A High-Fat Diet Increases the Characteristics of Gut Microbial Composition and the Intestinal Damage Associated with Non-Alcoholic Fatty Liver Disease
Abstract
:1. Introduction
2. Results
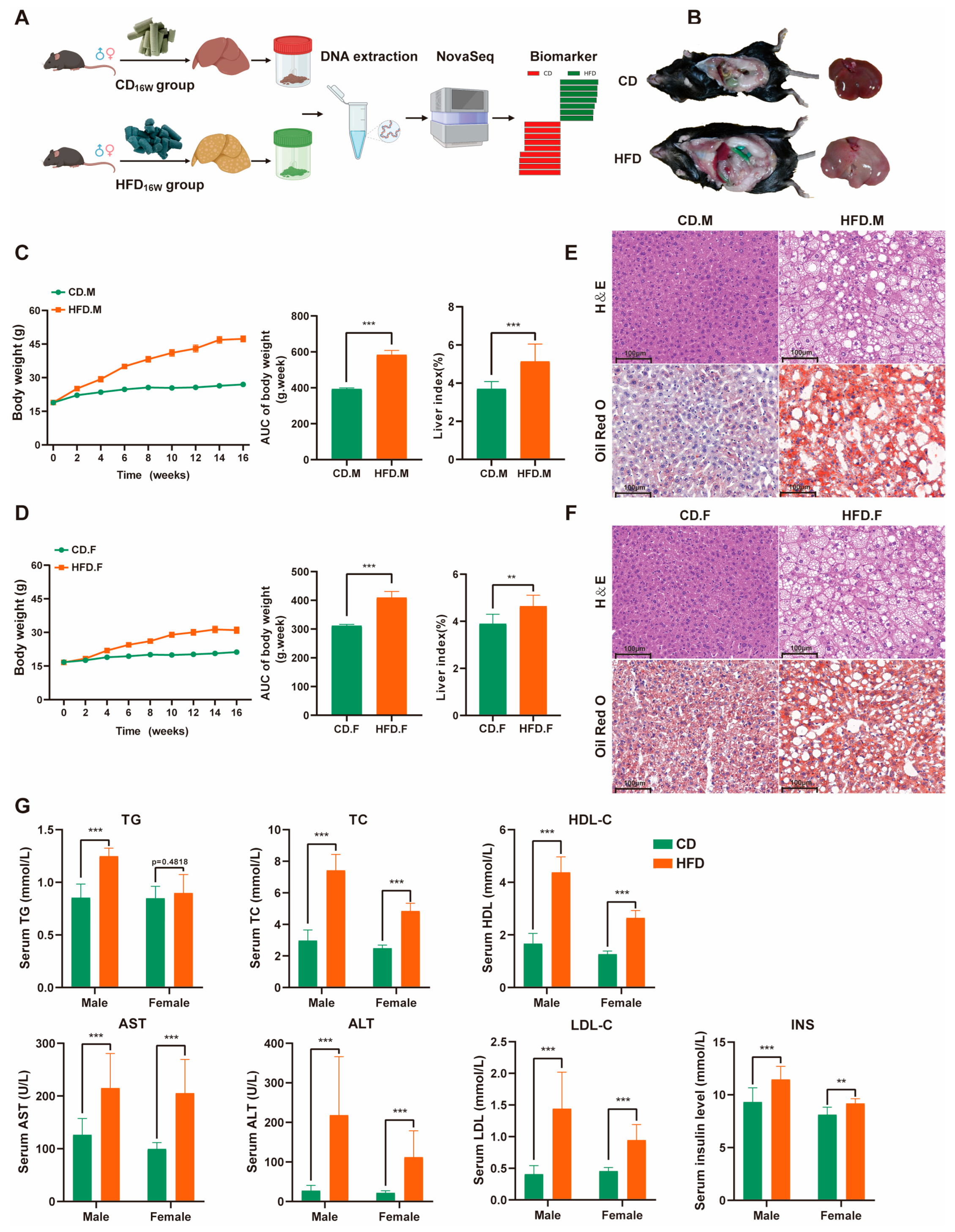
2.1. High-Fat Diet (HFD) Triggered Weight Gain and Lipid Accumulation
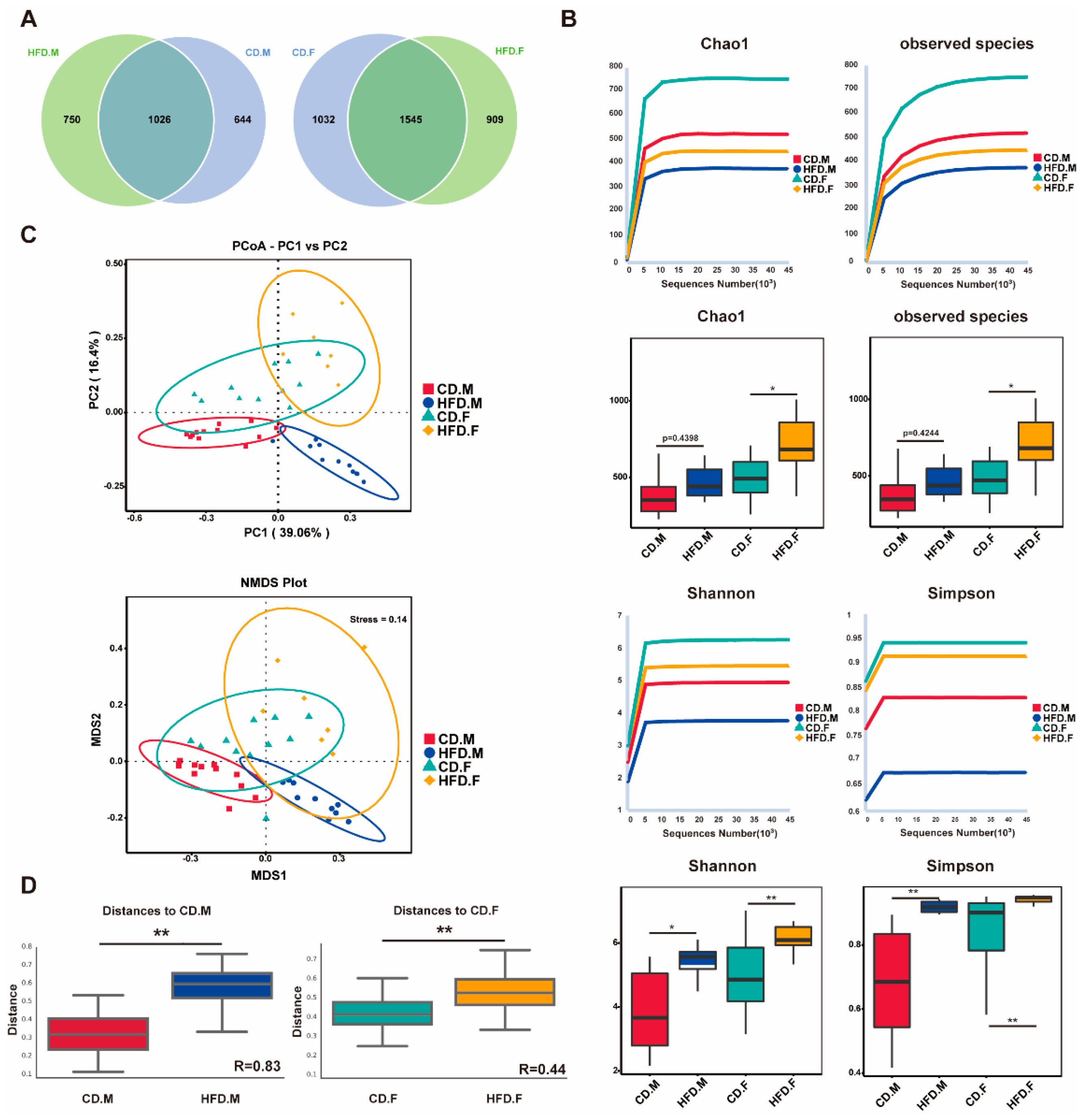
2.2. HFD Enhanced Biodiversity and Altered Gut Microbial Composition
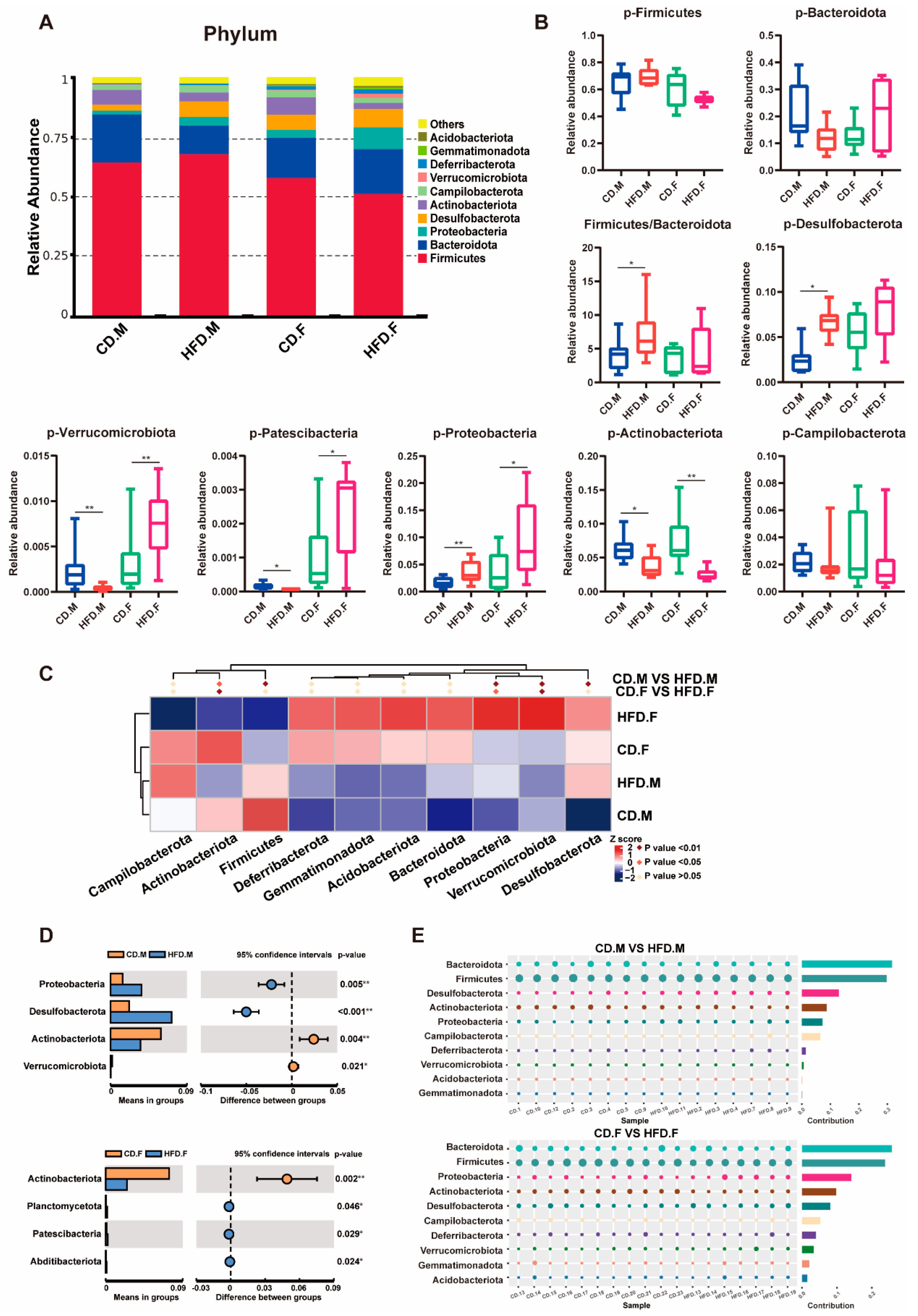
2.3. Relative Abundance of Bacteria at Phylum Level in the Intestinal Tract of CD and HFD Mice
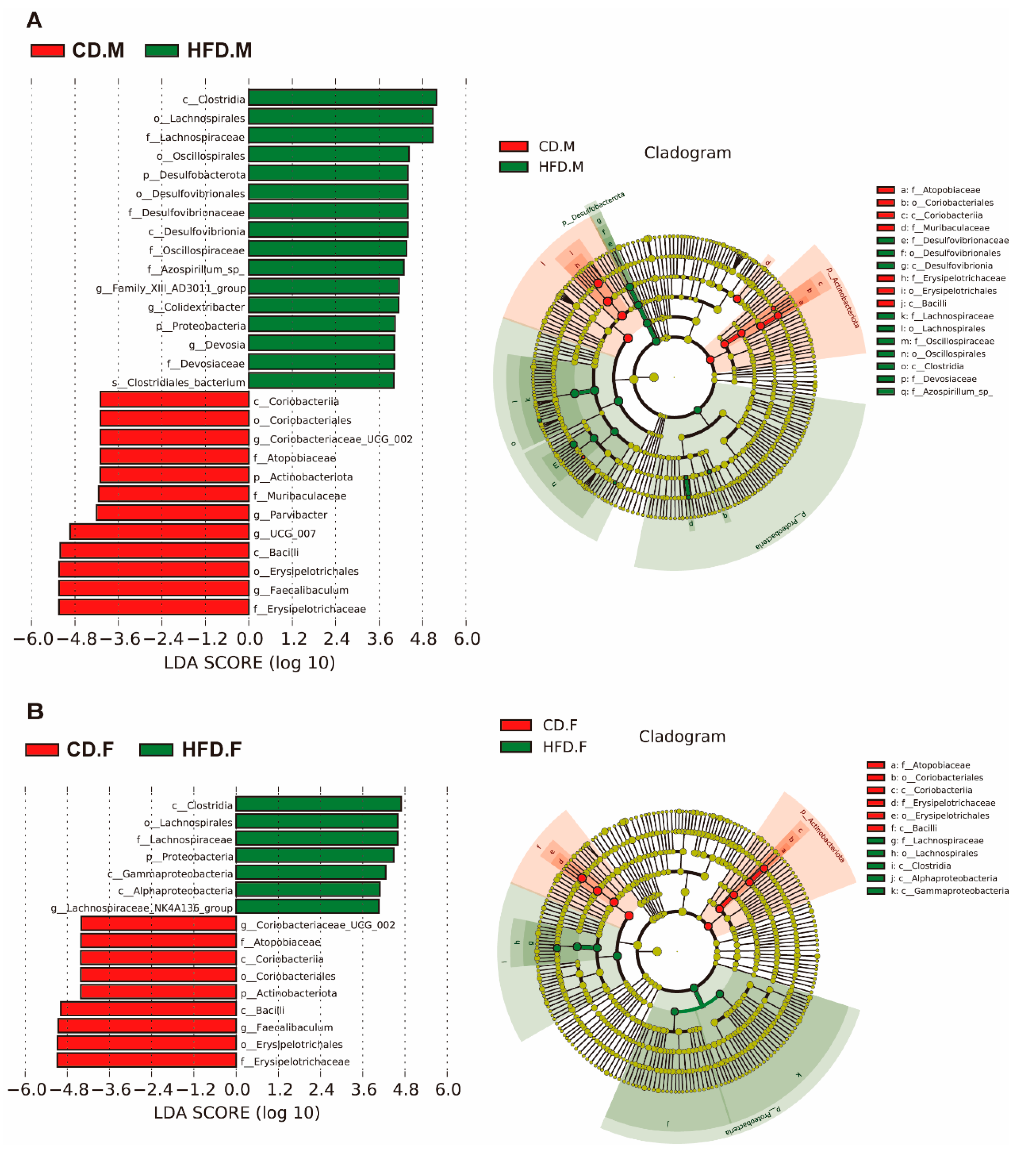
2.4. Relative Abundance of Bacteria at Genus Level in the Intestinal Tract of CD and HFD Mice
2.5. Biomarkers of Gut Microbiota in the Intestinal Tract of Mice with NAFLD
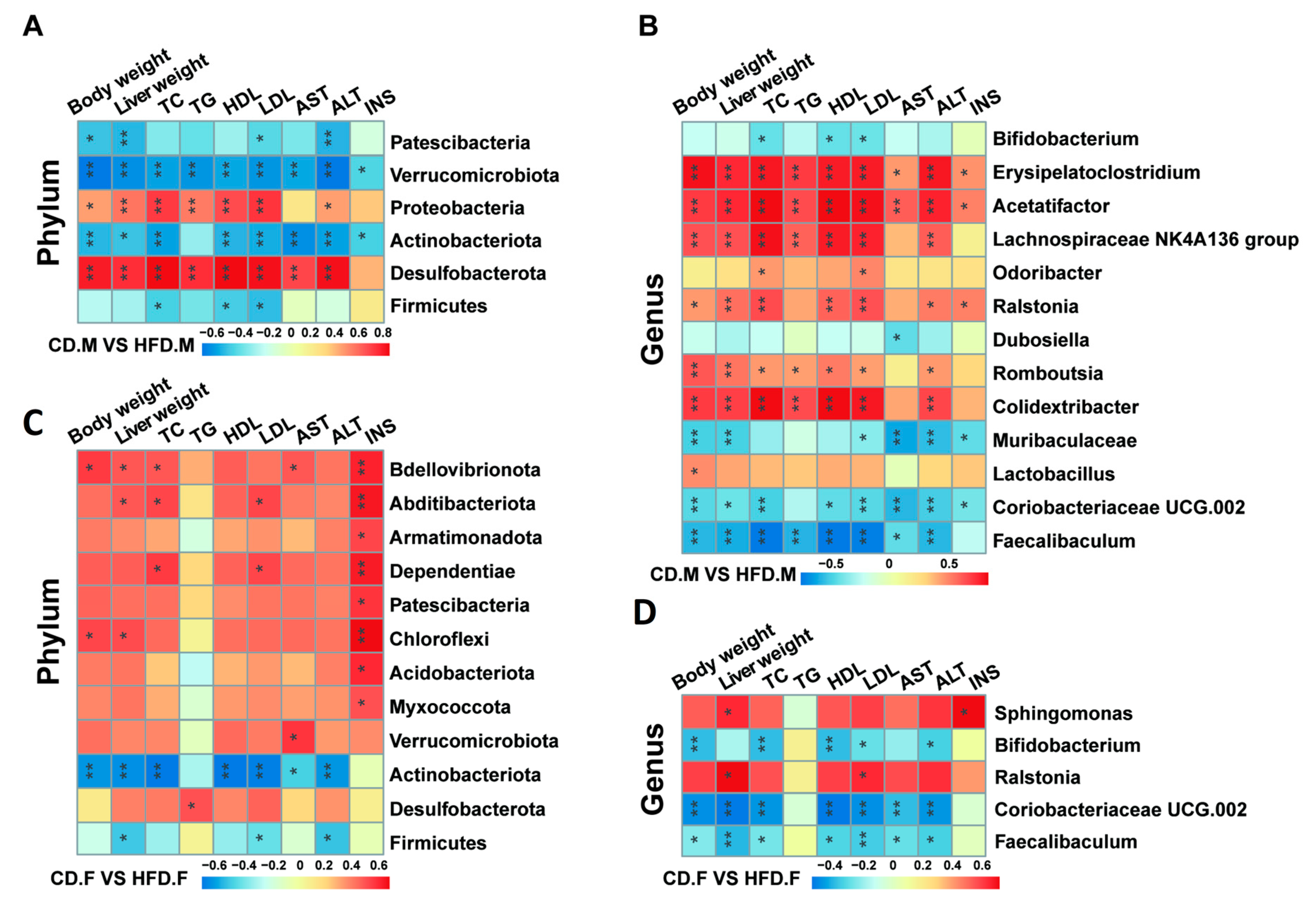
2.6. Correlation Analysis between Gut Microbiota and Indicators Related to NAFLD
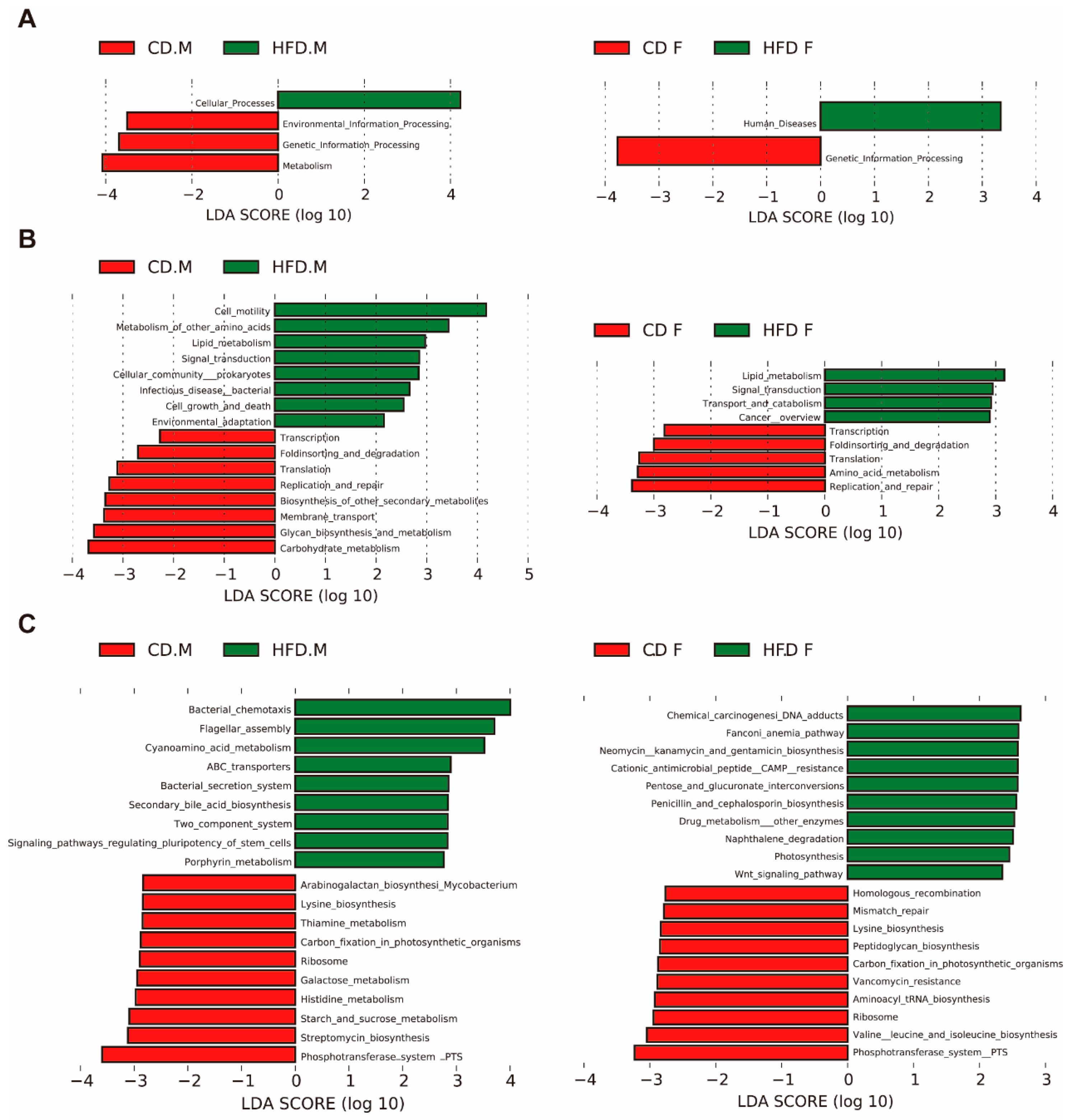
2.7. Predicted Function of Intestinal Microbiota
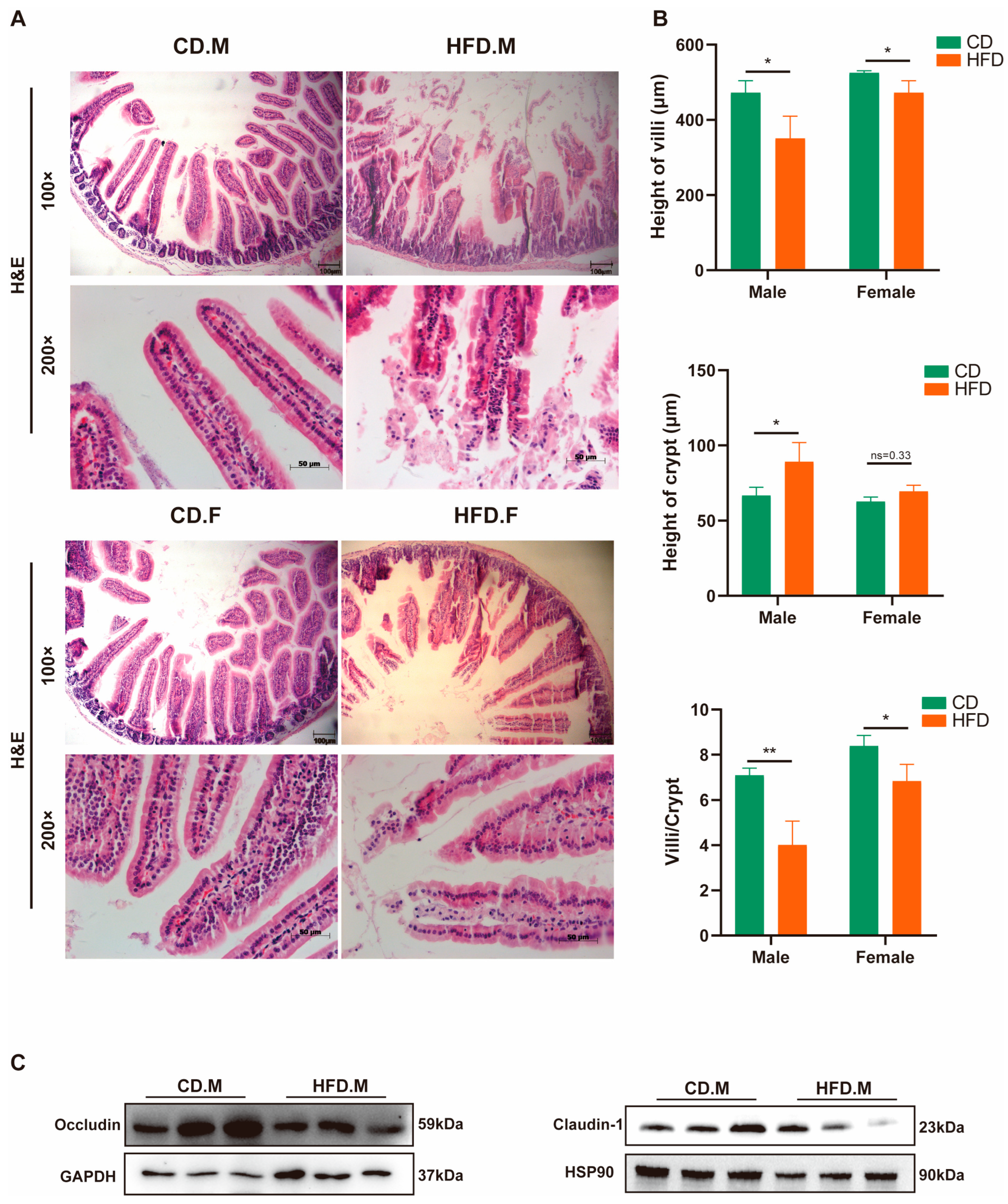
2.8. HFD Affects the Integrity and Permeability of the Intestinal Epithelium
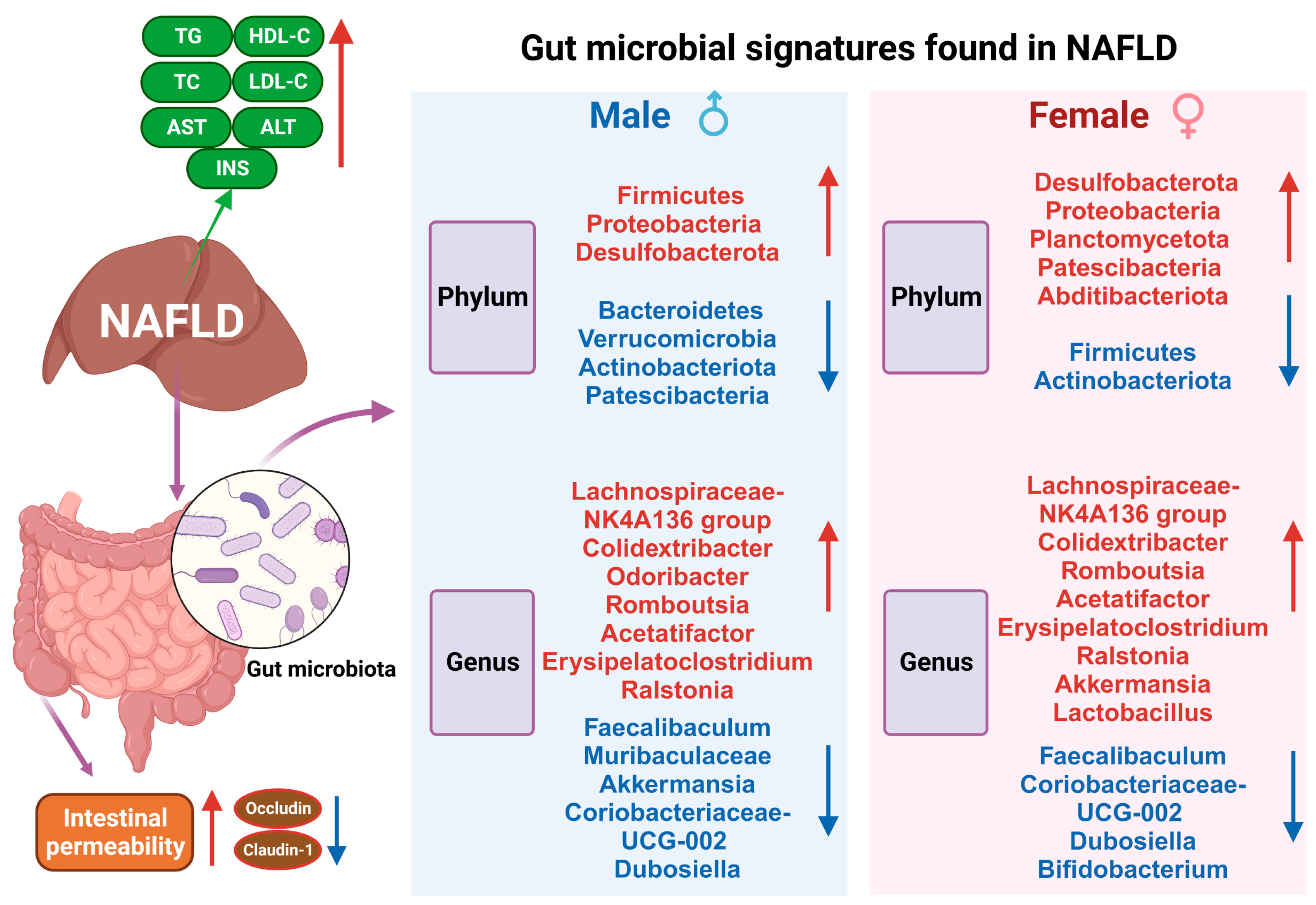
3. Discussion
4. Materials and Methods
4.1. Animal Feeding
4.2. Sample Collection
4.3. Biochemical Analysis of Serum
4.4. Gut Microbiota Sequencing
4.5. Western Blot Analysis
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petta, S.; Argano, C.; Colomba, D.; Cammà, C.; Di Marco, V.; Cabibi, D.; Tuttolomondo, A.; Marchesini, G.; Pinto, A.; Licata, G.; et al. Epicardial fat, cardiac geometry and cardiac function in patients with non-alcoholic fatty liver disease: Association with the severity of liver disease. J. Hepatol. 2015, 62, 928–933. [Google Scholar] [CrossRef]
- Pais, R.; Barritt, A.S.; Calmus, Y.; Scatton, O.; Runge, T.; Lebray, P.; Poynard, T.; Ratziu, V.; Conti, F. NAFLD and liver transplantation: Current burden and expected challenges. J. Hepatol. 2016, 65, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Pipitone, R.M.; Ciccioli, C.; Infantino, G.; La Mantia, C.; Parisi, S.; Tulone, A.; Pennisi, G.; Grimaudo, S.; Petta, S. MAFLD: A multisystem disease. Ther. Adv. Endocrinol. Metab. 2023, 14, 20420188221145549. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Lanti, C.; Riso, P.; Valenti, L. Nutritional therapy for nonalcoholic fatty liver disease. J. Nutr. Biochem. 2016, 29, 1–11. [Google Scholar] [CrossRef] [PubMed]
- EASL-EASD-EASO. Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. Diabetologia 2016, 59, 1121–1140. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.C.; Itsiopoulos, C.; Thodis, T.; Ward, G.; Trost, N.; Hofferberth, S.; O’Dea, K.; Desmond, P.V.; Johnson, N.A.; Wilson, A.M. The Mediterranean diet improves hepatic steatosis and insulin sensitivity in individuals with non-alcoholic fatty liver disease. J. Hepatol. 2013, 59, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.P.; Cunningham, R.P.; Dashek, R.J.; Mucinski, J.M.; Rector, R.S. A Fad too Far? Dietary Strategies for the Prevention and Treatment of NAFLD. Obesity 2020, 28, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Schwimmer, J.B.; Johnson, J.S.; Angeles, J.E.; Behling, C.; Belt, P.H.; Borecki, I.; Bross, C.; Durelle, J.; Goyal, N.P.; Hamilton, G.; et al. Microbiome Signatures Associated with Steatohepatitis and Moderate to Severe Fibrosis in Children with Nonalcoholic Fatty Liver Disease. Gastroenterology 2019, 157, 1109–1122. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef]
- Yu, J.S.; Youn, G.S.; Choi, J.; Kim, C.-H.; Kim, B.Y.; Yang, S.-J.; Lee, J.H.; Park, T.-S.; Kim, B.K.; Kim, Y.B.; et al. Lactobacillus lactis and Pediococcus pentosaceus-driven reprogramming of gut microbiome and metabolome ameliorates the progression of non-alcoholic fatty liver disease. Clin. Transl. Med. 2021, 11, e634. [Google Scholar] [CrossRef]
- Sharpton, S.R.; Ajmera, V.; Loomba, R. Emerging Role of the Gut Microbiome in Nonalcoholic Fatty Liver Disease: From Composition to Function. Clin. Gastroenterol. Hepatol. 2019, 17, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, S.; Nirmalkar, K.; Hoyo-Vadillo, C.; García-Espitia, M.; Ramírez-Sánchez, D.; García-Mena, J. Gut microbiome production of short-chain fatty acids and obesity in children. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Canfora, E.E.; Meex, R.C.R.; Venema, K.; Blaak, E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019, 15, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, X.; Xu, J.; Xue, C.; Xue, Y.; Wang, Y. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 2014, 118, 476–481. [Google Scholar] [CrossRef]
- Soderborg, T.K.; Clark, S.E.; Mulligan, C.E.; Janssen, R.C.; Babcock, L.; Ir, D.; Young, B.; Krebs, N.; Lemas, D.J.; Johnson, L.K.; et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat. Commun. 2018, 9, 4462. [Google Scholar] [CrossRef]
- Jiao, N.; Baker, S.S.; Chapa-Rodriguez, A.; Liu, W.; Nugent, C.A.; Tsompana, M.; Mastrandrea, L.; Buck, M.J.; Baker, R.D.; Genco, R.J.; et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 2018, 67, 1881–1891. [Google Scholar] [CrossRef]
- Yan, F.; Wang, Q.; Lu, M.; Chen, W.; Song, Y.; Jing, F.; Guan, Y.; Wang, L.; Lin, Y.; Bo, T.; et al. Thyrotropin increases hepatic triglyceride content through upregulation of SREBP-1c activity. J. Hepatol. 2014, 61, 1358–1364. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell Mol. Life Sci. 2018, 75, 3313–3327. [Google Scholar] [CrossRef]
- Jennison, E.; Byrne, C.D. The role of the gut microbiome and diet in the pathogenesis of non-alcoholic fatty liver disease. Clin. Mol. Hepatol. 2021, 27, 22–43. [Google Scholar] [CrossRef] [PubMed]
- Camporez, J.P.; Lyu, K.; Goldberg, E.L.; Zhang, D.; Cline, G.W.; Jurczak, M.J.; Dixit, V.D.; Petersen, K.F.; Shulman, G.I. Anti-inflammatory effects of oestrogen mediate the sexual dimorphic response to lipid-induced insulin resistance. J. Physiol. 2019, 597, 3885–3903. [Google Scholar] [CrossRef] [PubMed]
- Norheim, F.; Hui, S.T.; Kulahcioglu, E.; Mehrabian, M.; Cantor, R.M.; Pan, C.; Parks, B.W.; Lusis, A.J. Genetic and hormonal control of hepatic steatosis in female and male mice. J. Lipid Res. 2017, 58, 178–187. [Google Scholar] [CrossRef]
- Ganz, M.; Csak, T.; Szabo, G. High fat diet feeding results in gender specific steatohepatitis and inflammasome activation. World J. Gastroenterol. 2014, 20, 8525–8534. [Google Scholar] [CrossRef] [PubMed]
- Ferramosca, A.; Zara, V. Modulation of hepatic steatosis by dietary fatty acids. World J. Gastroenterol. 2014, 20, 1746–1755. [Google Scholar] [CrossRef] [PubMed]
- Watson, H.; Mitra, S.; Croden, F.C.; Taylor, M.; Wood, H.M.; Perry, S.L.; Spencer, J.A.; Quirke, P.; Toogood, G.J.; Lawton, C.L.; et al. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut 2018, 67, 1974–1983. [Google Scholar] [CrossRef]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef]
- Xu, J.; Bjursell, M.K.; Himrod, J.; Deng, S.; Carmichael, L.K.; Chiang, H.C.; Hooper, L.V.; Gordon, J.I. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 2003, 299, 2074–2076. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Schweiger, B.; Zadow, I.; Heckler, R.; Timm, H.; Pauli, G. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J. Clin. Microbiol. 2000, 38, 1552–1558. [Google Scholar] [CrossRef]
- Zoulim, F.; Locarnini, S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 2009, 137, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, M.; Pang, X.; Zhao, Y.; Wang, L.; Zhao, L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012, 6, 1848–1857. [Google Scholar] [CrossRef]
- Wang, K.; Peng, X.; Yang, A.; Huang, Y.; Tan, Y.; Qian, Y.; Lv, F.; Si, H. Effects of Diets With Different Protein Levels on Lipid Metabolism and Gut Microbes in the Host of Different Genders. Front. Nutr. 2022, 9, 940217. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-H.; Wang, J.; Chai, X.-Q.; Li, Z.-C.; Jiang, Y.-H.; Li, J.; Liu, X.; Fan, J.; Cai, J.-B.; Liu, F. The Intratumoral Bacterial Metataxonomic Signature of Hepatocellular Carcinoma. Microbiol. Spectr. 2022, 10, e0098322. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yi, X.; Yang, J.; Zhu, Z.; Wang, Y.; Liu, X.; Huang, X.; Wan, Y.; Fu, X.; Shu, W.; et al. Gut Microbiome Signatures in the Progression of Hepatitis B Virus-Induced Liver Disease. Front. Microbiol. 2022, 13, 916061. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Zhang, S.; Zhou, C.; Tang, X.; Li, C.; Wang, C.; Tang, X.; Suo, J.; Jia, Y.; El-Ashram, S.; et al. Influence of Eimeria falciformis Infection on Gut Microbiota and Metabolic Pathways in Mice. Infect. Immun. 2018, 86, e00073-18. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Li, W.; Zhang, B.; Yin, J.; Liuqi, S.; Wang, J.; Peng, B.; Wang, S. Fucoidan Ameliorated Dextran Sulfate Sodium-Induced Ulcerative Colitis by Modulating Gut Microbiota and Bile Acid Metabolism. J. Agric. Food Chem. 2022, 70, 14864–14876. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Xu, J.; Lian, F.; Yu, X.; Zhao, Y.; Xu, L.; Zhang, M.; Zhao, X.; Shen, J.; Wu, S.; et al. Structural Alteration of Gut Microbiota during the Amelioration of Human Type 2 Diabetes with Hyperlipidemia by Metformin and a Traditional Chinese Herbal Formula: A Multicenter, Randomized, Open Label Clinical Trial. mBio 2018, 9, e02392-17. [Google Scholar] [CrossRef]
- Miquel, S.; Martín, R.; Rossi, O.; Bermúdez-Humarán, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef]
- Guo, X.; Cao, X.; Fang, X.; Guo, A.; Li, E. Inhibitory effects of fermented Ougan (Citrus reticulata cv. Suavissima) juice on high-fat diet-induced obesity associated with white adipose tissue browning and gut microbiota modulation in mice. Food Funct. 2021, 12, 9300–9314. [Google Scholar]
- Xie, Y.; Zhou, G.; Wang, C.; Xu, X.; Li, C. Specific Microbiota Dynamically Regulate the Bidirectional Gut-Brain Axis Communications in Mice Fed Meat Protein Diets. J. Agric. Food Chem. 2019, 67, 1003–1017. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T.; Adley, C.C. Ralstonia pickettii: A persistent gram-negative nosocomial infectious organism. J. Hosp. Infect. 2006, 62, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Barouei, J.; Bendiks, Z.; Martinic, A.; Mishchuk, D.; Heeney, D.; Hsieh, Y.-H.; Kieffer, D.; Zaragoza, J.; Martin, R.; Slupsky, C.; et al. Microbiota, metabolome, and immune alterations in obese mice fed a high-fat diet containing type 2 resistant starch. Mol. Nutr. Food Res. 2017, 61, 1700184. [Google Scholar] [CrossRef] [PubMed]
- Obanda, D.; Page, R.; Guice, J.; Raggio, A.M.; Husseneder, C.; Marx, B.; Stout, R.W.; Welsh, D.A.; Taylor, C.M.; Luo, M.; et al. CD Obesity-Prone Rats, but not Obesity-Resistant Rats, Robustly Ferment Resistant Starch Without Increased Weight or Fat Accretion. Obesity 2018, 26, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Liu, D.; Gao, Q.; Ni, J.; Qian, L.; Ni, Y.; Fang, Q.; Jia, W.; Li, H. Bifidobacterium adolescentis Alleviates Liver Steatosis and Steatohepatitis by Increasing Fibroblast Growth Factor 21 Sensitivity. Front. Endocrinol. 2021, 12, 773340. [Google Scholar] [CrossRef] [PubMed]
- Zakham, F.; Pillonel, T.; Brunel, A.-S.; Zambelli, P.-Y.; Greub, G.; Croxatto, A.; Bertelli, C. Molecular diagnosis and enrichment culture identified a septic pseudoarthrosis due to an infection with Erysipelatoclostridium ramosum. Int. J. Infect. Dis. 2019, 81, 167–169. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Xu, L.; Jia, Y.; Xue, Z.; Zhang, M.; Phisalaphong, M.; Chen, H. Ultrasound-assisted modified pectin from unripe fruit pomace of raspberry (Rubus chingii Hu): Structural characterization and antioxidant activities. LWT-Food Sci. Technol. 2020, 134, 110007. [Google Scholar] [CrossRef]
- Lee, C.; Hong, S.N.; Paik, N.Y.; Kim, T.J.; Kim, E.R.; Chang, D.K.; Kim, Y.-H. CD1d Modulates Colonic Inflammation in NOD2−/− Mice by Altering the Intestinal Microbial Composition Comprising Acetatifactor muris. J. Crohn’s Colitis 2019, 13, 1081–1091. [Google Scholar] [CrossRef]
- Pfeiffer, N.; Desmarchelier, C.; Blaut, M.; Daniel, H.; Haller, D.; Clavel, T. Acetatifactor muris gen. nov., sp. nov., a novel bacterium isolated from the intestine of an obese mouse. Arch. Microbiol. 2012, 194, 901–907. [Google Scholar] [CrossRef]
- Matilla, M.A.; Krell, T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev. 2018, 42, fux052. [Google Scholar] [CrossRef]
- Sevrin, G.; Massier, S.; Chassaing, B.; Agus, A.; Delmas, J.; Denizot, J.; Billard, E.; Barnich, N. Adaptation of adherent-invasive E. coli to gut environment: Impact on flagellum expression and bacterial colonization ability. Gut Microbes 2020, 11, 364–380. [Google Scholar] [CrossRef]
- Li, W.; Zhang, H.; Assaraf, Y.G.; Zhao, K.; Xu, X.; Xie, J.; Yang, D.-H.; Chen, Z.-S. Overcoming ABC transporter-mediated multidrug resistance: Molecular mechanisms and novel therapeutic drug strategies. Drug Resist. Updates 2016, 27, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, W.J. The Phosphotransferase System in Solventogenic Clostridia. J. Mol. Microbiol. Biotechnol. 2015, 25, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Mouries, J.; Brescia, P.; Silvestri, A.; Spadoni, I.; Sorribas, M.; Wiest, R.; Mileti, E.; Galbiati, M.; Invernizzi, P.; Adorini, L.; et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 2019, 71, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.; Desai, C.; Iyer, S.S.; Thorn, N.E.; Kumar, P.; Liu, Y.; Smith, T.; Neish, A.S.; Li, H.; Tan, S.; et al. Loss of Junctional Adhesion Molecule A Promotes Severe Steatohepatitis in Mice on a Diet High in Saturated Fat, Fructose, and Cholesterol. Gastroenterology 2016, 151, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Kiso, S.; Yoshida, Y.; Chatani, N.; Kizu, T.; Hamano, M.; Tsubakio, M.; Takemura, T.; Ezaki, H.; Hayashi, N.; et al. Estrogen deficiency worsens steatohepatitis in mice fed high-fat and high-cholesterol diet. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G1031–G1043. [Google Scholar] [PubMed]
- Riant, E.; Waget, A.; Cogo, H.; Arnal, J.-F.; Burcelin, R.; Gourdy, P. Estrogens protect against high-fat diet-induced insulin resistance and glucose intolerance in mice. Endocrinology 2009, 150, 2109–2117. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 11. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Cai, J.; Wang, Y.; Liu, X.; Chen, X.; Wang, H.; Wu, Z.; Bao, W.; Fan, H.; Wu, S. A High-Fat Diet Increases the Characteristics of Gut Microbial Composition and the Intestinal Damage Associated with Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2023, 24, 16733. https://doi.org/10.3390/ijms242316733
Zhu X, Cai J, Wang Y, Liu X, Chen X, Wang H, Wu Z, Bao W, Fan H, Wu S. A High-Fat Diet Increases the Characteristics of Gut Microbial Composition and the Intestinal Damage Associated with Non-Alcoholic Fatty Liver Disease. International Journal of Molecular Sciences. 2023; 24(23):16733. https://doi.org/10.3390/ijms242316733
Chicago/Turabian StyleZhu, Xiaoyang, Jiajia Cai, Yifu Wang, Xinyu Liu, Xiaolei Chen, Haifei Wang, Zhengchang Wu, Wenbin Bao, Hairui Fan, and Shenglong Wu. 2023. "A High-Fat Diet Increases the Characteristics of Gut Microbial Composition and the Intestinal Damage Associated with Non-Alcoholic Fatty Liver Disease" International Journal of Molecular Sciences 24, no. 23: 16733. https://doi.org/10.3390/ijms242316733
APA StyleZhu, X., Cai, J., Wang, Y., Liu, X., Chen, X., Wang, H., Wu, Z., Bao, W., Fan, H., & Wu, S. (2023). A High-Fat Diet Increases the Characteristics of Gut Microbial Composition and the Intestinal Damage Associated with Non-Alcoholic Fatty Liver Disease. International Journal of Molecular Sciences, 24(23), 16733. https://doi.org/10.3390/ijms242316733








