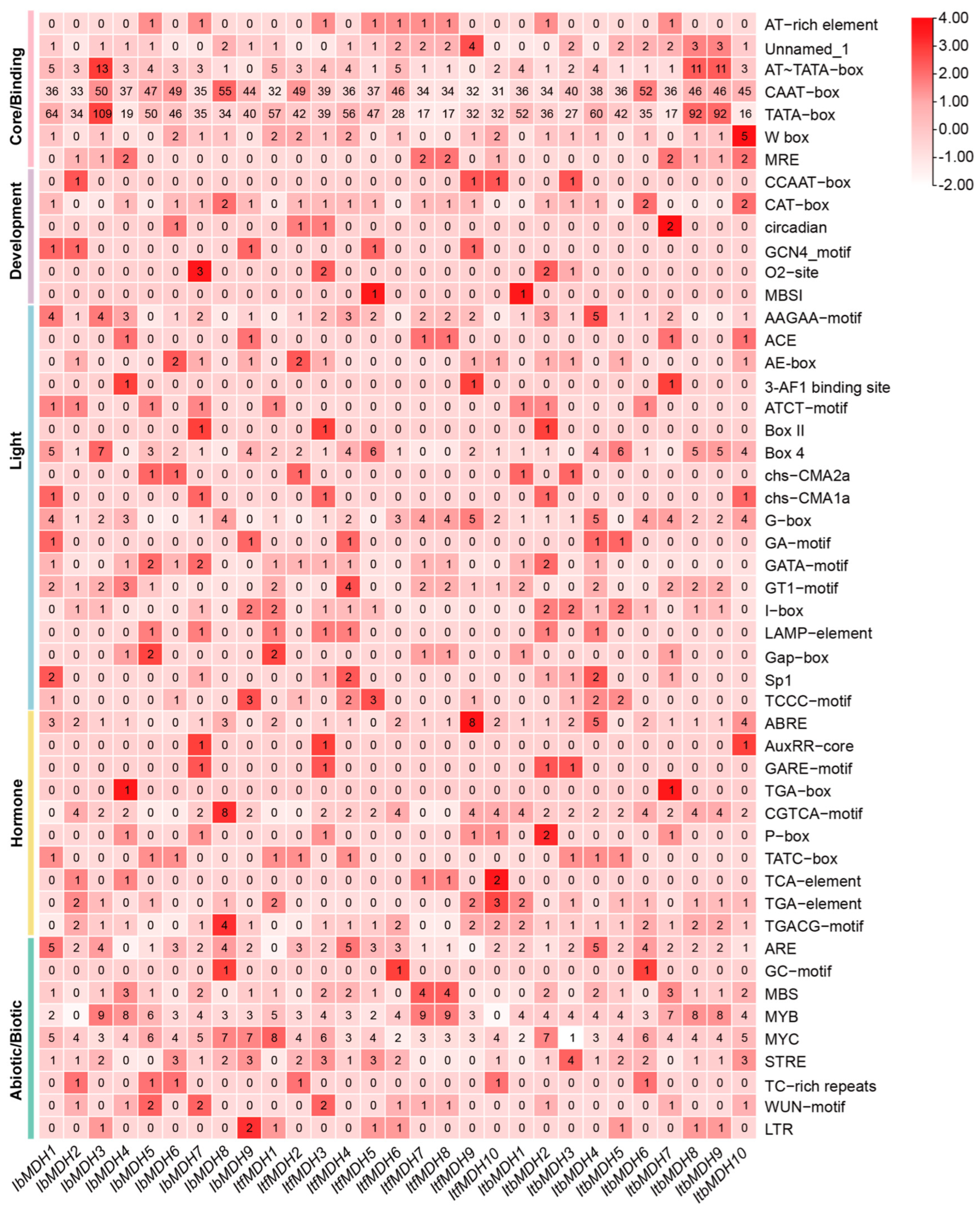
2.5. Putative Cis-Regulatory Element Analysis in the Promoter of MDHs in Sweet Potato and Its Two Diploid Wild Relatives
2.6. Expression Analysis of MDHs in Sweet Potato and Its Two Diploid Wild Relatives
2.6.1. Expression Analysis in Various Tissues
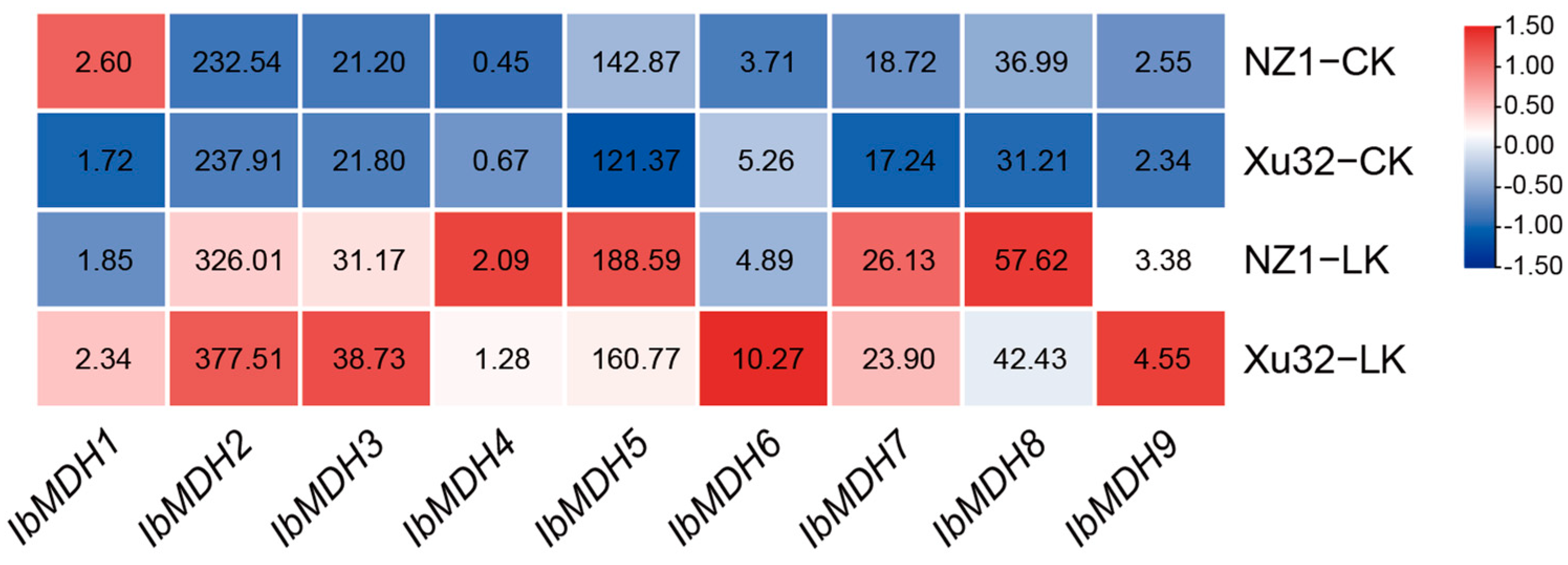
2.6.2. Expression Analysis under Potassium Deficiency in Sweet Potato
2.6.3. Expression Analysis under Hormone Stress
2.6.4. Expression Analysis under Cold Stress
2.6.5. Expression Analysis under Heat Stress
2.6.6. Expression Analysis under Salt and Drought Stresses
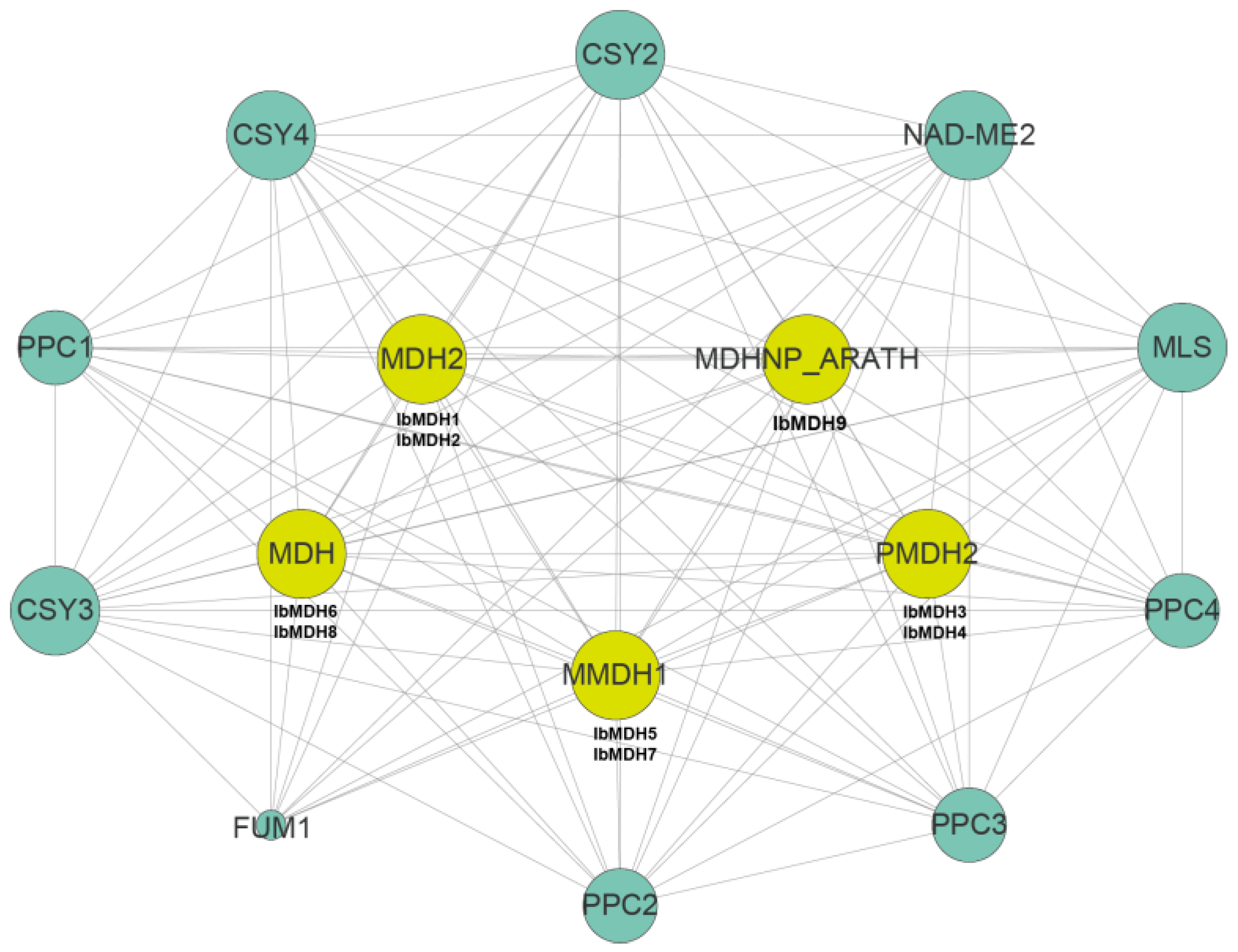
2.7. Protein Interactions Network of MDHs in Sweet Potato
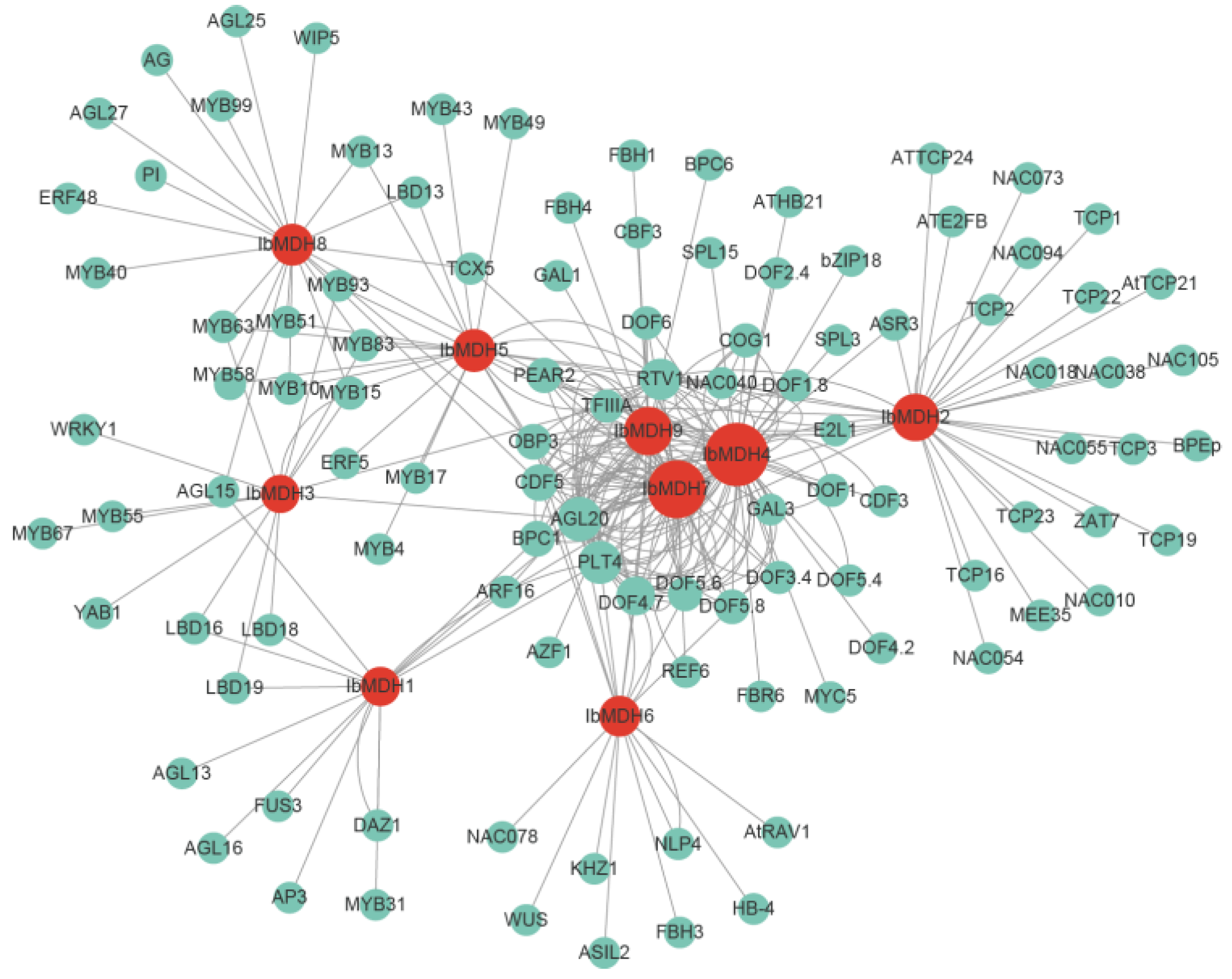
2.8. Transcript Factors Network of Sweet Potato MDH Genes
3. Discussion
3.1. Evolution of MDH Genes in Sweet Potato and Its Two Diploid Wild Relatives
3.2. Distinct Roles of MDH Genes in Biological Processes
4. Materials and Methods
4.1. Identification and Physicochemical Properties of MDH Family Members
4.2. Chromosome Distribution and Collinearity Analysis of MDH Genes
4.3. Synteny Analysis and Calculation of Ka/Ks Values of MDH Gene Homologous Pair
4.4. Phylogenetic Analysis of MDH Genes
4.5. Conserved Motifs and Gene Structure Analysis of MDH Genes
4.6. Cis-Regulatory Element Analysis of MDH Genes
4.7. Protein–Protein Interactions Analysis of Sweet Potato MDH Proteins
4.8. Transcription Factors Regulatory Network Analysis of Sweet Potato MDH Genes
4.9. Transcriptome Analysis
4.10. qRT-PCR Analysis of Sweet Potato MDH Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heber, U. Metabolite exchange between chloroplasts and cytoplasm. Annu. Rev. Plant Physiol. 1974, 25, 393–421. [Google Scholar] [CrossRef]
- Carr, P.D.; Verger, D.; Ashton, A.R.; Ollis, D.L. Chloroplast NADP-malate dehydrogenase: Structural basis of light-dependent regulation of activity by thiol oxidation and reduction. Structure 1999, 7, 461–475. [Google Scholar] [CrossRef]
- Tomaz, T.; Bagard, M.; Pracharoenwattana, I.; Lindén, P.; Lee, C.P.; Carroll, A.J.; Ströher, E.; Smith, S.M.; Gardeström, P.; Millar, A.H. Mitochondrial Malate Dehydrogenase Lowers Leaf Respiration and Alters Photorespiration and Plant Growth in Arabidopsis. Plant Physiol. 2010, 154, 1143–1157. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fu, Z.; Zhang, H.; Tian, R.; Yang, H.; Sun, C.; Wang, L.; Zhang, W.; Guo, Z.; Zhang, X.; et al. Cytosolic malate dehydrogenase 4 modulates cellular energetics and storage reserve accumulation in maize endosperm. Plant Biotechnol. J. 2020, 18, 2420–2435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, C.; Jia, R.; Yang, N.; Jin, L.; Zhu, L.; Ma, B.; Yao, Y.; Ma, F.; Li, M. Malate metabolism mediated by the cytoplasmic malate dehydrogenase gene MdcyMDH affects sucrose synthesis in apple fruit. Hortic. Res. 2022, 9, uhac194. [Google Scholar] [CrossRef]
- Lindén, P.; Keech, O.; Stenlund, H.; Gardeström, P.; Moritz, T. Reduced mitochondrial malate dehydrogenase activity has a strong effect on photorespiratory metabolism as revealed by 13 C labelling. J. Exp. Bot. 2016, 67, 3123–3135. [Google Scholar] [CrossRef]
- Selinski, J.; Scheibe, R. Malate valves: Old shuttles with new perspectives. Plant Biol. 2018, 21 (Suppl. S1), 21–30. [Google Scholar] [CrossRef]
- Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant. 2004, 120, 21–26. [Google Scholar] [CrossRef]
- Taniguchi, M.; Miyake, H. Redox-shuttling between chloroplast and cytosol: Integration of intra-chloroplast and extra-chloroplast metabolism. Curr. Opin. Plant Biol. 2012, 15, 252–260. [Google Scholar] [CrossRef]
- Schreier, T.B.; Cléry, A.; Schläfli, M.; Galbier, F.; Stadler, M.; Demarsy, E.; Albertini, D.; Maier, B.A.; Kessler, F.; Hörtensteiner, S.; et al. Plastidial NAD-Dependent Malate Dehydrogenase: A Moonlighting Protein Involved in Early Chloroplast Development through Its Interaction with an FtsH12-FtsHi Protease Complex. Plant Cell 2018, 30, 1745–1769. [Google Scholar] [CrossRef]
- Goward, C.R.; Nicholls, D.J. Malate dehydrogenase: A model for structure, evolution, and catalysis. Protein Sci. 1994, 3, 1883–1888. [Google Scholar] [CrossRef]
- Beeler, S.; Liu, H.; Stadler, M.; Schreier, T.; Eicke, S.; Lue, W.; Truernit, E.; Zeeman, S.C.; Chen, J.; Kötting, O. Plastidial NAD-Dependent Malate Dehydrogenase Is Critical for Embryo Development and Heterotrophic Metabolism in Arabidopsis. Plant Physiol. 2014, 164, 1175–1190. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Sun, X.; Yuan, J.; Zhao, Z.; Gao, J.; Wen, X.; Tang, F.; Kang, M.; Abliz, B.; et al. Genome-Wide Identification of MDH Family Genes and Their Association with Salt Tolerance in Rice. Plants 2022, 11, 1498. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Tang, K.; Liu, J. Comparative Genome-Wide Analysis of the Malate Dehydrogenase Gene Families in Cotton. PLoS ONE 2016, 11, e0166341. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Chen, Z.; Xie, B.; Guo, Q.; Chen, M.; Liang, C.; Bai, Z.; Wang, X.; Wang, H.; Liao, H.; et al. A phosphate starvation responsive malate dehydrogenase, GmMDH12 mediates malate synthesis and nodule size in soybean (Glycine max). Environ. Exp. Bot. 2021, 189, 104560. [Google Scholar] [CrossRef]
- Imran, M.; Munir, M.Z.; Ialhi, S.; Abbas, F.; Younus, M.; Ahmad, S.; Naeem, M.K.; Waseem, M.; Iqbal, A.; Gul, S.; et al. Identification and Characterization of Malate Dehydrogenases in Tomato (Solanum lycopersicum L.). Int. J. Mol. Sci. 2022, 23, 10028. [Google Scholar] [CrossRef] [PubMed]
- Yıldız, S.; Okay, A.; Büyük, İ. Defining the roles of PvMDH genes in response to salt stress and detailed characterization of the gene family. J. Plant Biochem. Biotechnol. 2022, 31, 380–393. [Google Scholar] [CrossRef]
- Ma, B.; Yuan, Y.; Gao, M.; Xing, L.; Li, C.; Li, M.; Ma, F. Genome-wide Identification, Classification, Molecular Evolution and Expression Analysis of Malate Dehydrogenases in Apple. Int. J. Mol. Sci. 2018, 19, 3312. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, J.; Zhang, C.; Wang, S.; Yang, M. Genome-wide investigation of malate dehydrogenase gene family in poplar (Populus trichocarpa) and their expression analysis under salt stress. Acta Physiol. Plant. 2021, 43, 28. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, W.; Yang, Q.; Zhang, Y.; Ma, X.; Li, M. Genome-Wide Characterization and Gene Expression Analyses of Malate Dehydrogenase (MDH) Genes in Low-Phosphorus Stress Tolerance of Chinese Fir (Cunninghamia lanceolata). Int. J. Mol. Sci. 2023, 24, 4414. [Google Scholar] [CrossRef]
- Sew, Y.S.; Ströher, E.; Fenske, R.; Millar, A.H. Loss of Mitochondrial Malate Dehydrogenase Activity Alters Seed Metabolism Impairing Seed Maturation and Post-Germination Growth in Arabidopsis. Plant Physiol. 2016, 171, 849–863. [Google Scholar] [CrossRef]
- Selinski, J.; König, N.; Wellmeyer, B.; Hanke, G.T.; Linke, V.; Neuhaus, H.E.; Scheibe, R. The Plastid-Localized NAD-Dependent Malate Dehydrogenase Is Crucial for Energy Homeostasis in Developing Arabidopsis thaliana Seeds. Mol. Plant 2014, 7, 170–186. [Google Scholar] [CrossRef]
- Cousins, A.B.; Pracharoenwattana, I.; Zhou, W.; Smith, S.M.; Badger, M.R. Peroxisomal Malate Dehydrogenase Is Not Essential for Photorespiration in Arabidopsis but Its Absence Causes an Increase in the Stoichiometry of Photorespiratory CO2 Release. Plant Physiol. 2008, 148, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Liszka, A.; Schimpf, R.; Cartuche Zaruma, K.I.; Buhr, A.; Seidel, T.; Walter, S.; Knuesting, J.; Dreyer, A.; Dietz, K.; Scheibe, R.; et al. Three cytosolic NAD-malate dehydrogenase isoforms of Arabidopsis thaliana: On the crossroad between energy fluxes and redox signaling. Biochem. J. 2020, 477, 3673–3693. [Google Scholar] [CrossRef] [PubMed]
- Heyno, E.; Innocenti, G.; Lemaire, S.D.; Issakidis-Bourguet, E.; Krieger-Liszkay, A. Putative role of the malate valve enzyme NADP–malate dehydrogenase in H2O2 signalling in Arabidopsis. Philos. Trans. R. Soc. B.-Biol. Sci. 2014, 369, 20130228. [Google Scholar] [CrossRef] [PubMed]
- Hebbelmann, I.; Selinski, J.; Wehmeyer, C.; Goss, T.; Voss, I.; Mulo, P.; Kangasjärvi, S.; Aro, E.; Oelze, M.; Dietz, K.; et al. Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J. Exp. Bot. 2012, 63, 1445–1459. [Google Scholar] [CrossRef]
- Huang, J.; Niazi, A.K.; Young, D.; Rosado, L.A.; Vertommen, D.; Bodra, N.; Abdelgawwad, M.R.; Vignols, F.; Wei, B.; Wahni, K.; et al. Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis. J. Exp. Bot. 2018, 69, 3491–3505. [Google Scholar] [CrossRef]
- Guo, Y.; Song, Y.; Zheng, H.; Zhang, Y.; Guo, J.; Sui, N. NADP-Malate Dehydrogenase of Sweet Sorghum Improves Salt Tolerance of Arabidopsis thaliana. J. Agric. Food Chem. 2018, 66, 5992–6002. [Google Scholar] [CrossRef]
- Kandoi, D.; Mohanty, S.; Tripathy, B.C. Overexpression of plastidic maize NADP-malate dehydrogenase (ZmNADP-MDH) in Arabidopsis thaliana confers tolerance to salt stress. Protoplasma 2018, 255, 547–563. [Google Scholar] [CrossRef]
- Wang, Q.J.; Sun, H.; Dong, Q.L.; Sun, T.Y.; Jin, Z.X.; Hao, Y.J.; Yao, Y.X. The enhancement of tolerance to salt and cold stresses by modifying the redox state and salicylic acid content via the cytosolic malate dehydrogenase gene in transgenic apple plants. Plant Biotechnol. J. 2016, 14, 1986–1997. [Google Scholar] [CrossRef]
- Yao, Y.; Dong, Q.; Zhai, H.; You, C.; Hao, Y. The functions of an apple cytosolic malate dehydrogenase gene in growth and tolerance to cold and salt stresses. Plant Physiol. Biochem. 2011, 49, 257–264. [Google Scholar] [CrossRef]
- Shi, Y.; Feng, J.; Wang, L.; Liu, Y.; He, D.; Sun, Y.; Luo, Y.; Jin, C.; Zhang, Y. OsMDH12: A Peroxisomal Malate Dehydrogenase Regulating Tiller Number and Salt Tolerance in Rice. Plants 2023, 12, 3558. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.A.; Li, Q.; Ge, X.Y.; Yang, C.L.; Luo, X.L.; Zhang, A.H.; Xiao, J.L.; Tian, Y.C.; Xia, G.X.; Chen, X.Y.; et al. The mitochondrial malate dehydrogenase 1 gene GhmMDH1 is involved in plant and root growth under phosphorus deficiency conditions in cotton. Sci. Rep. 2015, 5, 10343. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, M.; Temple, S.J.; Allan, D.L.; Vance, C.P.; Samac, D.A. Overexpression of Malate Dehydrogenase in Transgenic Alfalfa Enhances Organic Acid Synthesis and Confers Tolerance to Aluminum. Plant Physiol. 2001, 127, 1836–1844. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, Y.; Yi, Q.; Li, K.; Yu, Y.; Chen, L. Overexpression of malate dehydrogenase in transgenic tobacco leaves: Enhanced malate synthesis and augmented Al-resistance. Acta Physiol. Plant. 2010, 32, 1209–1220. [Google Scholar] [CrossRef]
- Wang, S.; Nie, S.; Zhu, F. Chemical constituents and health effects of sweet potato. Food Res. Int. 2016, 89, 90–116. [Google Scholar] [CrossRef]
- Yang, J.; Moeinzadeh, M.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.; Zheng, J.; Sun, Z.; Fan, W.; et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef]
- Liu, Q. Improvement for agronomically important traits by gene engineering in sweetpotato. Breed. Sci. 2017, 67, 15–26. [Google Scholar] [CrossRef]
- Wu, S.; Lau, K.H.; Cao, Q.; Hamilton, J.P.; Sun, H.; Zhou, C.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.; et al. Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, Q.; Liu, S.; Liu, Z.; Bian, X.; Yu, T. De novo transcriptome sequencing and gene expression profiling of sweetpotato leaves during low temperature stress. Plant Biotechnol. Rep. 2023, 1–14. [Google Scholar] [CrossRef]
- Dai, Z.; Yan, P.; He, S.; Jia, L.; Wang, Y.; Liu, Q.; Zhai, H.; Zhao, N.; Gao, S.; Zhang, H. Genome-Wide Identification and Expression Analysis of SWEET Family Genes in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2022, 23, 15848. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhang, H.; Gao, S.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. Genome-Wide Identification and Expression Analysis of the Sucrose Synthase Gene Family in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2023, 24, 12493. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Gao, X.; Dong, Z.; Yi, J.; An, L. Improved phosphorus acquisition by tobacco through transgenic expression of mitochondrial malate dehydrogenase from Penicillium oxalicum. Plant Cell Rep. 2012, 31, 49–56. [Google Scholar] [CrossRef]
- Gregory, A.L.; Hurley, B.A.; Tran, H.T.; Valentine, A.J.; She, Y.; Knowles, V.L.; Plaxton, W.C. In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem. J. 2009, 420, 57–65. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Pagano, E.A.; Chueca, A.; López-Gorgé, J. Expression of thioredoxins f and m, and of their targets fructose-1,6-bisphosphatase and NADP-malate dehydrogenase, in pea plants grown under normal and light/temperature stress conditions. J. Exp. Bot. 2000, 51, 1299–1307. [Google Scholar] [CrossRef]
- Nan, N.; Wang, J.; Shi, Y.; Qian, Y.; Jiang, L.; Huang, S.; Liu, Y.; Wu, Y.; Liu, B.; Xu, Z.Y. Rice plastidial NAD-dependent malate dehydrogenase 1 negatively regulates salt stress response by reducing the vitamin B6 content. Plant Biotechnol. J. 2019, 18, 172–184. [Google Scholar] [CrossRef]
- Sánchez, R.; Flores, A.; Cejudo, F.J. Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta 2006, 223, 901–909. [Google Scholar] [CrossRef]
- Feria, A.B.; Bosch, N.; Sánchez, A.; Nieto-Ingelmo, A.I.; de la Osa, C.; Echevarría, C.; García-Mauriño, S.; Monreal, J.A. Phosphoenolpyruvate carboxylase (PEPC) and PEPC-kinase (PEPC-k) isoenzymes in Arabidopsis thaliana: Role in control and abiotic stress conditions. Planta 2016, 244, 901–913. [Google Scholar] [CrossRef]
- Chang, Y.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.; Duan, C. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Sere, D.; Martin, A. Epigenetic regulation: Another layer in plant nutrition. Plant Signal. Behav. 2020, 15, 1686236. [Google Scholar] [CrossRef] [PubMed]
- Fedorin, D.N.; Eprintsev, A.T.; Igamberdiev, A.U. Light Regulation of Tricarboxylic Acid Cycle Isoenzymes in Plants. Russ. J. Plant Physiol. 2022, 69, 589–596. [Google Scholar] [CrossRef]
- Eprintsev, A.T.; Fedorin, D.N.; Anokhina, G.B.; Gataullina, M.O. Role of Epigenetic Mechanisms in Regulating the Activity of 2-OGDH and MDH in Maize Leaves (Zea mays L.) during Hypoxia. Russ. J. Plant Physiol. 2021, 68, 331–336. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Krejci, A.; Hupp, T.R.; Lexa, M.; Vojtesek, B.; Muller, P. Hammock: A hidden Markov model-based peptide clustering algorithm to identify protein-interaction consensus motifs in large datasets. Bioinformatics 2016, 32, 9–16. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Ma, G.; Teixeira Da Silva, J.A.; Qiu, L.; Xu, J.; Zhou, H.; Wei, M.; Xiong, J.; Li, M.; Zhou, S.; et al. Genome-Wide Identification and Analysis of NAC Transcription Factor Family in Two Diploid Wild Relatives of Cultivated Sweet Potato Uncovers Potential NAC Genes Related to Drought Tolerance. Front. Genet. 2021, 12, 744220. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.; Meng, Y.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; He, M.; Zeng, Z.; Chen, Y.; Hanna, A.; Zhu, H. Genome-Wide Identification and Expression Analysis of the MADS-Box Gene Family in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Genet. 2021, 12, 750137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, T.; Yu, J.; Hong, H.; Liu, S.; Guo, F.; Ma, H.; Zhang, J.; Zhu, M.; Meng, X. Genome-wide survey and expression analysis of Dof transcription factor family in sweetpotato shed light on their promising functions in stress tolerance. Front. Plant Sci. 2023, 14, 1140727. [Google Scholar] [CrossRef]



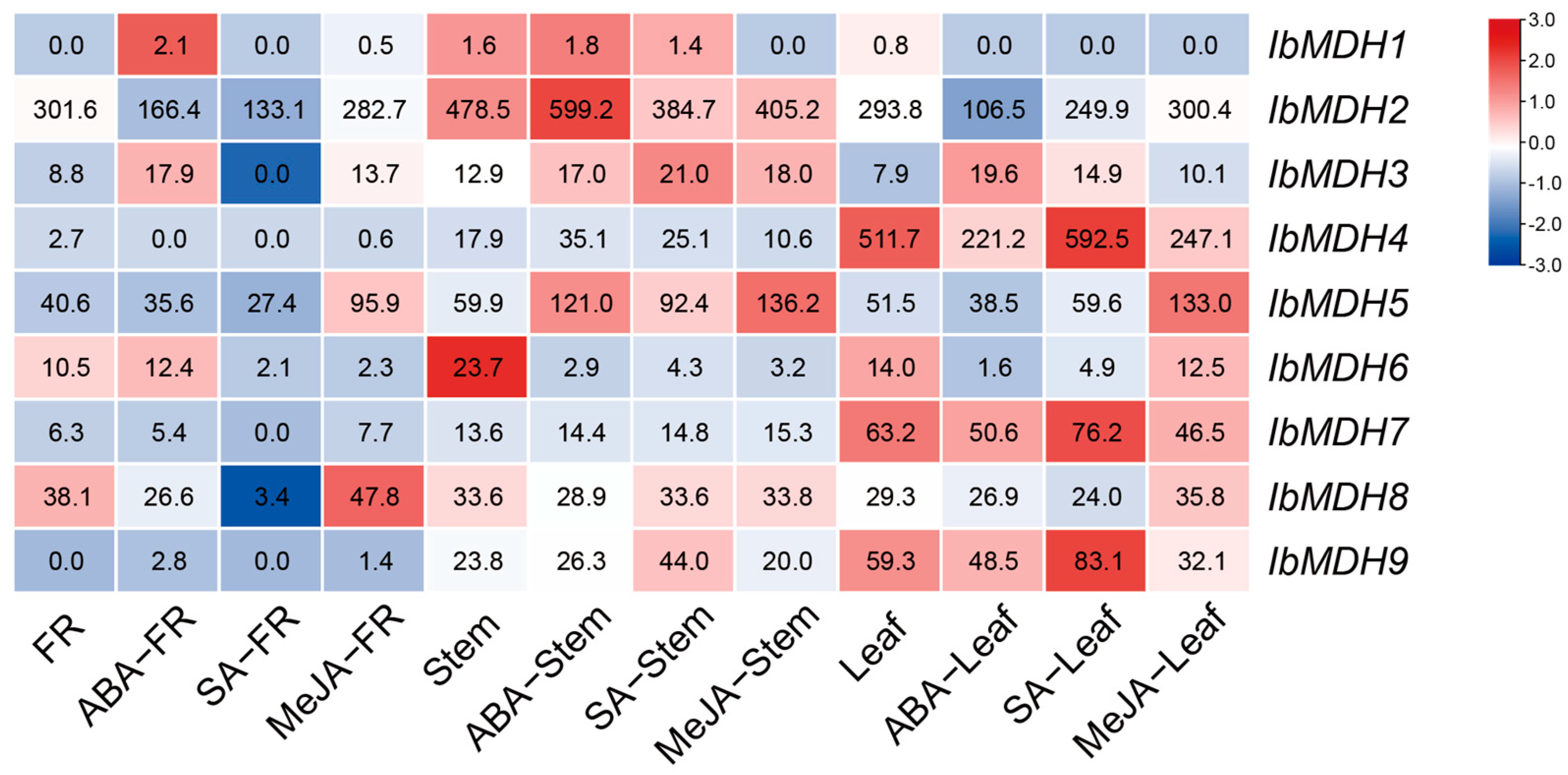
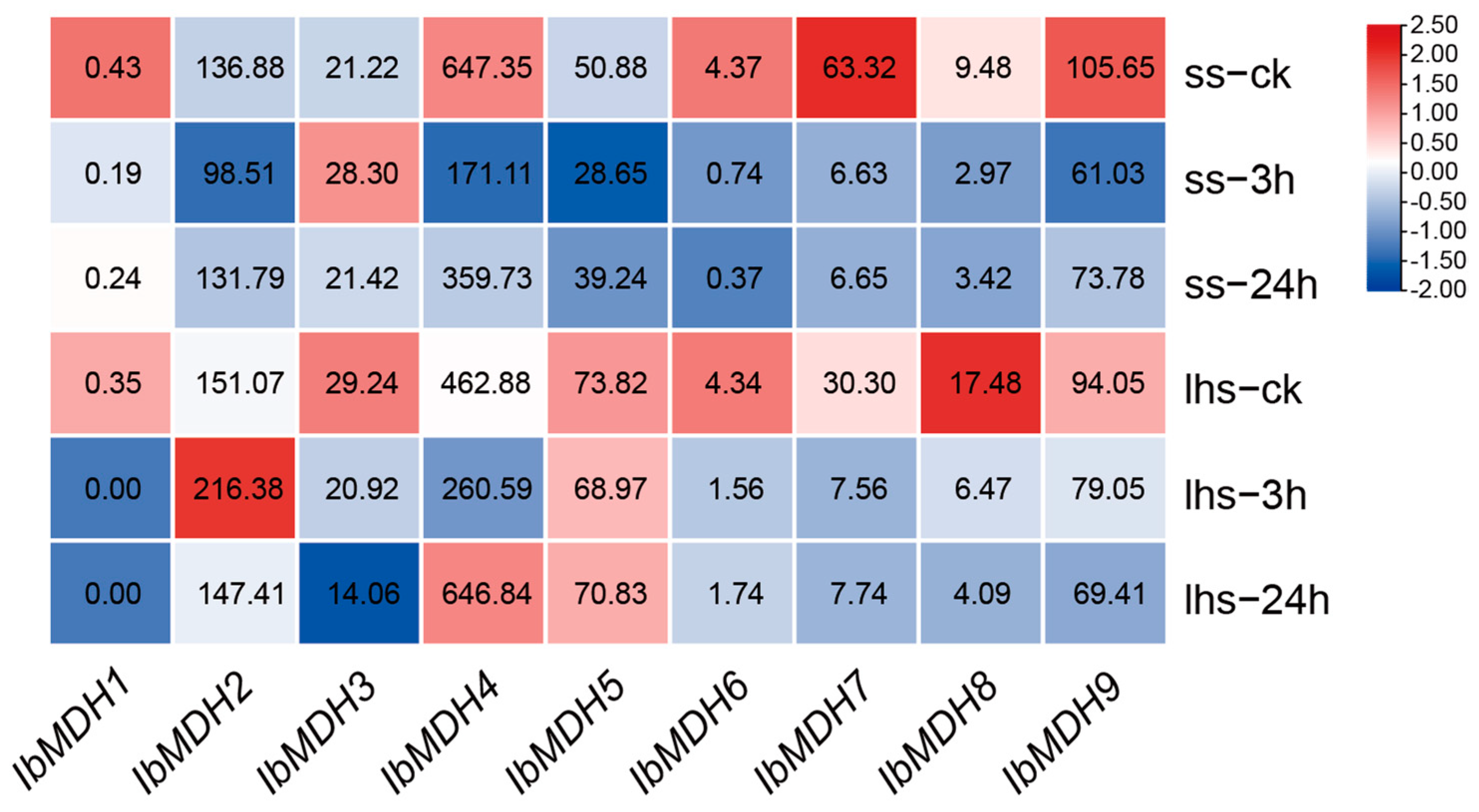
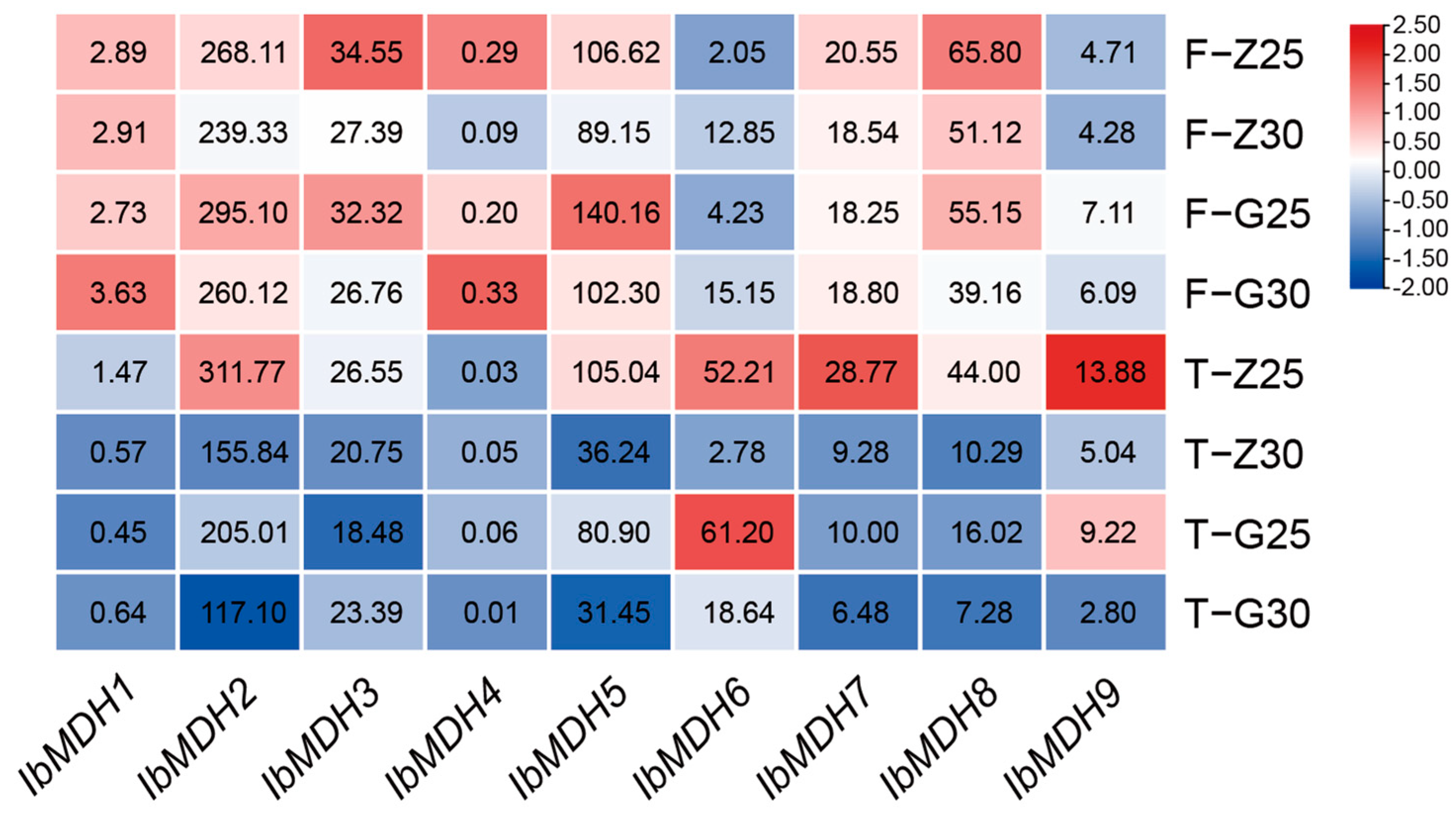
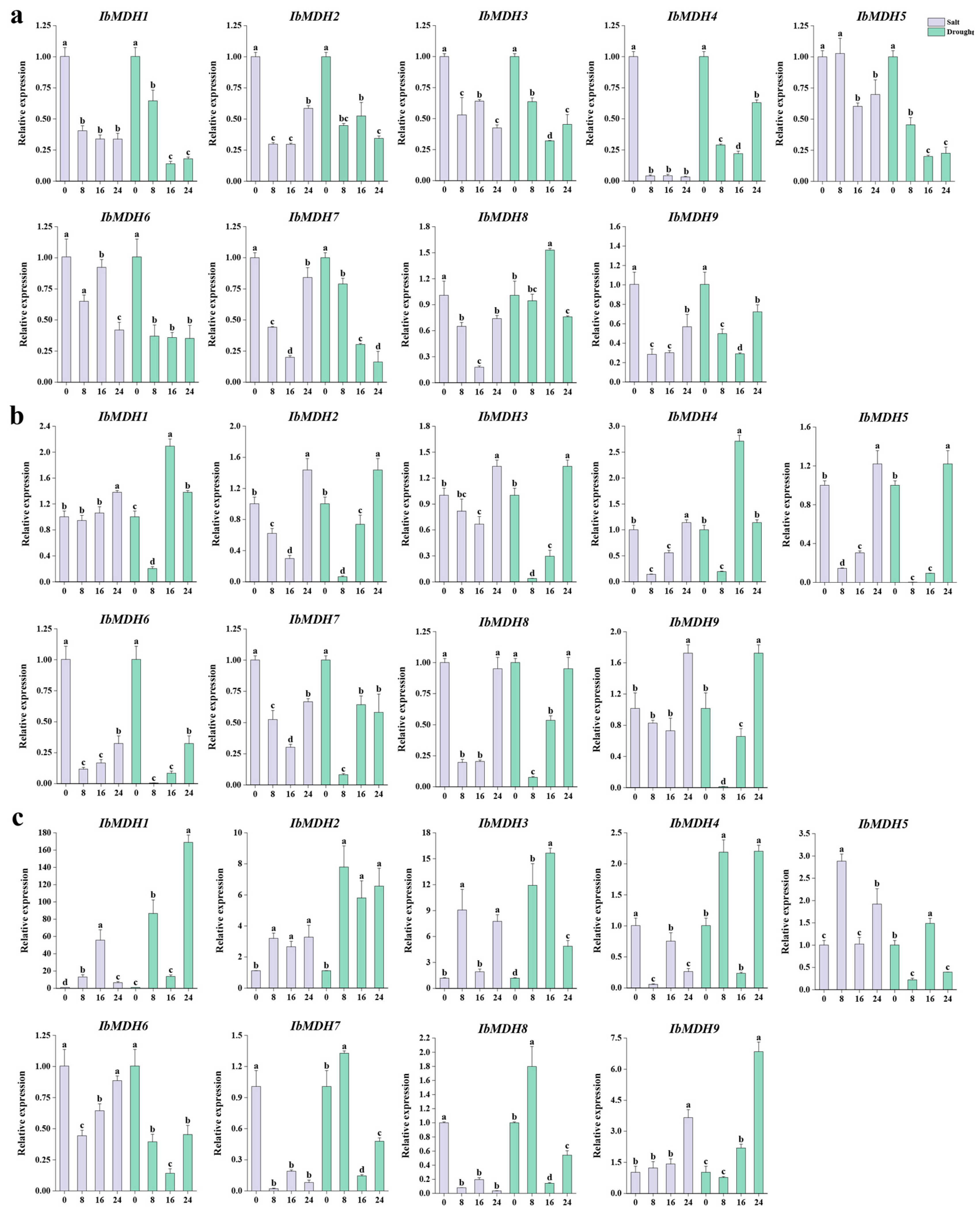
| Gene Name | Accession Number | Genomic Length (bp) | CDS Length (bp) | Protein Size (aa) | MW (kDa) | Isoelectric Point (pI) | Instability Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| IbMDH1 | OR359888 | 3360 | 1086 | 361 | 39.72 | 6.27 | 42.77 | −0.008 | Cytoplasm |
| IbMDH2 | OR359889 | 2474 | 981 | 326 | 34.59 | 6.1 | 34.3 | 0.077 | Chloroplast |
| IbMDH3 | OR359890 | 3567 | 999 | 332 | 34.69 | 7.54 | 32.43 | 0.211 | Cytoplasm |
| IbMDH4 | OR359891 | 2227 | 1068 | 355 | 37.27 | 7.57 | 30 | 0.196 | Chloroplast |
| IbMDH5 | OR359893 | 2749 | 1035 | 344 | 35.98 | 8.58 | 35.63 | 0.126 | Chloroplast |
| IbMDH6 | OR359895 | 4394 | 1161 | 386 | 41.12 | 8.45 | 41.48 | −0.006 | Chloroplast |
| IbMDH7 | OR359896 | 2726 | 1041 | 346 | 35.98 | 8.74 | 34.19 | 0.133 | Mitochondrion |
| IbMDH8 | OR359898 | 2040 | 1239 | 412 | 43.69 | 6.55 | 41.85 | 0.034 | Chloroplast |
| IbMDH9 | OR359899 | 4842 | 1326 | 441 | 48.18 | 7.07 | 27.27 | −0.182 | Chloroplast |
| ItfMDH1 | OR359902 | 2973 | 1035 | 344 | 35.99 | 8.79 | 36.16 | 0.122 | Mitochondrion |
| ItfMDH2 | OR359900 | 4025 | 1203 | 400 | 42.57 | 7.57 | 40.29 | 0.017 | Chloroplast |
| ItfMDH3 | OR359911 | 3058 | 1017 | 338 | 35.22 | 8.89 | 36.09 | 0.117 | Mitochondrion |
| ItfMDH4 | OR359906 | 2234 | 1095 | 364 | 39.92 | 6.47 | 42.56 | 0.015 | Chloroplast |
| ItfMDH5 | OR359908 | 5058 | 1329 | 442 | 48.25 | 6.73 | 28.78 | −0.172 | Chloroplast |
| ItfMDH6 | OR359912 | 2416 | 1239 | 412 | 43.68 | 6.55 | 41.97 | 0.052 | Chloroplast |
| ItfMDH7 | OR359913 | 2457 | 1011 | 336 | 35.52 | 8.76 | 30.44 | 0.176 | Mitochondrion |
| ItfMDH8 | OR359901 | 2457 | 1065 | 354 | 37.27 | 8.4 | 29.26 | 0.177 | Mitochondrion |
| ItfMDH9 | OR359905 | 3647 | 1074 | 357 | 37.51 | 8.56 | 34.95 | 0.127 | Chloroplast |
| ItfMDH10 | OR359904 | 2777 | 999 | 332 | 35.53 | 6.11 | 32.35 | 0.045 | Cytoplasm |
| ItbMDH1 | OR359916 | 2978 | 1035 | 344 | 35.99 | 8.79 | 36.16 | 0.122 | Mitochondrion |
| ItbMDH2 | OR359926 | 2999 | 1017 | 338 | 35.21 | 8.54 | 38.46 | 0.133 | Mitochondrion |
| ItbMDH3 | OR359919 | 5829 | 1203 | 400 | 42.58 | 8.43 | 38.83 | 0 | Chloroplast |
| ItbMDH4 | OR359925 | 2558 | 1095 | 364 | 39.93 | 6.27 | 42.24 | 0.013 | Cytoplasm |
| ItbMDH5 | OR359923 | 5094 | 1329 | 442 | 48.28 | 6.48 | 28.82 | −0.172 | Chloroplast |
| ItbMDH6 | OR359915 | 2307 | 1239 | 412 | 43.71 | 6.96 | 42.71 | 0.043 | Chloroplast |
| ItbMDH7 | OR359914 | 2536 | 1065 | 354 | 37.28 | 8.4 | 29.49 | 0.177 | Mitochondrion |
| ItbMDH8 | OR359917 | 3552 | 1074 | 357 | 37.42 | 8.36 | 32.55 | 0.149 | Chloroplast |
| ItbMDH9 | OR359924 | 3580 | 1074 | 357 | 37.42 | 8.36 | 32.55 | 0.149 | Chloroplast |
| ItbMDH10 | OR359918 | 2756 | 999 | 332 | 35.54 | 6.11 | 31.37 | 0.045 | Cytoplasm |
| Seq_1 | Seq_2 | Ka | Ks | Ka/Ks | T (Mya) | Duplication Type | Type of Selection |
|---|---|---|---|---|---|---|---|
| IbMDH5 | IbMDH7 | 0.12 | 0.91 | 0.13 | 57.2 | Segmental | Purify selection |
| IbMDH6 | IbMDH8 | 0.11 | 0.74 | 0.14 | 46.4 | Segmental | Purify selection |
| ItfMDH1 | ItfMDH3 | 0.05 | 0.81 | 0.06 | 50.3 | Segmental | Purify selection |
| ItbMDH1 | ItbMDH2 | 0.04 | 0.78 | 0.06 | 48.9 | Segmental | Purify selection |
| ItbMDH3 | ItbMDH6 | 0.09 | 0.73 | 0.13 | 45.4 | Segmental | Purify selection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Shi, L.; Lin, X.; Tang, B.; Xing, M.; Zhu, H. Genome-Wide Identification and Expression Analysis of Malate Dehydrogenase Gene Family in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2023, 24, 16549. https://doi.org/10.3390/ijms242316549
Li Z, Shi L, Lin X, Tang B, Xing M, Zhu H. Genome-Wide Identification and Expression Analysis of Malate Dehydrogenase Gene Family in Sweet Potato and Its Two Diploid Relatives. International Journal of Molecular Sciences. 2023; 24(23):16549. https://doi.org/10.3390/ijms242316549
Chicago/Turabian StyleLi, Zhenqin, Lei Shi, Xiongjian Lin, Binquan Tang, Meng Xing, and Hongbo Zhu. 2023. "Genome-Wide Identification and Expression Analysis of Malate Dehydrogenase Gene Family in Sweet Potato and Its Two Diploid Relatives" International Journal of Molecular Sciences 24, no. 23: 16549. https://doi.org/10.3390/ijms242316549
APA StyleLi, Z., Shi, L., Lin, X., Tang, B., Xing, M., & Zhu, H. (2023). Genome-Wide Identification and Expression Analysis of Malate Dehydrogenase Gene Family in Sweet Potato and Its Two Diploid Relatives. International Journal of Molecular Sciences, 24(23), 16549. https://doi.org/10.3390/ijms242316549






