Stem Cell Responsiveness to Imatinib in Chronic Myeloid Leukemia
Abstract
:1. Introduction
2. Results
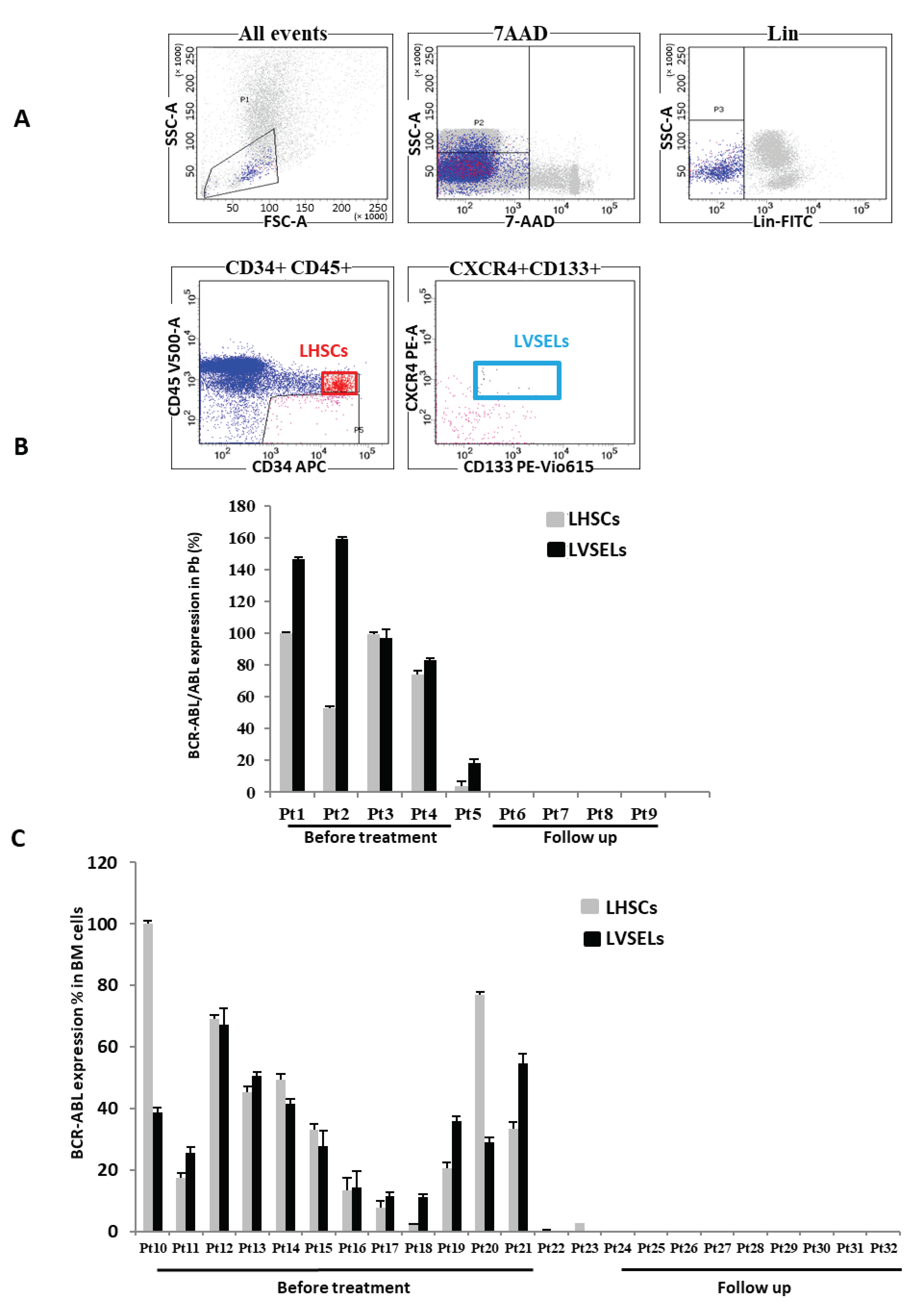
2.1. BCR-ABL Expression in Leukemic LHSCs and LVSELs
2.2. Quantification of Lin−CD34+ Stem Cell Populations’ Responsiveness to IM
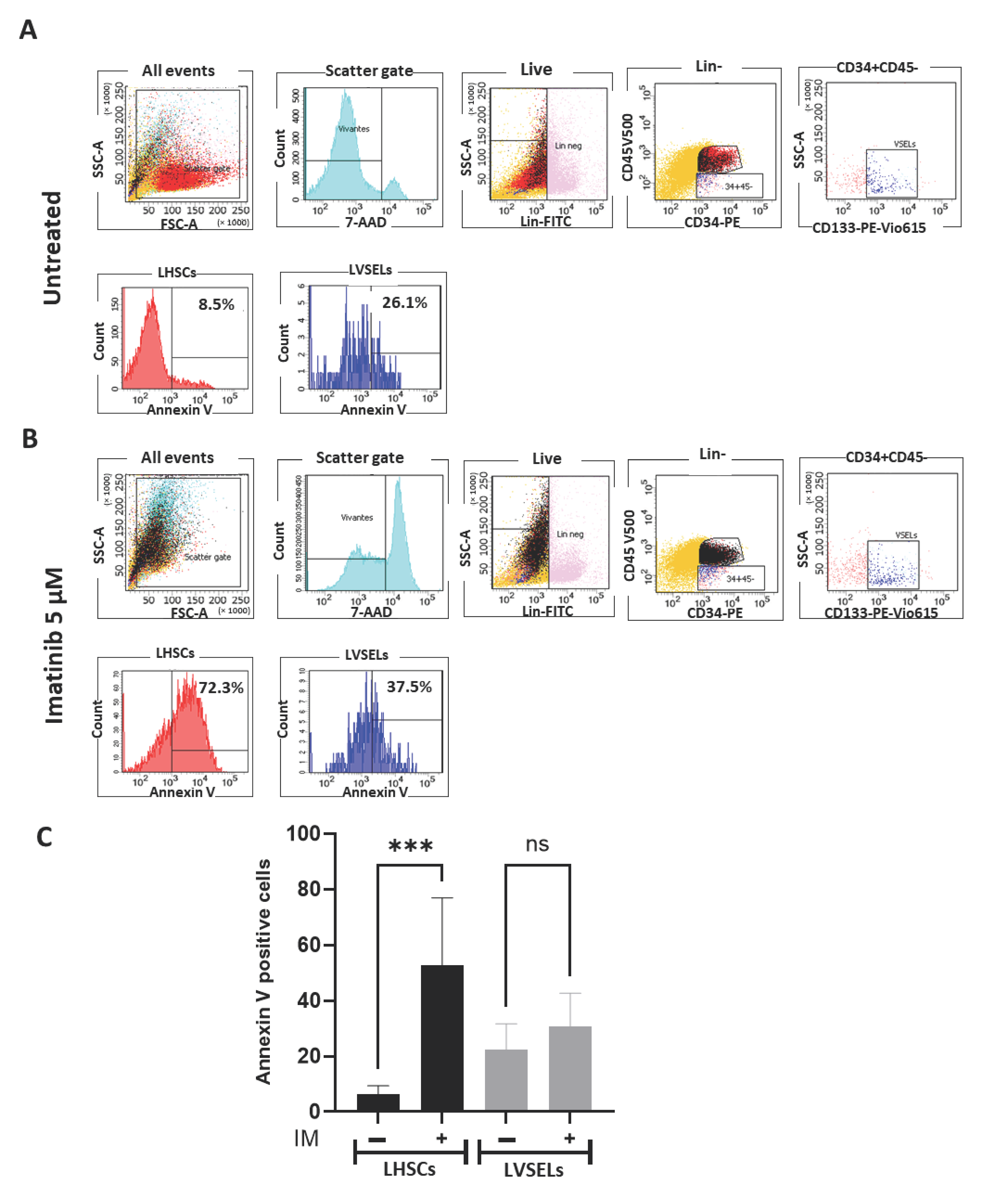
2.3. Quantification of Stem Cell Survival
2.4. Quantification of Gene Expression
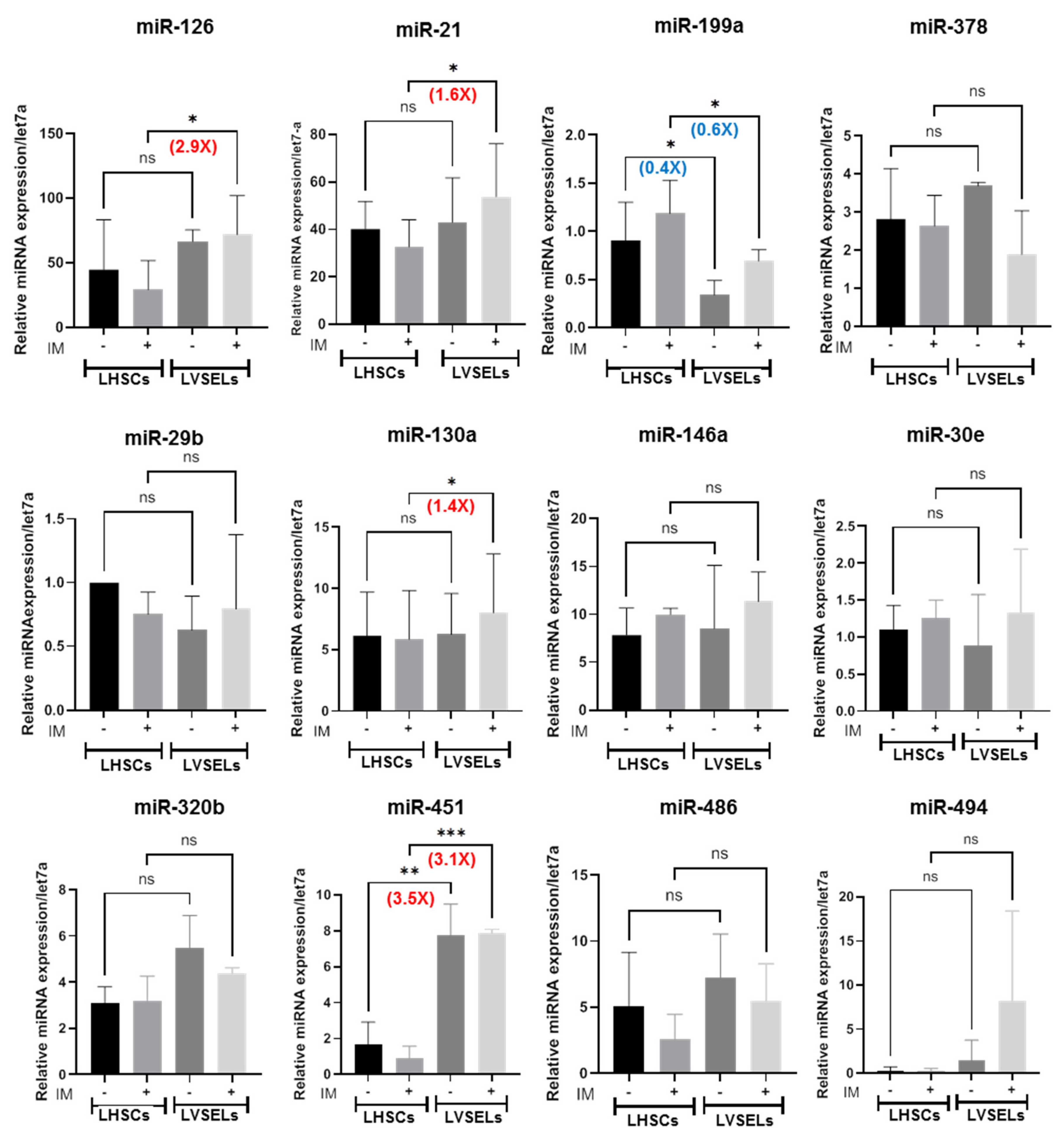
2.5. miRNA Expression in Lin−CD34+CD45+ and Lin−CD34+CD45− CML Stem Cells
3. Discussion
4. Materials and Methods
4.1. Stem Cell Expansion
4.2. Flow Cytometry Analysis
4.3. RNA Isolation and Quantitative RT-PCR
4.4. MicroRNA Quantification
4.5. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cortes, J.; Pavlovsky, C.; Saußele, S. Chronic Myeloid Leukaemia. Lancet 2021, 398, 1914–1926. [Google Scholar] [CrossRef] [PubMed]
- von Bubnoff, N.; Schneller, F.; Peschel, C.; Duyster, J. BCR-ABL Gene Mutations in Relation to Clinical Resistance of Philadelphia-Chromosome-Positive Leukaemia to STI571: A Prospective Study. Lancet 2002, 359, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Amarante-Mendes, G.P.; Rana, A.; Datoguia, T.S.; Hamerschlak, N.; Brumatti, G. BCR-ABL1 Tyrosine Kinase Complex Signaling Transduction: Challenges to Overcome Resistance in Chronic Myeloid Leukemia. Pharmaceutics 2022, 14, 215. [Google Scholar] [CrossRef]
- Kucia, M.; Reca, R.; Campbell, F.R.; Zuba-Surma, E.; Majka, M.; Ratajczak, J.; Ratajczak, M.Z. A Population of Very Small Embryonic-like (VSEL) CXCR4(+)SSEA-1(+)Oct-4+ Stem Cells Identified in Adult Bone Marrow. Leukemia 2006, 20, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z. Why Are Hematopoietic Stem Cells so ‘Sexy’? On a Search for Developmental Explanation. Leukemia 2017, 31, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Corbin, A.S.; Agarwal, A.; Loriaux, M.; Cortes, J.; Deininger, M.W.; Druker, B.J. Human Chronic Myeloid Leukemia Stem Cells Are Insensitive to Imatinib despite Inhibition of BCR-ABL Activity. J. Clin. Investig. 2011, 121, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; McDonald, T.; Lin, A.; Chakraborty, S.; Huang, Q.; Snyder, D.S.; Bhatia, R. Persistence of Leukemia Stem Cells in Chronic Myelogenous Leukemia Patients in Prolonged Remission with Imatinib Treatment. Blood 2011, 118, 5565–5572. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.E.A.; Wong, J.J.-L.; Rasko, J.E.J. MicroRNAs in Myeloid Malignancies. Br. J. Haematol. 2013, 162, 162–176. [Google Scholar] [CrossRef]
- Litwińska, Z.; Machaliński, B. miRNAs in Chronic Myeloid Leukemia: Small Molecules, Essential Function. Leuk. Lymphoma 2017, 58, 1297–1305. [Google Scholar] [CrossRef]
- Rudich, A.; Garzon, R.; Dorrance, A. Non-Coding RNAs Are Implicit in Chronic Myeloid Leukemia Therapy Resistance. Int. J. Mol. Sci. 2022, 23, 12271. [Google Scholar] [CrossRef]
- Salati, S.; Salvestrini, V.; Carretta, C.; Genovese, E.; Rontauroli, S.; Zini, R.; Rossi, C.; Ruberti, S.; Bianchi, E.; Barbieri, G.; et al. Deregulated Expression of miR-29a-3p, miR-494-3p and miR-660-5p Affects Sensitivity to Tyrosine Kinase Inhibitors in CML Leukemic Stem Cells. Oncotarget 2017, 8, 49451–49469. [Google Scholar] [CrossRef] [PubMed]
- Maj, M.; Schneider, G.; Ratajczak, J.; Suszynska, M.; Kucia, M.; Ratajczak, M.Z. The Cell Cycle- and Insulin-Signaling-Inhibiting miRNA Expression Pattern of Very Small Embryonic-like Stem Cells Contributes to Their Quiescent State. Exp. Biol. Med. 2015, 240, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Lahlil, R.; Scrofani, M.; Barbet, R.; Tancredi, C.; Aries, A.; Hénon, P. VSELs Maintain Their Pluripotency and Competence to Differentiate after Enhanced Ex Vivo Expansion. Stem Cell Rev. Rep. 2018, 14, 510–524. [Google Scholar] [CrossRef] [PubMed]
- Nakai, S.; Masaki, T.; Shiratori, Y.; Ohgi, T.; Morishita, A.; Kurokohchi, K.; Watanabe, S.; Kuriyama, S. Expression of P57(KIP2) in Hepatocellular Carcinoma: Relationship between Tumor Differentiation and Patient Survival. Int. J. Oncol. 2002, 20, 769–775. [Google Scholar] [PubMed]
- Borriello, A.; Caldarelli, I.; Bencivenga, D.; Cucciolla, V.; Oliva, A.; Usala, E.; Danise, P.; Ronzoni, L.; Perrotta, S.; Della Ragione, F. p57Kip2 Is a Downstream Effector of BCR-ABL Kinase Inhibitors in Chronic Myelogenous Leukemia Cells. Carcinogenesis 2011, 32, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, P.; Nyman, U.; Hajji, N.; Joseph, B. The Cell Cycle Inhibitor p57Kip2 Promotes Cell Death via the Mitochondrial Apoptotic Pathway. Cell Death Differ. 2007, 14, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Fukunaka, A.; Hagihara, M.; Watanabe, K.; Kamino, S.; Kambe, T.; Enomoto, S.; Hiromura, M. Essential Role of the Zinc Transporter ZIP9/SLC39A9 in Regulating the Activations of Akt and Erk in B-Cell Receptor Signaling Pathway in DT40 Cells. PLoS ONE 2013, 8, e58022. [Google Scholar] [CrossRef]
- Zhang, B.; Nguyen, L.X.T.; Li, L.; Zhao, D.; Kumar, B.; Wu, H.; Lin, A.; Pellicano, F.; Hopcroft, L.; Su, Y.-L.; et al. Bone Marrow Niche Trafficking of miR-126 Controls the Self-Renewal of Leukemia Stem Cells in Chronic Myelogenous Leukemia. Nat. Med. 2018, 24, 450–462. [Google Scholar] [CrossRef]
- Ferreira, A.F.; Moura, L.G.; Tojal, I.; Ambrósio, L.; Pinto-Simões, B.; Hamerschlak, N.; Calin, G.A.; Ivan, C.; Covas, D.T.; Kashima, S.; et al. ApoptomiRs Expression Modulated by BCR-ABL Is Linked to CML Progression and Imatinib Resistance. Blood Cells Mol. Dis. 2014, 53, 47–55. [Google Scholar] [CrossRef]
- Etuknwa, A.; Daniels, K.; Eib, C. Sustainable Return to Work: A Systematic Review Focusing on Personal and Social Factors. J. Occup. Rehabil. 2019, 29, 679–700. [Google Scholar] [CrossRef]
- Habib, E.M.; Nosiar, N.A.; Eid, M.A.; Taha, A.M.; Sherief, D.E.; Hassan, A.E.; Abdel Ghafar, M.T. Circulating miR-146a Expression Predicts Early Treatment Response to Imatinib in Adult Chronic Myeloid Leukemia. J. Investig. Med. 2021, 69, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-H.; Liu, A.-J.; Ho, K.-H.; Chiu, Y.-T.; Anne Lin, Z.-H.; Lee, Y.-T.; Shih, C.-M.; Chen, K.-C. microRNA-199a/b-5p Enhance Imatinib Efficacy via Repressing WNT2 Signaling-Mediated Protective Autophagy in Imatinib-Resistant Chronic Myeloid Leukemia Cells. Chem. Biol. Interact. 2018, 291, 144–151. [Google Scholar] [CrossRef]
- Martins, J.R.B.; Moraes, L.N.; Cury, S.S.; Capannacci, J.; Carvalho, R.F.; Nogueira, C.R.; Hokama, N.K.; Hokama, P.O.M. MiR-125a-3p and MiR-320b Differentially Expressed in Patients with Chronic Myeloid Leukemia Treated with Allogeneic Hematopoietic Stem Cell Transplantation and Imatinib Mesylate. Int. J. Mol. Sci. 2021, 22, 10216. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, B.M. Consent Including Sexual Dysfunction and Pain with Sexual Activity after Inguinal Hernia Repair? J. Am. Coll. Surg. 2020, 230, 1123–1124. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-S.; Li, L.; Li, L.; Chu, S.; Shiang, K.-D.; Li, M.; Sun, H.-Y.; Xu, J.; Xiao, F.-J.; Sun, G.; et al. MicroRNA-486 Regulates Normal Erythropoiesis and Enhances Growth and Modulates Drug Response in CML Progenitors. Blood 2015, 125, 1302–1313. [Google Scholar] [CrossRef]
- Testa, U.; Saulle, E.; Castelli, G.; Pelosi, E. Endothelial Progenitor Cells in Hematologic Malignancies. Stem Cell Investig. 2016, 3, 26. [Google Scholar] [CrossRef]
- Fang, B.; Zheng, C.; Liao, L.; Han, Q.; Sun, Z.; Jiang, X.; Zhao, R.C.H. Identification of Human Chronic Myelogenous Leukemia Progenitor Cells with Hemangioblastic Characteristics. Blood 2005, 105, 2733–2740. [Google Scholar] [CrossRef]
- Gunsilius, E.; Duba, H.C.; Petzer, A.L.; Kähler, C.M.; Grünewald, K.; Stockhammer, G.; Gabl, C.; Dirnhofer, S.; Clausen, J.; Gastl, G. Evidence from a Leukaemia Model for Maintenance of Vascular Endothelium by Bone-Marrow-Derived Endothelial Cells. Lancet 2000, 355, 1688–1691. [Google Scholar] [CrossRef]
- Holyoake, T.; Jiang, X.; Eaves, C.; Eaves, A. Isolation of a Highly Quiescent Subpopulation of Primitive Leukemic Cells in Chronic Myeloid Leukemia. Blood 1999, 94, 2056–2064. [Google Scholar] [CrossRef]
- Holyoake, T.L.; Vetrie, D. The Chronic Myeloid Leukemia Stem Cell: Stemming the Tide of Persistence. Blood 2017, 129, 1595–1606. [Google Scholar] [CrossRef]
- Gerber, J.M.; Qin, L.; Kowalski, J.; Smith, B.D.; Griffin, C.A.; Vala, M.S.; Collector, M.I.; Perkins, B.; Zahurak, M.; Matsui, W.; et al. Characterization of Chronic Myeloid Leukemia Stem Cells. Am. J. Hematol. 2011, 86, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.M. Primitive, Quiescent, Philadelphia-Positive Stem Cells from Patients with Chronic Myeloid Leukemia Are Insensitive to STI571 in Vitro. Blood 2002, 99, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Copland, M.; Hamilton, A.; Elrick, L.J.; Baird, J.W.; Allan, E.K.; Jordanides, N.; Barow, M.; Mountford, J.C.; Holyoake, T.L. Dasatinib (BMS-354825) Targets an Earlier Progenitor Population than Imatinib in Primary CML but Does Not Eliminate the Quiescent Fraction. Blood 2006, 107, 4532–4539. [Google Scholar] [CrossRef] [PubMed]
- Stoklosa, T.; Poplawski, T.; Koptyra, M.; Nieborowska-Skorska, M.; Basak, G.; Slupianek, A.; Rayevskaya, M.; Seferynska, I.; Herrera, L.; Blasiak, J.; et al. BCR/ABL Inhibits Mismatch Repair to Protect from Apoptosis and Induce Point Mutations. Cancer Res. 2008, 68, 2576–2580. [Google Scholar] [CrossRef] [PubMed]
- Skorski, T. BCR/ABL, DNA Damage and DNA Repair: Implications for New Treatment Concepts. Leuk. Lymphoma 2008, 49, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Creff, J.; Besson, A. Functional Versatility of the CDK Inhibitor p57Kip2. Front. Cell Dev. Biol. 2020, 8, 584590. [Google Scholar] [CrossRef] [PubMed]
- Rangatia, J.; Bonnet, D. Transient or Long-Term Silencing of BCR-ABL Alone Induces Cell Cycle and Proliferation Arrest, Apoptosis and Differentiation. Leukemia 2006, 20, 68–76. [Google Scholar] [CrossRef]
- Mirza, M.A.B.; Guru, S.A.; Abdullah, S.M.; Rizvi, A.; Saxena, A. microRNA-21 Expression as Prognostic and Therapeutic Response Marker in Chronic Myeloid Leukaemia Patients. Asian Pac. J. Cancer Prev. 2019, 20, 2379–2383. [Google Scholar] [CrossRef]
- Alves, R.; Gonçalves, A.C.; Jorge, J.; Marques, G.; Luís, D.; Ribeiro, A.B.; Freitas-Tavares, P.; Oliveiros, B.; Almeida, A.M.; Sarmento-Ribeiro, A.B. MicroRNA Signature Refine Response Prediction in CML. Sci. Rep. 2019, 9, 9666. [Google Scholar] [CrossRef]
- Wang, W.-Z.; Pu, Q.-H.; Lin, X.-H.; Liu, M.-Y.; Wu, L.-R.; Wu, Q.-Q.; Chen, Y.-H.; Liao, F.-F.; Zhu, J.-Y.; Jin, X.-B. Silencing of miR-21 Sensitizes CML CD34+ Stem/Progenitor Cells to Imatinib-Induced Apoptosis by Blocking PI3K/AKT Pathway. Leuk. Res. 2015, 39, 1117–1124. [Google Scholar] [CrossRef]
- Li, Q.; Wu, Y.; Zhang, J.; Yi, T.; Li, W. MicroRNA-130a Regulates Cell Malignancy by Targeting RECK in Chronic Myeloid Leukemia. Am. J. Transl. Res. 2016, 8, 955–967. [Google Scholar] [PubMed]
- Abdulmawjood, B.; Costa, B.; Roma-Rodrigues, C.; Baptista, P.V.; Fernandes, A.R. Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far? Int. J. Mol. Sci. 2021, 22, 12516. [Google Scholar] [CrossRef] [PubMed]
- Joshi, D.; Chandrakala, S.; Korgaonkar, S.; Ghosh, K.; Vundinti, B.R. Down-Regulation of miR-199b Associated with Imatinib Drug Resistance in 9q34.1 Deleted BCR/ABL Positive CML Patients. Gene 2014, 542, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cui, M.-M.; Zhu, H.-Z.; Fu, P.-Y.; Wang, G.-C.; Huang, L. MiR-199a-3p Overexpression Suppressed Cell Proliferation and Sensitized Chronic Myeloid Leukaemia Cells to Imatinib by Inhibiting mTOR Signalling. Acta Haematol. 2022, 145, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Volinia, S.; Liu, C.-G.; Fernandez-Cymering, C.; Palumbo, T.; Pichiorri, F.; Fabbri, M.; Coombes, K.; Alder, H.; Nakamura, T.; et al. MicroRNA Signatures Associated with Cytogenetics and Prognosis in Acute Myeloid Leukemia. Blood 2008, 111, 3183–3189. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wu, D.; Yi, J.; Yi, Y.; Zhu, X.; Qiu, H.; Kong, R.; Lin, J.; Qian, J.; Deng, Z. MiR-378 Promoted Cell Proliferation and Inhibited Apoptosis by Enhanced Stem Cell Properties in Chronic Myeloid Leukemia K562 Cells. Biomed. Pharmacother. 2019, 112, 108623. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Tao, K.; Xiao, Q.; Huang, Z.; Zhong, L.; Cao, W.; Wen, J.; Feng, W. miR-29b Suppresses CML Cell Proliferation and Induces Apoptosis via Regulation of BCR/ABL1 Protein. Exp. Cell Res. 2013, 319, 1094–1101. [Google Scholar] [CrossRef]
- Bhartiya, D.; Jha, N.; Tripathi, A.; Tripathi, A. Very Small Embryonic-like Stem Cells Have the Potential to Win the Three-Front War on Tissue Damage, Cancer, and Aging. Front. Cell Dev. Biol. 2023, 10, 1061022. [Google Scholar] [CrossRef]



| Reference Range (M/F) # | Diagnosis (n = 16) | Follow Up (n = 20) | |
|---|---|---|---|
| Gender (M/F) | - | (10/6) | (14/6) |
| Age (year) | - | 57 ± 16 | 56 ± 15 |
| WBC (×109/L) | 3.9–10.9/4.9–12.68 | 92 ± 76 | 6 ± 2 |
| RBC (×1012/L) | 4.44–5.61/3.92–5.08 | 4 ± 1 | 4 ± 1 |
| Hgb (g/dL) | 13.5–16.9/11.9–14.6 | 12 ± 2 | 13 ± 3 |
| hematocrit | 40.0–49.4/36.6–44.0 | 38 ± 5 | 38 ± 10 |
| PLT (×109/L) | 166–308/173–390 | 586 ± 560 | 204 ± 91 |
| Primer Sequences | |
|---|---|
| BCR ABL M Fw | TCC GCT GAC CAT CAA TAA GGA |
| ABL Rv | CAC TCA GAC CCT GAG GCT CAA |
| Probe BCRABL | [6-FAM] CCC TTC AGC GGC CAG TAG CAT CTG A [BHQ1] |
| ABL1 Fw | TGG AGA TAA CAC TCT AAG CAT AAC TAA AGG |
| ABL2 Rv | GAT GTA GTT GCT TGG GAC CCA |
| Probe ABL | [6-FAM] CCA TTT TTG GTT TGG GCT TCA CAC CAT T [BHQ1] |
| KI67 Fw | ATTGAACCTGCGGAAGAGCTGA |
| KI67 Rv | GGAGCGCAGGGATATTCCCTTA |
| P57 Kip2 Fw | GCGGCGATCAAGAAGCTGT |
| P57 Kip2 Rv | TGGCGAAGAAATCGGAGATCA |
| p21 Cip1 Fw | TGTCCGTCAGAACCCATGC |
| p21 Cip1 Rv | AAAGTCGAAGTTCCATCGCTC |
| GAPDH Fw | CATCGCTCAGACACCATGG |
| GAPDH Rv | ATGTAGTTGAGGTCAATGAAGGG |
| β2-microglobulin Fw | TGACTTTGTCACAGCCCAAGATA |
| β2-microglobulin Rv | AATGCGGCATCTTCAAACCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lahlil, R.; Aries, A.; Scrofani, M.; Zanetti, C.; Hennequin, D.; Drénou, B. Stem Cell Responsiveness to Imatinib in Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2023, 24, 16671. https://doi.org/10.3390/ijms242316671
Lahlil R, Aries A, Scrofani M, Zanetti C, Hennequin D, Drénou B. Stem Cell Responsiveness to Imatinib in Chronic Myeloid Leukemia. International Journal of Molecular Sciences. 2023; 24(23):16671. https://doi.org/10.3390/ijms242316671
Chicago/Turabian StyleLahlil, Rachid, Anne Aries, Maurice Scrofani, Céline Zanetti, Desline Hennequin, and Bernard Drénou. 2023. "Stem Cell Responsiveness to Imatinib in Chronic Myeloid Leukemia" International Journal of Molecular Sciences 24, no. 23: 16671. https://doi.org/10.3390/ijms242316671






