Pyruvate Kinase Differentially Alters Metabolic Signatures during Head and Neck Carcinogenesis
Abstract
1. Introduction
2. Results
2.1. Differential Impacts of PKM2 in HNSC Progression and Prognosis
2.2. PKM2 Loss Facilitated HNSC Cell Growth
2.3. PKM2 Silencing Led to Decreased Cell Motility in HNSC Cells
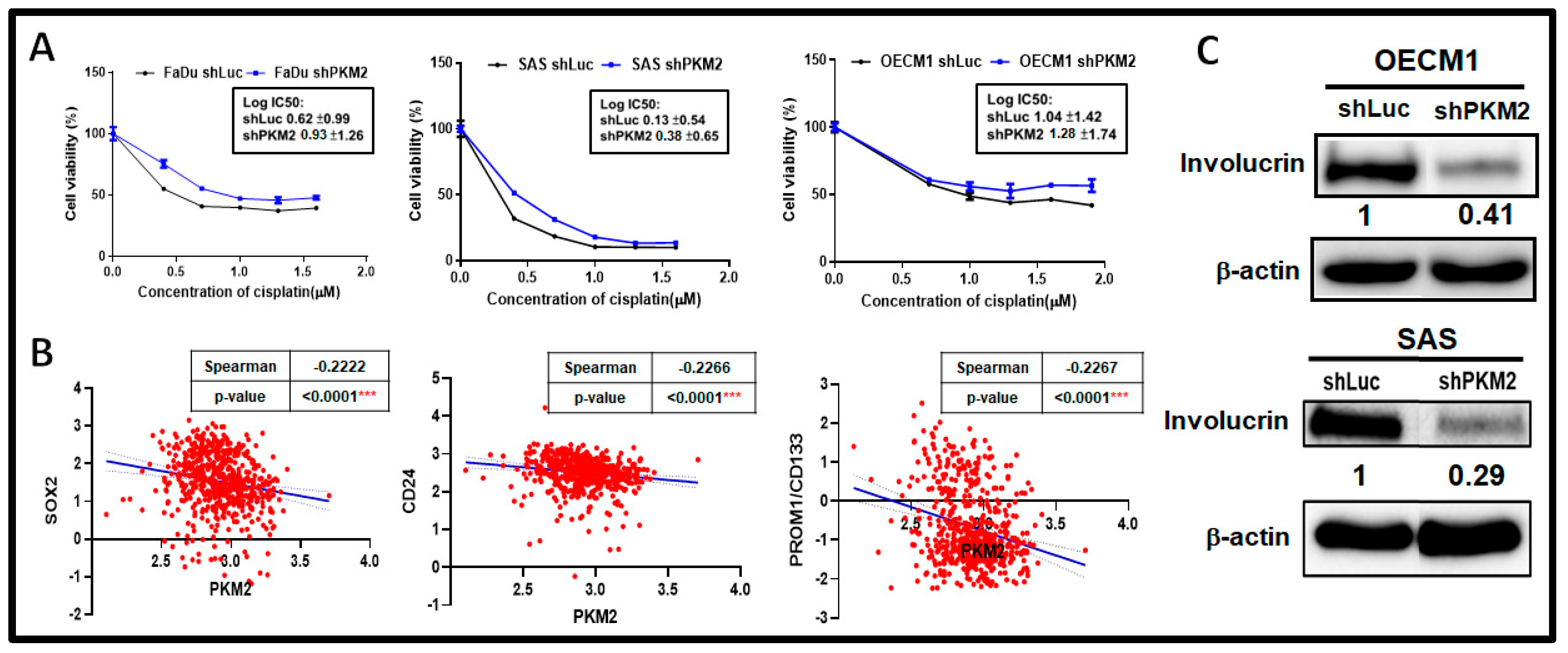
2.4. PKM2 Loss Modulated Chemosensitivity in HNSC Cells
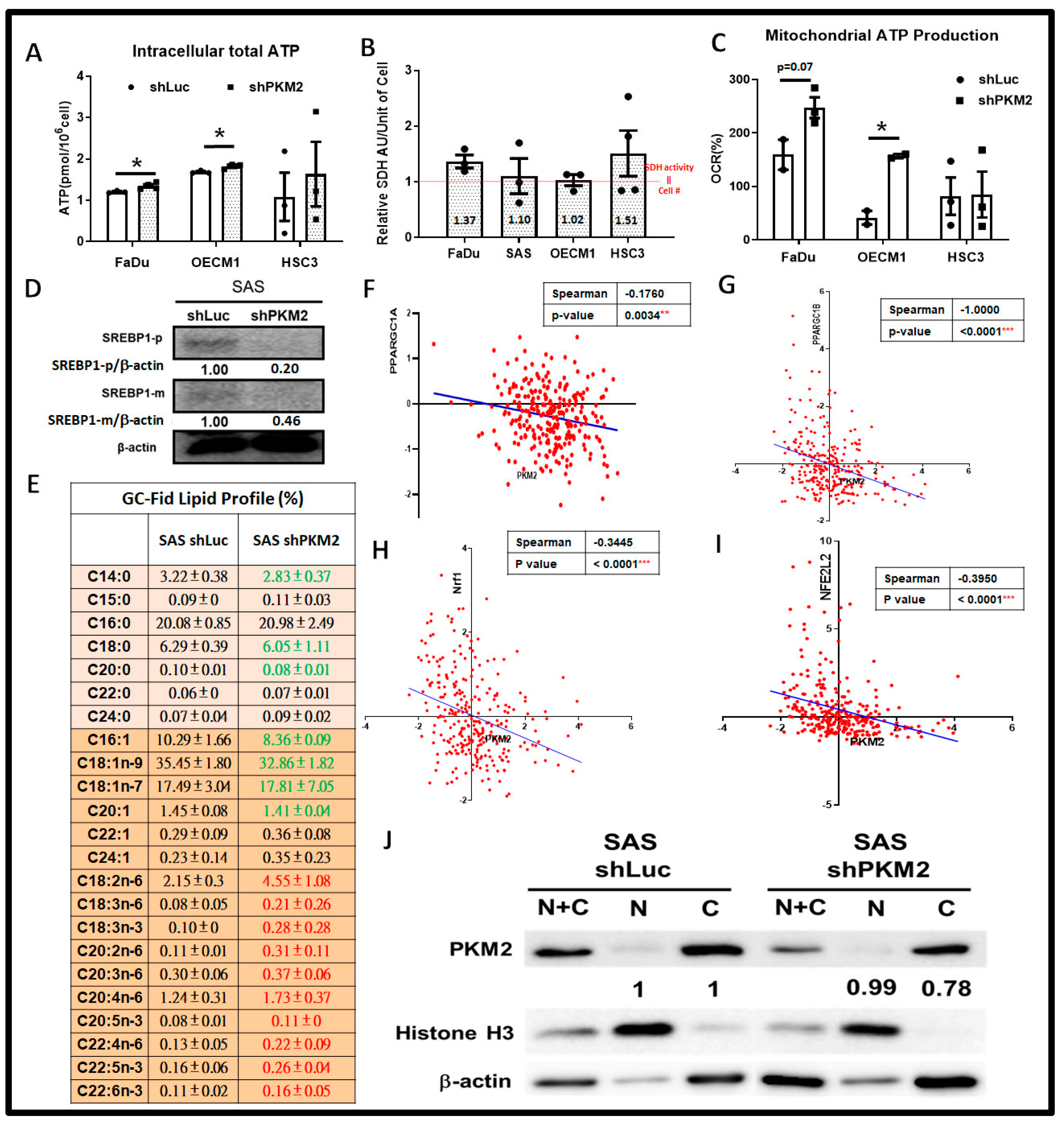
2.5. Metabolic Shift Was Evident in Response to PKM2 Loss
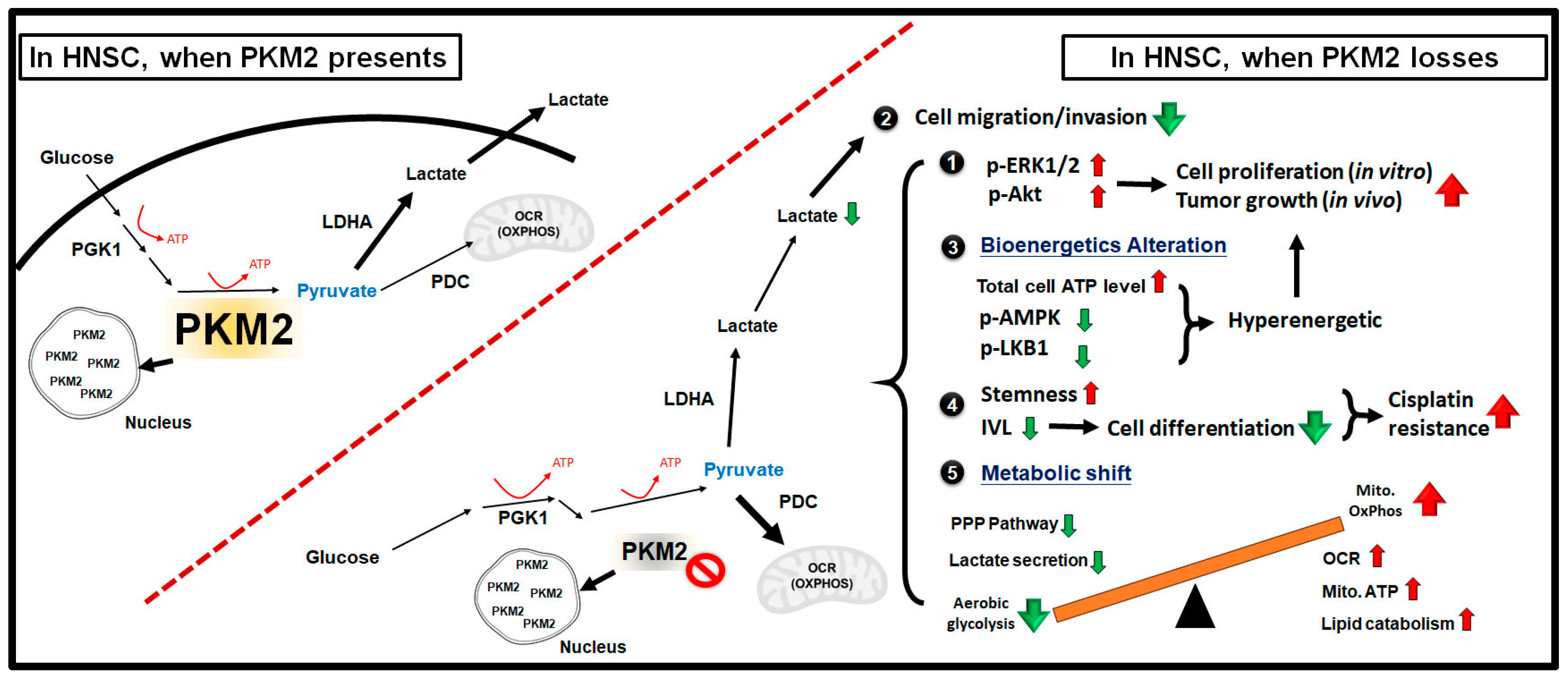
3. Discussion
4. Materials and Methods
4.1. Chemicals, HNSC Cells, Animal and Human HNSC Tissues
4.2. HNSC Datasets
4.3. Establishment of PKM2-Deficient HNSC Cells
4.4. Cellular and Molecular Analyses
4.5. Immunohistochemistry Staining Analysis
4.6. XF Metabolic Assay
4.7. Measurement of Extracellular Lactate, Intracellular Pyruvate/ATP and Glucose Uptake
4.8. Lipidomic Analysis
4.9. Animal Procedures
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Sciacovelli, M.; Gaude, E.; Hilvo, M.; Frezza, C. The metabolic alterations of cancer cells. Methods Enzymol. 2014, 542, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Flores, R.E.; Poff, A.M.; D’Agostino, D.P. Cancer as a metabolic disease: Implications for novel therapeutics. Carcinogenesis 2014, 35, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, Y. Cancer metabolism and intervention therapy. Mol. Biomed. 2021, 2, 5. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Shestov, A.A.; Liu, X.; Ser, Z.; Cluntun, A.A.; Hung, Y.P.; Huang, L.; Kim, D.; Le, A.; Yellen, G.; Albeck, J.G.; et al. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. Elife 2014, 3, e03342. [Google Scholar] [CrossRef]
- Slavov, N.; Budnik, B.A.; Schwab, D.; Airoldi, E.M.; van Oudenaarden, A. Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell Rep. 2014, 7, 705–714. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- van Beek, J. The dynamic side of the Warburg effect: Glycolytic intermediate storage as buffer for fluctuating glucose and O (2) supply in tumor cells. F1000Research 2018, 7, 1177. [Google Scholar] [CrossRef] [PubMed]
- Fukushi, A.; Kim, H.D.; Chang, Y.C.; Kim, C.H. Revisited Metabolic Control and Reprogramming Cancers by Means of the Warburg Effect in Tumor Cells. Int. J. Mol. Sci. 2022, 23, 10037. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Yu, Z. Phosphoglycerate kinase 1 (PGK1) in cancer: A promising target for diagnosis and therapy. Life Sci. 2020, 256, 117863. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Shen, P.; Ni, Y.; Han, X. The basic functions of phosphoglycerate kinase 1 and its roles in cancer and other diseases. Eur. J. Pharmacol. 2022, 920, 174835. [Google Scholar] [CrossRef]
- Gou, R.; Hu, Y.; Liu, O.; Dong, H.; Gao, L.; Wang, S.; Zheng, M.; Li, X.; Lin, B. PGK1 Is a Key Target for Anti-Glycolytic Therapy of Ovarian Cancer: Based on the Comprehensive Analysis of Glycolysis-Related Genes. Front. Oncol. 2021, 11, 682461. [Google Scholar] [CrossRef]
- Hsu, M.C.; Hung, W.C. Pyruvate kinase M2 fuels multiple aspects of cancer cells: From cellular metabolism, transcriptional regulation to extracellular signaling. Mol. Cancer 2018, 17, 35. [Google Scholar] [CrossRef]
- Gupta, V.; Bamezai, R.N. Human pyruvate kinase M2: A multifunctional protein. Protein Sci. 2010, 19, 2031–2044. [Google Scholar] [CrossRef]
- Zhang, Z.; Deng, X.; Liu, Y.; Liu, Y.; Sun, L.; Chen, F. PKM2, function and expression and regulation. Cell Biosci. 2019, 9, 52. [Google Scholar] [CrossRef]
- Desai, S.; Ding, M.; Wang, B.; Lu, Z.; Zhao, Q.; Shaw, K.; Yung, W.K.; Weinstein, J.N.; Tan, M.; Yao, J. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget 2014, 5, 8202–8210. [Google Scholar] [CrossRef]
- Imamura, K.; Tanaka, T. Multimolecular forms of pyruvate kinase from rat and other mammalian tissues. I. Electrophoretic studies. J. Biochem. 1972, 71, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, S.; Yu, D. Protein kinase function of pyruvate kinase M2 and cancer. Cancer Cell Int. 2020, 20, 523. [Google Scholar] [CrossRef] [PubMed]
- Dombrauckas, J.D.; Santarsiero, B.D.; Mesecar, A.D. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 2005, 44, 9417–9429. [Google Scholar] [CrossRef] [PubMed]
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Alquraishi, M.; Puckett, D.L.; Alani, D.S.; Humidat, A.S.; Frankel, V.D.; Donohoe, D.R.; Whelan, J.; Bettaieb, A. Pyruvate kinase M2: A simple molecule with complex functions. Free Radic. Biol. Med. 2019, 143, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Mao, Q.; Xia, W.; Xu, Y.; Wang, J.; Xu, L.; Jiang, F. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol. Lett. 2016, 11, 1980–1986. [Google Scholar] [CrossRef]
- Zahra, K.; Dey, T.; Ashish; Mishra, S.P.; Pandey, U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front. Oncol. 2020, 10, 159. [Google Scholar] [CrossRef]
- Fung, K.Y.; Tabor, B.; Buckley, M.J.; Priebe, I.K.; Purins, L.; Pompeia, C.; Brierley, G.V.; Lockett, T.; Gibbs, P.; Tie, J.; et al. Blood-based protein biomarker panel for the detection of colorectal cancer. PLoS ONE 2015, 10, e0120425. [Google Scholar] [CrossRef]
- Israelsen, W.J.; Dayton, T.L.; Davidson, S.M.; Fiske, B.P.; Hosios, A.M.; Bellinger, G.; Li, J.; Yu, Y.; Sasaki, M.; Horner, J.W.; et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 2013, 155, 397–409. [Google Scholar] [CrossRef]
- Dayton, T.L.; Gocheva, V.; Miller, K.M.; Israelsen, W.J.; Bhutkar, A.; Clish, C.B.; Davidson, S.M.; Luengo, A.; Bronson, R.T.; Jacks, T.; et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 2016, 30, 1020–1033. [Google Scholar] [CrossRef]
- Mody, M.D.; Rocco, J.W.; Yom, S.S.; Haddad, R.I.; Saba, N.F. Head and neck cancer. Lancet 2021, 398, 2289–2299. [Google Scholar] [CrossRef]
- Boschert, V.; Teusch, J.; Muller-Richter, U.D.A.; Brands, R.C.; Hartmann, S. PKM2 Modulation in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 775. [Google Scholar] [CrossRef]
- Yuan, C.; Li, Z.; Wang, Y.; Qi, B.; Zhang, W.; Ye, J.; Wu, H.; Jiang, H.; Song, L.N.; Yang, J.; et al. Overexpression of metabolic markers PKM2 and LDH5 correlates with aggressive clinicopathological features and adverse patient prognosis in tongue cancer. Histopathology 2014, 65, 595–605. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Zhang, Y.; Zhu, Y.; Yuan, C.; Qi, B.; Zhang, W.; Wang, D.; Ding, X.; Wu, H.; et al. Overexpression of pyruvate kinase M2 associates with aggressive clinicopathological features and unfavorable prognosis in oral squamous cell carcinoma. Cancer Biol. Ther. 2015, 16, 839–845. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.; Li, Y.; Jiang, L.; Ren, X.; Cheng, B.; Xia, J. Prognostic value of glycolysis markers in head and neck squamous cell carcinoma: A meta-analysis. Aging 2021, 13, 7284–7299. [Google Scholar] [CrossRef] [PubMed]
- Kurihara-Shimomura, M.; Sasahira, T.; Nakashima, C.; Kuniyasu, H.; Shimomura, H.; Kirita, T. The Multifarious Functions of Pyruvate Kinase M2 in Oral Cancer Cells. Int. J. Mol. Sci. 2018, 19, 2907. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Bhagat, S.D.; Gupta, A.; Samaiya, A.; Srivastava, A.; Shukla, S. Dietary-phytochemical mediated reversion of cancer-specific splicing inhibits Warburg effect in head and neck cancer. BMC Cancer 2019, 19, 1031. [Google Scholar] [CrossRef]
- Jing, C.; Qu, X.; Li, Z.; Wu, C.; Zhao, M.; Wang, Y.; Sun, S.; Zhang, S.; Chen, J.; Qiao, Y.; et al. EGFRwt/vIII-PKM2-beta-catenin cascade affects proliferation and chemo-sensitivity in head and neck squamous cell carcinoma. Am. J. Cancer Res. 2017, 7, 2491–2502. [Google Scholar] [PubMed]
- Gupta, A.; Ajith, A.; Singh, S.; Panday, R.K.; Samaiya, A.; Shukla, S. PAK2-c-Myc-PKM2 axis plays an essential role in head and neck oncogenesis via regulating Warburg effect. Cell Death Dis. 2018, 9, 825. [Google Scholar] [CrossRef]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Singh, A.K.; Singh, A.K.; Rawat, K.; Banerjee, J.; Agnihotri, V.; Upadhaya, D. Critical pathways of oral squamous cell carcinoma: Molecular biomarker and therapeutic intervention. Med. Oncol. 2022, 39, 30. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Hsieh, Y.T.; Huang, J.M.; Liu, C.J.; Chuang, L.T.; Huang, P.C.; Kuo, T.Y.; Chia, H.Y.; Chou, C.Y.; Chang, C.W.; et al. Determination of Pyruvate Metabolic Fates Modulates Head and Neck Tumorigenesis. Neoplasia 2019, 21, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Li, W.C.; Huang, C.H.; Hsieh, Y.T.; Chen, T.Y.; Cheng, L.H.; Chen, C.Y.; Liu, C.J.; Chen, H.M.; Huang, C.L.; Lo, J.F.; et al. Regulatory Role of Hexokinase 2 in Modulating Head and Neck Tumorigenesis. Front. Oncol. 2020, 10, 176. [Google Scholar] [CrossRef]
- Carneiro, A.; Silveira, I.C.D.; Rezende, A.S.; Silva, B.R.O.; Crema, V.O. Tyrosine kinase inhibitor TKI-258 inhibits cell motility in oral squamous cell carcinoma in vitro. J. Oral Pathol. Med. 2017, 46, 484–488. [Google Scholar] [CrossRef]
- Yamada, S.; Yanamoto, S.; Kawasaki, G.; Mizuno, A.; Nemoto, T.K. Overexpression of cortactin increases invasion potential in oral squamous cell carcinoma. Pathol. Oncol. Res. 2010, 16, 523–531. [Google Scholar] [CrossRef]
- Goetze, K.; Walenta, S.; Ksiazkiewicz, M.; Kunz-Schughart, L.A.; Mueller-Klieser, W. Lactate enhances motility of tumor cells and inhibits monocyte migration and cytokine release. Int. J. Oncol. 2011, 39, 453–463. [Google Scholar] [CrossRef]
- Usami, Y.; Satake, S.; Nakayama, F.; Matsumoto, M.; Ohnuma, K.; Komori, T.; Semba, S.; Ito, A.; Yokozaki, H. Snail-associated epithelial-mesenchymal transition promotes oesophageal squamous cell carcinoma motility and progression. J. Pathol. 2008, 215, 330–339. [Google Scholar] [CrossRef]
- Chen, Y.S.; Huang, W.L.; Chang, S.H.; Chang, K.W.; Kao, S.Y.; Lo, J.F.; Su, P.F. Enhanced filopodium formation and stem-like phenotypes in a novel metastatic head and neck cancer cell model. Oncol. Rep. 2013, 30, 2829–2837. [Google Scholar] [CrossRef][Green Version]
- Morfouace, M.; Lalier, L.; Oliver, L.; Cheray, M.; Pecqueur, C.; Cartron, P.F.; Vallette, F.M. Control of glioma cell death and differentiation by PKM2-Oct4 interaction. Cell Death Dis. 2014, 5, e1036. [Google Scholar] [CrossRef]
- Yang, R.; Liu, B.; Yang, M.; Xu, F.; Wu, S.; Zhao, S. Lumiflavin Reduces Cisplatin Resistance in Cancer Stem-Like Cells of OVCAR-3 Cell Line by Inducing Differentiation. Front. Oncol. 2022, 12, 859275. [Google Scholar] [CrossRef] [PubMed]
- Kulsum, S.; Sudheendra, H.V.; Pandian, R.; Ravindra, D.R.; Siddappa, G.; Nisheena, R.; Chevour, P.; Ramachandran, B.; Sagar, M.; Jayaprakash, A.; et al. Cancer stem cell mediated acquired chemoresistance in head and neck cancer can be abrogated by aldehyde dehydrogenase 1 A1 inhibition. Mol. Carcinog. 2017, 56, 694–711. [Google Scholar] [CrossRef] [PubMed]
- Christofk, H.R.; Vander Heiden, M.G.; Wu, N.; Asara, J.M.; Cantley, L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008, 452, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, H.; Yang, J.J.; Liu, X.; Liu, Z.R. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 2012, 45, 598–609. [Google Scholar] [CrossRef]
- Yang, W.; Xia, Y.; Ji, H.; Zheng, Y.; Liang, J.; Huang, W.; Gao, X.; Aldape, K.; Lu, Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 2011, 480, 118–122. [Google Scholar] [CrossRef]
- Cortes-Cros, M.; Hemmerlin, C.; Ferretti, S.; Zhang, J.; Gounarides, J.S.; Yin, H.; Muller, A.; Haberkorn, A.; Chene, P.; Sellers, W.R.; et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc. Natl. Acad. Sci. USA 2013, 110, 489–494. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Christofk, H.R.; Schuman, E.; Subtelny, A.O.; Sharfi, H.; Harlow, E.E.; Xian, J.; Cantley, L.C. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem. Pharmacol. 2010, 79, 1118–1124. [Google Scholar] [CrossRef]
- Yoo, B.C.; Ku, J.L.; Hong, S.H.; Shin, Y.K.; Park, S.Y.; Kim, H.K.; Park, J.G. Decreased pyruvate kinase M2 activity linked to cisplatin resistance in human gastric carcinoma cell lines. Int. J. Cancer 2004, 108, 532–539. [Google Scholar] [CrossRef]
- Martinez-Balibrea, E.; Plasencia, C.; Gines, A.; Martinez-Cardus, A.; Musulen, E.; Aguilera, R.; Manzano, J.L.; Neamati, N.; Abad, A. A proteomic approach links decreased pyruvate kinase M2 expression to oxaliplatin resistance in patients with colorectal cancer and in human cell lines. Mol. Cancer Ther. 2009, 8, 771–778. [Google Scholar] [CrossRef]
- Lin, Y.; Lv, F.; Liu, F.; Guo, X.; Fan, Y.; Gu, F.; Gu, J.; Fu, L. High Expression of Pyruvate Kinase M2 Is Associated with Chemosensitivity to Epirubicin and 5-Fluorouracil in Breast Cancer. J. Cancer 2015, 6, 1130–1139. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lu, Z. Pyruvate kinase M2 at a glance. J. Cell Sci. 2015, 128, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.P.; Luo, L.J.; Chen, H.Z.; Chen, Q.T.; Bian, X.L.; Wu, S.F.; Zhou, J.X.; Zhao, W.X.; Liu, J.M.; Wang, X.M.; et al. Ectosomal PKM2 Promotes HCC by Inducing Macrophage Differentiation and Remodeling the Tumor Microenvironment. Mol. Cell 2020, 78, 1192–1206. [Google Scholar] [CrossRef] [PubMed]
- Adem, S.; Comakli, V.; Uzun, N. Pyruvate kinase activators as a therapy target: A patent review 2011–2017. Expert Opin. Ther. Pat. 2018, 28, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Chang, W.J.; Chen, C.Y.; Sun, F.J.; Cheng, H.W.; Chen, T.Y.; Lin, S.C.; Li, W.C. Dynamic cellular and molecular modulations of diabetes mediated head and neck carcinogenesis. Oncotarget 2015, 6, 29268–29284. [Google Scholar] [CrossRef]
- Demokan, S.; Chuang, A.Y.; Chang, X.; Khan, T.; Smith, I.M.; Pattani, K.M.; Dasgupta, S.; Begum, S.; Khan, Z.; Liegeois, N.J.; et al. Identification of guanine nucleotide-binding protein gamma-7 as an epigenetically silenced gene in head and neck cancer by gene expression profiling. Int. J. Oncol. 2013, 42, 1427–1436. [Google Scholar] [CrossRef]
- Pyeon, D.; Newton, M.A.; Lambert, P.F.; den Boon, J.A.; Sengupta, S.; Marsit, C.J.; Woodworth, C.D.; Connor, J.P.; Haugen, T.H.; Smith, E.M.; et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007, 67, 4605–4619. [Google Scholar] [CrossRef]
- Chuang, L.T.; Glew, R.H.; Li, C.C.; VanderJagt, D.J.; Broyles, J.S.; Ray, G.M.; Shah, V.O. Comparison of the fatty acid composition of the serum phospholipids of controls, prediabetics and adults with type 2 diabetes. J. Diabetes Mellit. 2012, 2, 393–401. [Google Scholar] [CrossRef][Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, P.-C.; Chang, C.-W.; Lin, Y.-C.; Chen, C.-Y.; Chen, T.-Y.; Chuang, L.-T.; Liu, C.-J.; Huang, C.-L.; Li, W.-C. Pyruvate Kinase Differentially Alters Metabolic Signatures during Head and Neck Carcinogenesis. Int. J. Mol. Sci. 2023, 24, 16639. https://doi.org/10.3390/ijms242316639
Huang P-C, Chang C-W, Lin Y-C, Chen C-Y, Chen T-Y, Chuang L-T, Liu C-J, Huang C-L, Li W-C. Pyruvate Kinase Differentially Alters Metabolic Signatures during Head and Neck Carcinogenesis. International Journal of Molecular Sciences. 2023; 24(23):16639. https://doi.org/10.3390/ijms242316639
Chicago/Turabian StyleHuang, Pei-Chun, Ching-Wen Chang, Yu-Cheng Lin, Chang-Yi Chen, Tsai-Ying Chen, Lu-Te Chuang, Chung-Ji Liu, Chien-Ling Huang, and Wan-Chun Li. 2023. "Pyruvate Kinase Differentially Alters Metabolic Signatures during Head and Neck Carcinogenesis" International Journal of Molecular Sciences 24, no. 23: 16639. https://doi.org/10.3390/ijms242316639
APA StyleHuang, P.-C., Chang, C.-W., Lin, Y.-C., Chen, C.-Y., Chen, T.-Y., Chuang, L.-T., Liu, C.-J., Huang, C.-L., & Li, W.-C. (2023). Pyruvate Kinase Differentially Alters Metabolic Signatures during Head and Neck Carcinogenesis. International Journal of Molecular Sciences, 24(23), 16639. https://doi.org/10.3390/ijms242316639





