Genome-Wide and Transcriptome Analysis of Jacalin-Related Lectin Genes in Barley and the Functional Characterization of HvHorcH in Low-Nitrogen Tolerance in Arabidopsis
Abstract
1. Introduction
2. Results
2.1. Identification of JRL Genes in Barley
2.2. Structural Conservation Analysis of the Jacalin Domains in Barley
2.3. Gene Structure, Conserved Motif, and Domain Analysis of HvJRLs
2.4. Chromosomal Distribution and Gene Duplication of HvJRL Genes
2.5. Analysis of cis-Acting Elements in HvJRL Promoters
2.6. Evolutionary Analysis of JRL Gene Family
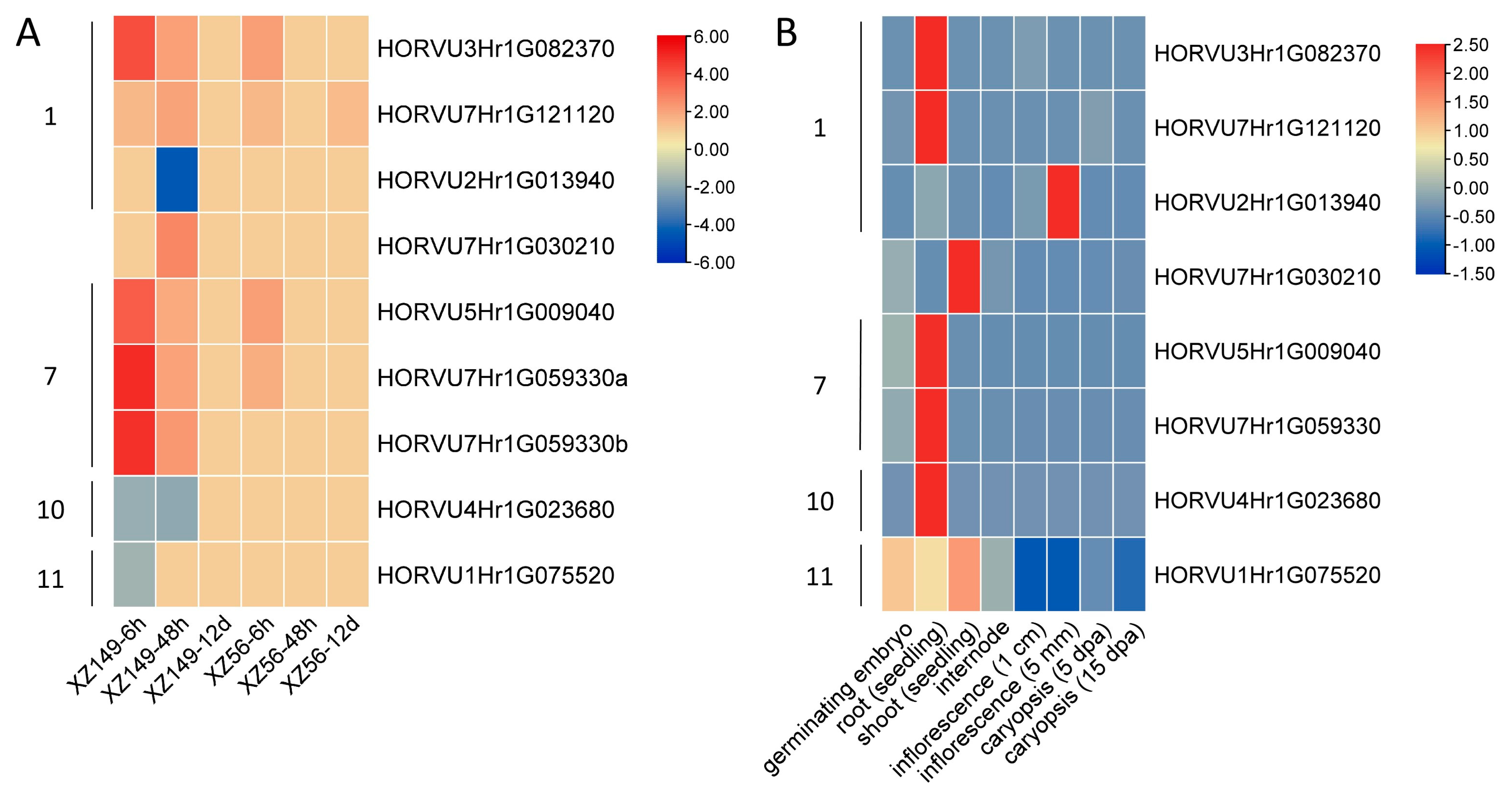
2.7. Expression Profiles of the HvJRLs in Response to Low-Nitrogen Stress
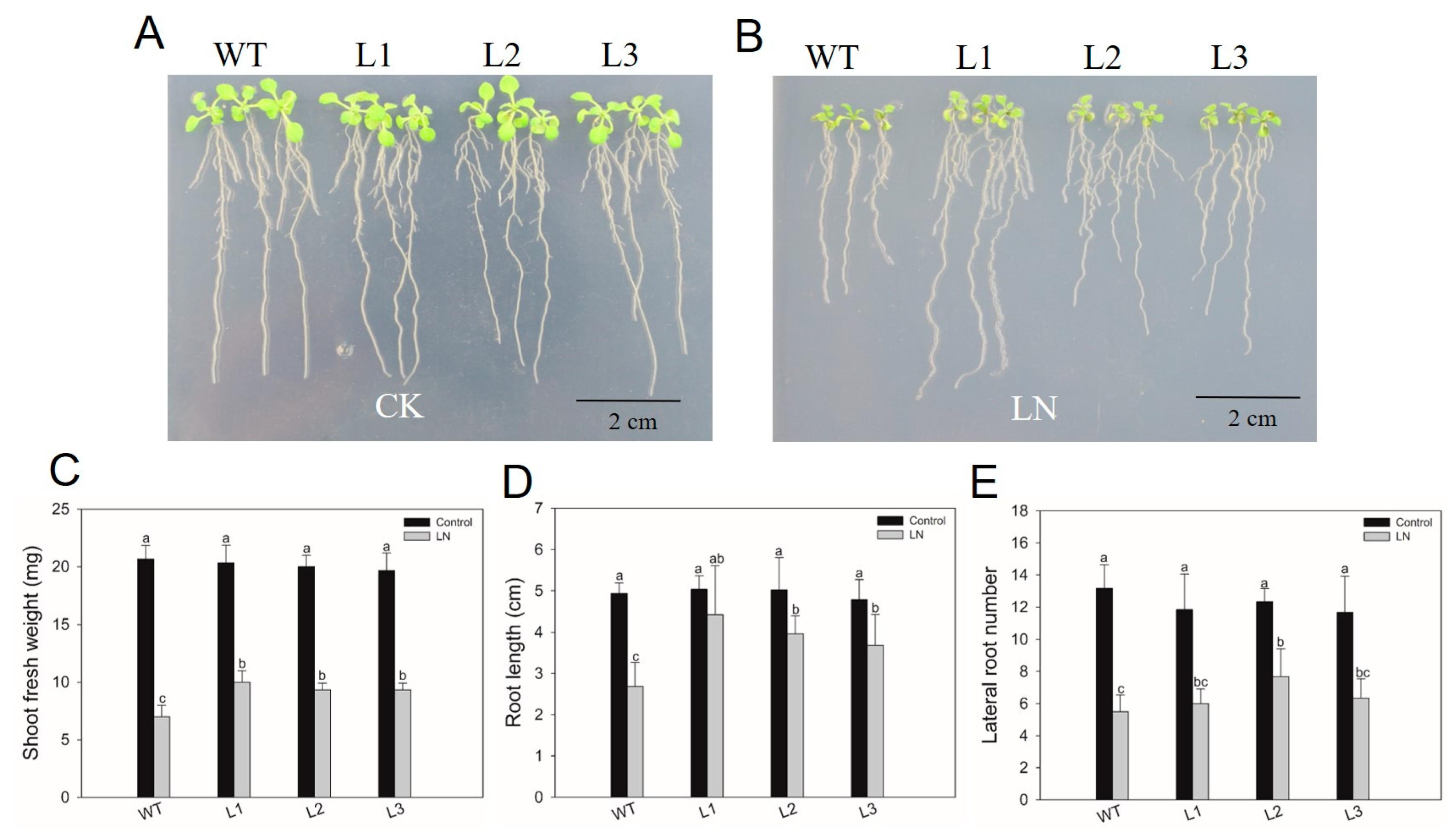
2.8. HvHorcH Enhanced Low-Nitrogen Tolerance in Transgenic Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Identification of JRL Genes in Barley
4.2. Sequence Alignment and Phylogenetic Analysis
4.3. Gene Structure, Conserved Motif, and Domain Analysis
4.4. Chromosomal Distribution and Duplication of HvJRL Genes
4.5. cis-Elements in the Promoter Regions of HvJRLs
4.6. Vector Construction and Arabidopsis Transformation
4.7. Plant Materials and Treatments
4.8. Transcriptome Analysis of HvJRL Genes in Response to LN Stress
4.9. Tissue Expression Profile of JRL Gene Family in Barley
4.10. Real-Time PCR
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Jung, I.J.; Ahn, J.W.; Jung, S.; Hwang, J.E.; Hong, M.J.; Choi, H.I.; Kim, J.B. Overexpression of rice jacalin-related mannose-binding lectin (OsJAC1) enhances resistance to ionizing radiation in Arabidopsis. BMC Plant Biol. 2019, 19, 561. [Google Scholar] [CrossRef]
- Han, Y.J.; Zhong, Z.H.; Song, L.L.; Stefan, O.; Wang, Z.H.; Lu, G.L. Evolutionary analysis of plant jacalin-related lectins (JRLs) family and expression of rice JRLs in response to Magnaporthe oryzae. J. Integr. Agric. 2018, 17, 1252–1266. [Google Scholar] [CrossRef]
- Song, M.; Xu, W.Q.; Xiang, Y.; Jia, H.Y.; Zhang, L.X.; Ma, Z.Q. Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 2014, 84, 95–110. [Google Scholar] [CrossRef]
- Sharon, N. Lectins: Carbohydrate-specific reagents and biological recognition molecules. J. Biol. Chem. 2007, 282, 2753–2764. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Hause, B.; Van Damme, E.J. The galactose-binding and mannose-binding jacalin-related lectins are located in different sub-cellular compartments. FEBS Lett. 2000, 477, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Lannoo, N.; Peumans, W.J. Plant lectins. Adv. Bot. Res. 2008, 48, 107–209. [Google Scholar]
- Raval, S.; Gowda, S.B.; Singh, D.D.; Chandra, N.R. A database analysis of jacalin-like lectins: Sequence-structure-function relationships. Glycobiology 2004, 14, 1247–1263. [Google Scholar] [CrossRef]
- Bunn-Moreno, M.M.; Campos-Neto, A. Lectin(s) extracted from seeds of Artocarpus integrifolia (jackfruit): Potent and selective stimulator(s) of distinct human T and B cell function. J. Immunol. 1981, 127, 427–429. [Google Scholar] [CrossRef]
- Nagano, A.J.; Fukao, Y.; Fujiwara, M.; Nishimura, M.; Hara-Nishimura, I. Antagonistic jacalin-related lectins regulate the size of ER body-type b-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 969–980. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, B.; Chen, J.; Jiao, Y.; Guo, H.; Liu, S.; Ramakrishnan, M.; Qi, G. Genome-wide identification of JRL genes in Moso Bamboo and their expression profiles in response to multiple hormones and abiotic stresses. Front. Plant Sci. 2022, 12, 809666. [Google Scholar] [CrossRef]
- Yong, W.D.; Xu, Y.Y.; Xu, W.Z.; Wang, X.; Li, N.; Wu, J.S.; Liang, T.B.; Chong, K.; Xu, Z.H.; Tan, K.H.; et al. Vernalization induced flowering in wheat is mediated by a lectin-like gene VER2. Planta 2003, 217, 261–270. [Google Scholar] [CrossRef]
- Xing, L.; Li, J.; Xu, Y.; Xu, Z.; Chong, K. Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility. PLoS ONE 2009, 3, e4854. [Google Scholar] [CrossRef]
- Jiang, J.; Xu, Y.; Chong, K. Overexpression of OsJAC1, a lectin gene, suppresses the coleoptile and stem elongation in rice. J. Integr. Plant Biol. 2007, 49, 230–237. [Google Scholar] [CrossRef]
- Xiao, J.; Li, C.; Xu, S.; Xing, L.; Xu, Y.; Chong, K. JACALIN-LECTIN LIKE1 regulates the nuclear accumulation of GLYCINE-RICH RNA-BINDING PROTEIN7, influencing the RNA processing of FLOWERING LOCUS C antisense transcripts and flowering time in Arabidopsis. Plant Physiol. 2015, 3, 2102–2117. [Google Scholar]
- Yamaji, Y.; Maejima, K.; Komatsu, K.; Shiraishi, T.; Okano, Y.; Himeno, M.; Sugawara, K.; Neriya, Y.; Minato, N.; Miura, C.; et al. Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell. 2012, 24, 778–793. [Google Scholar] [CrossRef]
- Xiang, Y.; Song, M.; Wei, Z.Y.; Tong, J.; Zhang, L.X.; Xiao, L.; Ma, Z.Q.; Wang, Y. A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J. Exp. Bot. 2011, 62, 5471–5483. [Google Scholar] [CrossRef] [PubMed]
- Subramanyam, S.; Smith, D.F.; Clemens, J.C.; Webb, M.A.; Sardesai, N.; Williams, C.E. Functional characterization of HFR-1, a high mannose N-glycan-specific wheat lectin induced by Hessian fly larvae. Plant Physiol. 2008, 147, 1412–1426. [Google Scholar] [CrossRef] [PubMed]
- Subramanyam, S.; Sardesai, N.; Puthoff, D.P.; Meyer, J.M.; Nemacheck, J.A.; Gonzalo, M.; Williams, C.E. Expression of two wheat defense-response genes, Hfr-1 and Wci-1, under biotic and abiotic stresses. Plant Sci. 2006, 170, 90–103. [Google Scholar] [CrossRef]
- Qin, Q.M.; Zhang, Q.; Zhao, W.S.; Wang, Y.Y.; Peng, Y.L. Identification of a lectin gene induced in rice in response to Magnaporthe grisea infection. Acta Bot. Sin. 2003, 45, 76–81. [Google Scholar]
- Atalah, B.A.; Smagghe, G.; Damme, E.V.J.P.S. Orysata, a jacalin-related lectin from rice, could protect plants against biting-chewing and piercing-sucking insects. Plant Sci. 2014, 221–222, 21–28. [Google Scholar] [CrossRef]
- Weidenbach, D.; Esch, L.; Moller, C.; Hensel, G.; Kumlehn, J.; Hofle, C.; Huckelhoven, R.; Schaffrath, U. Polarized defense against fungal pathogens is mediated by the jacalin-related lectin domain of modular Poaceae-specific proteins. Mol. Plant 2016, 9, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Claes, B.; Dekeyser, R.; Villarroel, R.; Van den Bulcke, M.; Bauw, G.; Van Montagu, M.; Caplanet, A. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 1990, 2, 19–27. [Google Scholar] [PubMed]
- Filho, G.A.S.; Ferreira, B.S.; Dias, J.M.; Queiroz, K.S.; Branco, A.T.; Bressan-Smith, R.E.; Oliveira, J.G.; Garcia, A.B. Accumulation of SALT protein in rice plants as a response to environmental stresses. Plant Sci. 2003, 164, 623–628. [Google Scholar]
- Paul, S.; Gayen, D.; Datta, S.K.; Datta, S.K. Dissecting root proteome of transgenic rice cultivars unravels metabolic alterations and accumulation of novel stress responsive proteins under drought stress. Plant Sci. 2015, 234, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Sahid, S.; Roy, C.; Shee, D.; Datta, R.; Paul, S. Jacalin domain-containing protein OsSalT interacts with OsDREB2A and OsNAC1 to impart drought stress tolerance in planta. Environ. Exp. Bot. 2021, 183, 104362. [Google Scholar] [CrossRef]
- Feng, H.; Xu, W.Z.; Lin, H.H.; Chong, K. Transcriptional regulation of wheat VER2 promoter in rice in response to abscisic acid, jasmonate, and light. J. Genet. Genom. 2009, 6, 371–377. [Google Scholar] [CrossRef]
- Abebe, T.; Skadsen, R.W.; Kaeppler, H.F. A proximal upstream sequence controls tissue-specific expression of Lem2, a salicylate-inducible barley lectin-like gene. Planta 2005, 221, 170–183. [Google Scholar] [CrossRef]
- Zhang, W.; Peumans, W.J.; Barre, A.; Astoul, C.H.; Rovira, P.; Rougé, P.; Proost, P.; Truffa-Bachi, P.; Jalali, A.A.; Van Damme, E.J.M. Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 2000, 210, 970–978. [Google Scholar]
- He, X.; Li, L.; Xu, H.; Xi, J.; Cao, X.; Xu, H. A rice jacalin-related mannose-binding lectin gene, OsJRL, enhances Escherichia coli viability under high salinity stress and improves salinity tolerance of rice. Plant Biol. 2017, 19, 257–261. [Google Scholar] [CrossRef]
- Gao, Q.; Yin, X.; Wang, F.; Hu, S.; Liu, W.; Chen, L.; Dai, X.; Liang, M. OsJRL40, a jacalin-related lectin gene, promotes salt stress tolerance in rice. Int. J. Mol. Sci. 2023, 24, 7441. [Google Scholar] [CrossRef]
- Witzel, K.; Matros, A.; Bertsch, U.; Aftab, T.; Rutten, T.; Ramireddy, E.; Melzer, M.; Kunze, G.; Mock, H. The jacalin-related lectin HvHorcH is involved in the physiological response of barley roots to salt stress. Int. J. Mol. Sci. 2021, 22, 10248. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, C.; Yao, J.; Zhang, Y.L.; Zhang, Y.N.; Deng, S.; Zhao, N.; Sa, G.; Zhou, X.; Lu, C.; et al. Populus euphratica JRL mediates ABA response, ionic and ROS homeostasis in Arabidopsis under salt stress. Int. J. Mol. Sci. 2019, 20, 815. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Zhang, W.; Peumans, W.J. Induction of cytoplasmic mannose-binding jacalin-related lectins is a common phenomenon in cereals treated with jasmonate methylester. Commun. Agric. Appl. Biol. Sci. 2004, 69, 23–31. [Google Scholar] [PubMed]
- Grunwald, I.; Heinig, I.; Thole, H.H.; Neumann, D.; Kahmann, U.; Kloppstech, K.; Gau, A.E. Purification and characterization of a jacalin-related, coleoptile specific lectin from Hordeum vulgare. Planta 2007, 226, 225–234. [Google Scholar] [CrossRef]
- Wang, X.M.; Ma, Q.H. Characterization of a jasmonate- regulated wheat protein related to a beta-glucosidase aggregating factor. Plant Physiol. Biochem. 2005, 2, 185–192. [Google Scholar] [CrossRef]
- Quan, X.; Zeng, J.; Han, Z.; Zhang, G. Ionomic and physiological responses to low nitrogen stress in Tibetan wild and cultivated barley. Plant Physiol. Biochem. 2017, 111, 257–265. [Google Scholar] [CrossRef]
- Quan, X.; Zeng, J.; Ye, L.; Chen, G.; Han, Z.; Shah, J.M.; Zhang, G. Transcriptome profiling analysis for two Tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol. 2016, 16, 30. [Google Scholar] [CrossRef]
- Quan, X.; Zeng, J.; Chen, G.; Zhang, G. Transcriptomic analysis reveals adaptive strategies to chronic low nitrogen in Tibetan wild barley. BMC Plant Biol. 2019, 19, 68. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Li, L.; Luan, S. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 2004, 134, 1248–1267. [Google Scholar] [CrossRef]
- Marchler, B.A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, 222–226. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, 458–460. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; Castro, E.D.; Flegel, V.; Fortier, A.; Gasteiger, E.; Ioannidis, V. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams, C.C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- Bourne, Y.; Zamboni, V.; Barre, A.; Peumans, W.J.; Van Damme, E.J.; Rougé, P. Helianthus tuberosus lectin reveals a widespread scaffold for mannose-binding lectins. Structure 1999, 7, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.H.; Tian, B.; Li, Y.L. Overexpression of a wheat jasmonate-regulated lectin increases pathogen resistance. Biochimie 2010, 92, 187–193. [Google Scholar] [CrossRef]
- Ma, Q.H.; Zhen, W.B.; Liu, Y.C. Jacalin domain in wheat jasmonate-regulated protein Ta-JA1 confers agglutinating activity and pathogen resistance. Biochimie 2013, 2, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Teraoka, T.; Yamanaka, H.; Harashima, A.; Kunisaki, A.; Takahashi, H.; Hosolawa, D. Novel mannose-binding rice lectin composed of some isolectins and its relation to a stress-inducible salT gene. Plant Cell Physiol. 2000, 41, 258–267. [Google Scholar] [CrossRef]
- Kittur, F.S.; Lalgondar, M.; Yu, H.Y.; Bevan, D.R.; Esen, A. Maize beta-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for beta-glucosidase aggregation. J. Biol. Chem. 2007, 10, 7299–7311. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Girin, T.; Tillard, P.; Lepetit, M.; Gojon, A. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006, 140, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Kapoor, S.; Tyagi, A.K. Analysis of transcriptional and upstream regulatory sequence activity of two environmental stress-inducible genes, NBS-Str1 and BLEC-Str8, of rice. Transgenic Res. 2012, 21, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Bourne, Y.; Roig-Zamboni, V.; Barre, A.; Peumans, W.J.; Astoul, C.H.; Van Damme, E.J.; Rougé, P. The crystal structure of the Calystegia sepium agglutinin reveals a novel quaternary arrangement of lectin subunits with a beta-prism fold. J. Biol. Chem. 2004, 279, 527–533. [Google Scholar] [CrossRef]
- Liu, X.Y.; Li, H.; Zhang, W. The lectin from Musa paradisiaca binds with the capsid protein of tobacco mosaic virus and prevents viral infection. Biotechnol. Biotec Eq. 2014, 3, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, S.; Kito-Nakamura, K.; Matsuoka, K.; Morikami, A.; Nakamura, K. A major jasmonate-inducible protein of sweet potato, ipomoelin, is an ABA-independent wound-inducible protein. Plant Cell Physiol. 1997, 38, 643–652. [Google Scholar] [CrossRef][Green Version]
- Chang, W.C.; Liu, K.L.; Hsu, F.C.; Jeng, S.T.; Cheng, Y.S. Ipomoelin, a jacalin-related lectin with a compact tetrameric association and versatile carbohydrate binding properties regulated by its N terminus. PLoS ONE 2012, 7, e40618. [Google Scholar] [CrossRef]
- Chen, P.Y.; De Schutter, K.; Pauwels, J.; Gevaert, K.; Van Damme, E.J.M.; Smagghe, G. The lectin Orysata induces phosphatase-mediated and carbohydrate-independent aggregation of insect cells. J. Insect Physiol. 2021, 131, 104241. [Google Scholar] [CrossRef]
- Lee, R.H.; Wang, C.H.; Huang, L.T.; Chen, S.C.G. Leaf senescence in rice plants: Cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001, 52, 1117–1122. [Google Scholar] [CrossRef]
- Sofie, V.H.; Kristof, D.S.; Lore, E.; Mariya, T.; Dang, L.; Els, V.D. Comparative study of lectin domains in model species: New insights into evolutionary dynamics. Int. J. Mol. Sci. 2017, 18, 1136–1164. [Google Scholar]
- Jayaprakash, N.G.; Singh, A.; Vivek, R.; Yadav, S.; Pathak, S.; Trivedi, J.; Jayaraman, N.; Nandi, D.; Mitra, D.; Surolia, A. The barley lectin, horcolin, binds high-mannose glycans in a multivalent fashion, enabling high-affinity, specific inhibition of cellular HIV infection. J. Biol. Chem. 2020, 295, 12111–12129. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Gu, Z.; Cavalcanti, A.; Chen, F.C.; Bouman, P.; Li, W.H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Hu, H.; Zhu, B.; Jin, X.; Wu, F.; Zhang, G. Genotypic variations of N use efficiency in Tibetan wild and cultivated barleys. J. Zhejiang Univ. Agric. Life Sci. 2014, 40, 155–164. [Google Scholar]
- Kiba, T.; Feria-Bourrellier, A.B.; Lafouge, F.; Lezhneva, L.; Boutet-Mercey, S.; Orsel, M.; Brehaut, V.; Miller, A.; Daniel-Vedele, F.; Sakakibara, H.; et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 2012, 24, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.V.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.X.; Waugh, R.; Brown, J.W.S.; Schulman, A.; Langridge, P.; Platzer, M.; Fincher, G.B.; Muehlbauer, G.J.; Sato, K.; Close, T.J.; et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–716. [Google Scholar] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
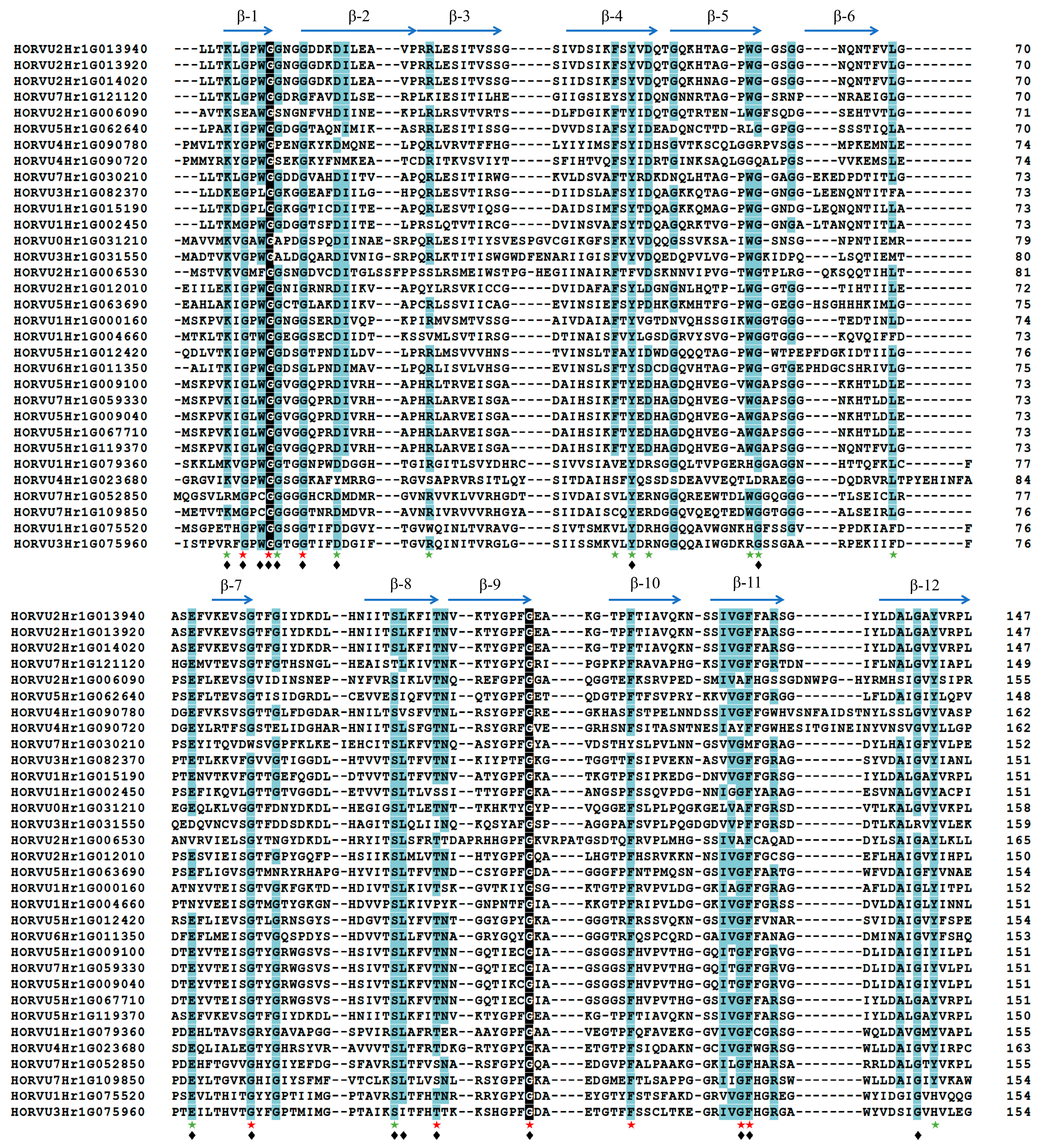
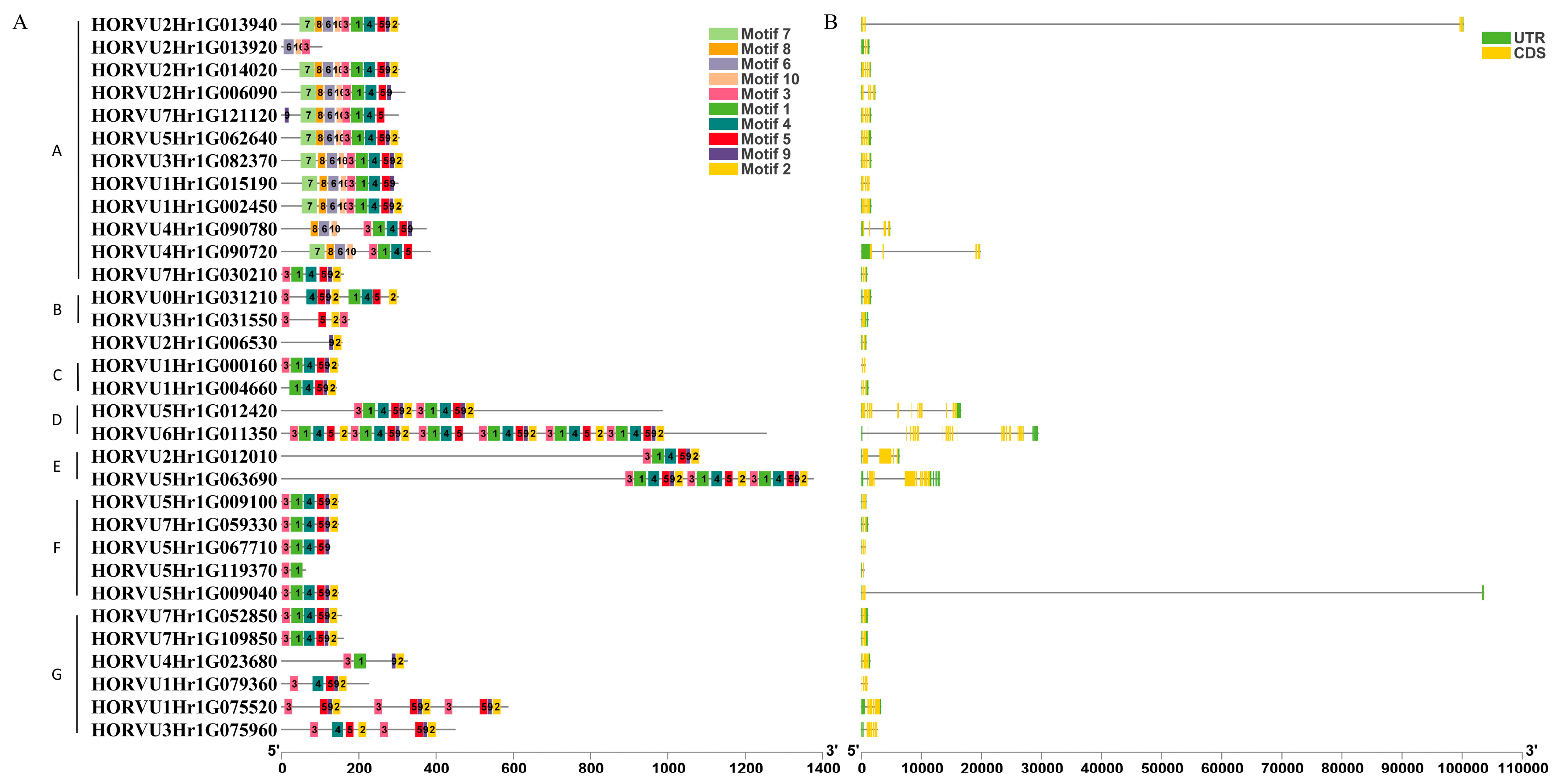
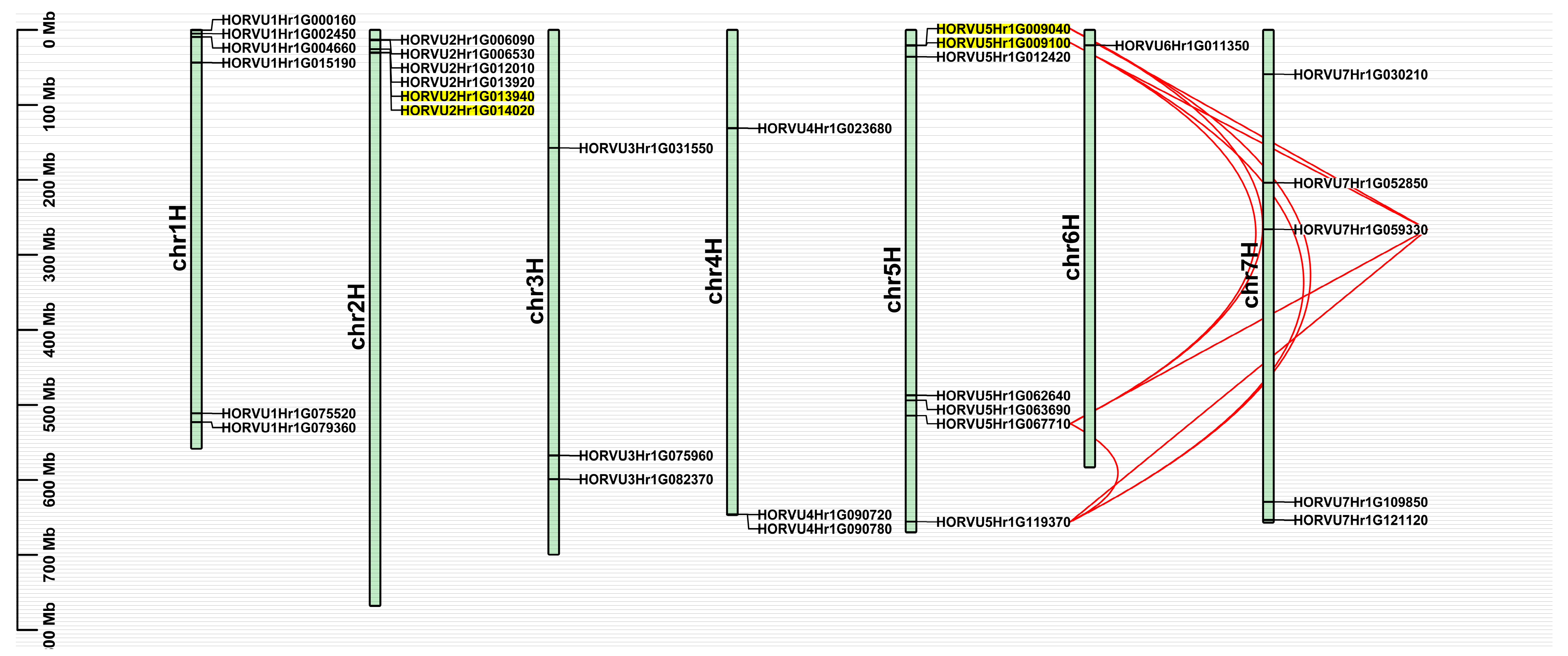

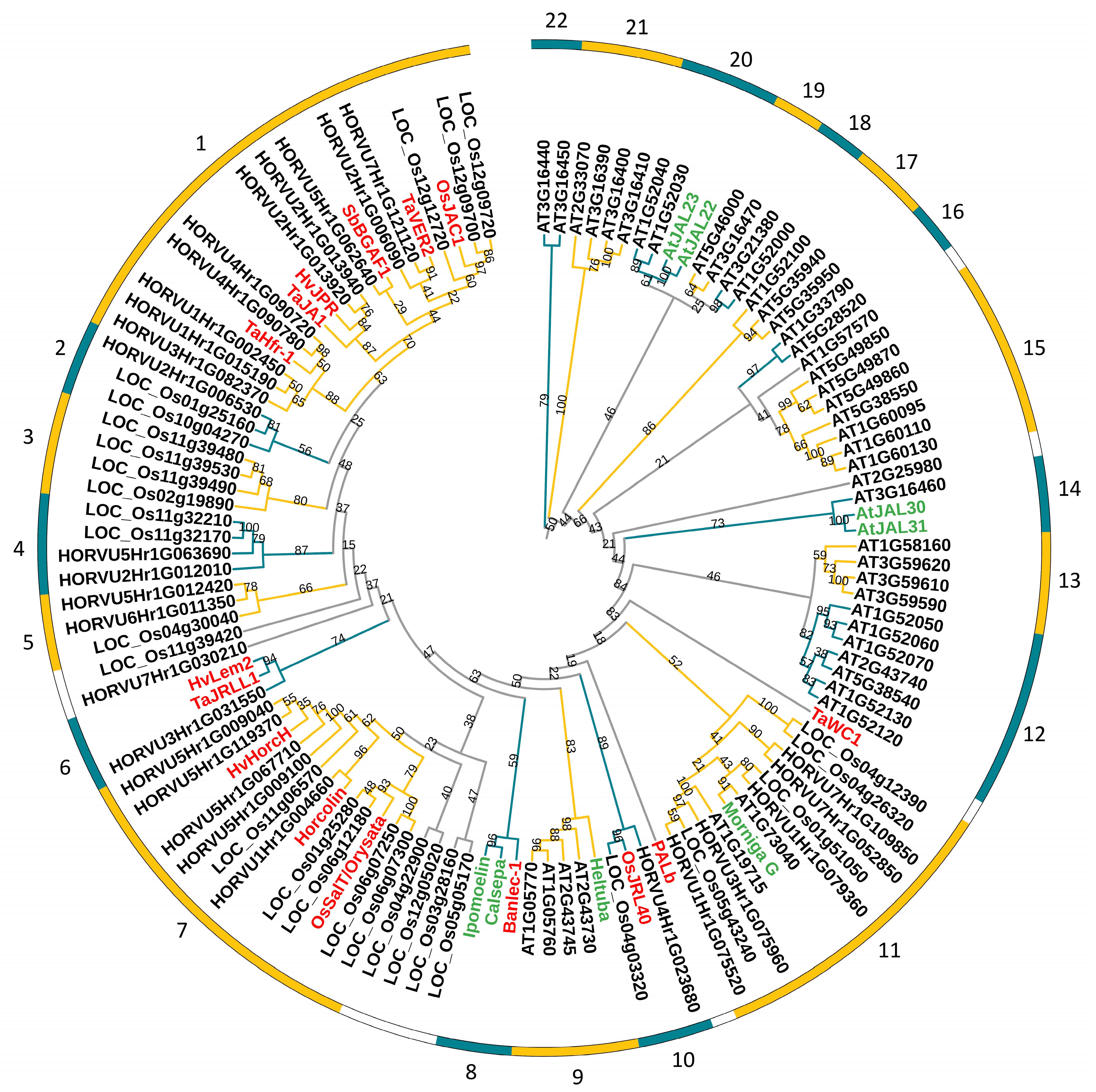
| Gene ID | Chromosome Location | Amino Acids | Molecular Weight (kDa) | pI |
|---|---|---|---|---|
| HORVU0Hr1G031210/LEM2 | chrUn:186390238-186406458 | 302 | 32.10 | 5.78 |
| HORVU1Hr1G000160/Horcolin | chr1H:475026-476135 | 146 | 15.05 | 7.95 |
| HORVU1Hr1G002450 | chr1H:4960525-4962190 | 314 | 33.16 | 6.94 |
| HORVU1Hr1G004660 | chr1H:9609641-9610836 | 142 | 14.94 | 4.95 |
| HORVU1Hr1G015190 | chr1H:43686713-43688076 | 301 | 32.66 | 5.57 |
| HORVU1Hr1G075520 | chr1H:511072663-511076165 | 586 | 63.44 | 6.64 |
| HORVU1Hr1G079360 | chr1H:522823378-522824959 | 208 | 22.34 | 9.22 |
| HORVU2Hr1G006090 | chr2H:12993110-12995463 | 319 | 35.34 | 6 |
| HORVU2Hr1G006530 | chr2H:13676587-13677516 | 156 | 16.89 | 9.34 |
| HORVU2Hr1G012010 | chr2H:25563354-25569794 | 1082 | 122.10 | 6.55 |
| HORVU2Hr1G013920 | chr2H:30286228-30287832 | 104 | 11.10 | 5.23 |
| HORVU2Hr1G013940 | chr2H:30372662-30472925 | 304 | 32.69 | 5.64 |
| HORVU2Hr1G014020/HvJPR | chr2H:30577014-30745532 | 304 | 32.66 | 6.17 |
| HORVU3Hr1G031550 | chr3H:157387209-157388376 | 175 | 18.77 | 4.31 |
| HORVU3Hr1G075960 | chr3H:567163924-567167067 | 449 | 48.41 | 9.18 |
| HORVU3Hr1G082370 | chr3H:598849424-598851100 | 314 | 33.72 | 5.98 |
| HORVU4Hr1G023680 | chr4H:131046735-131048211 | 325 | 35.11 | 9.64 |
| HORVU4Hr1G0907204 | chr4H:645950575-645970388 | 385 | 41.87 | 8.25 |
| HORVU4Hr1G090780 | chr4H:645995092-646003566 | 374 | 40.25 | 9 |
| HORVU5Hr1G009040 | chr5H:20488797-20637689 | 147 | 15.55 | 6.42 |
| HORVU5Hr1G009100 | chr5H:20927441-21001231 | 147 | 15.64 | 6.21 |
| HORVU5Hr1G012420 | chr5H:35699858-35716432 | 986 | 109.11 | 6.15 |
| HORVU5Hr1G062640 | chr5H:487199094-487200722 | 304 | 33.06 | 5.24 |
| HORVU5Hr1G063690 | chr5H:493882556-493895687 | 1376 | 153.70 | 6.46 |
| HORVU5Hr1G067710 | chr5H:514246154-514246870 | 125 | 13.21 | 6.21 |
| HORVU5Hr1G119370 | chr5H:655562953-655564277 | 62 | 6.65 | 6.96 |
| HORVU6Hr1G011350 | chr6H:20611501-20640921 | 1254 | 134.57 | 6.75 |
| HORVU7Hr1G030210 | chr7H:59140290-59141279 | 160 | 17.25 | 5.03 |
| HORVU7Hr1G052850 | chr7H:203881126-203882208 | 155 | 16.63 | 6.5 |
| HORVU7Hr1G059330/HvHorcH | chr7H:265851060-265852199 | 147 | 15.58 | 6.02 |
| HORVU7Hr1G109850 | chr7H:629167675-629168735 | 160 | 17.45 | 7.72 |
| HORVU7Hr1G121120 | chr7H:653151090-653152795 | 302 | 32.80 | 7.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quan, X.; Meng, C.; Xie, C.; Sun, H.; Xu, B.; Santos Bermudez, R.; He, W. Genome-Wide and Transcriptome Analysis of Jacalin-Related Lectin Genes in Barley and the Functional Characterization of HvHorcH in Low-Nitrogen Tolerance in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 16641. https://doi.org/10.3390/ijms242316641
Quan X, Meng C, Xie C, Sun H, Xu B, Santos Bermudez R, He W. Genome-Wide and Transcriptome Analysis of Jacalin-Related Lectin Genes in Barley and the Functional Characterization of HvHorcH in Low-Nitrogen Tolerance in Arabidopsis. International Journal of Molecular Sciences. 2023; 24(23):16641. https://doi.org/10.3390/ijms242316641
Chicago/Turabian StyleQuan, Xiaoyan, Chen Meng, Chunjuan Xie, Huifang Sun, Boyang Xu, Ramon Santos Bermudez, and Wenxing He. 2023. "Genome-Wide and Transcriptome Analysis of Jacalin-Related Lectin Genes in Barley and the Functional Characterization of HvHorcH in Low-Nitrogen Tolerance in Arabidopsis" International Journal of Molecular Sciences 24, no. 23: 16641. https://doi.org/10.3390/ijms242316641
APA StyleQuan, X., Meng, C., Xie, C., Sun, H., Xu, B., Santos Bermudez, R., & He, W. (2023). Genome-Wide and Transcriptome Analysis of Jacalin-Related Lectin Genes in Barley and the Functional Characterization of HvHorcH in Low-Nitrogen Tolerance in Arabidopsis. International Journal of Molecular Sciences, 24(23), 16641. https://doi.org/10.3390/ijms242316641






