Dynamic Interplay of Metabolic and Transcriptional Responses in Shrimp during Early and Late Infection Stages of Enterocytozoon hepatopenaei (EHP)
Abstract
:1. Introduction
2. Results
2.1. Shrimp PCR Analysis and EHP Quantification Results
2.2. Transcriptome Analysis of Hepatopancreas in Litopenaeus vannamei after EHP Infection
2.3. Sample Relationship Analysis
2.4. Identification and Analysis of DEGs
2.5. Functional Annotation Analysis of DEGs
2.6. Pathway Enrichment Analysis of DEGs
2.7. Validation of Transcriptome Data by qRT-PCR
2.8. QC Metabolome Sample Principal Component Analysis
2.9. Identification of Significantly Different Metabolites and KEGG Enrichment Analysis
2.10. Cluster Analysis of Differential Metabolites
2.11. Transcriptome and Metabolome Correlation Analysis
2.11.1. Effect of EHP on Purine Metabolism during Early Infection
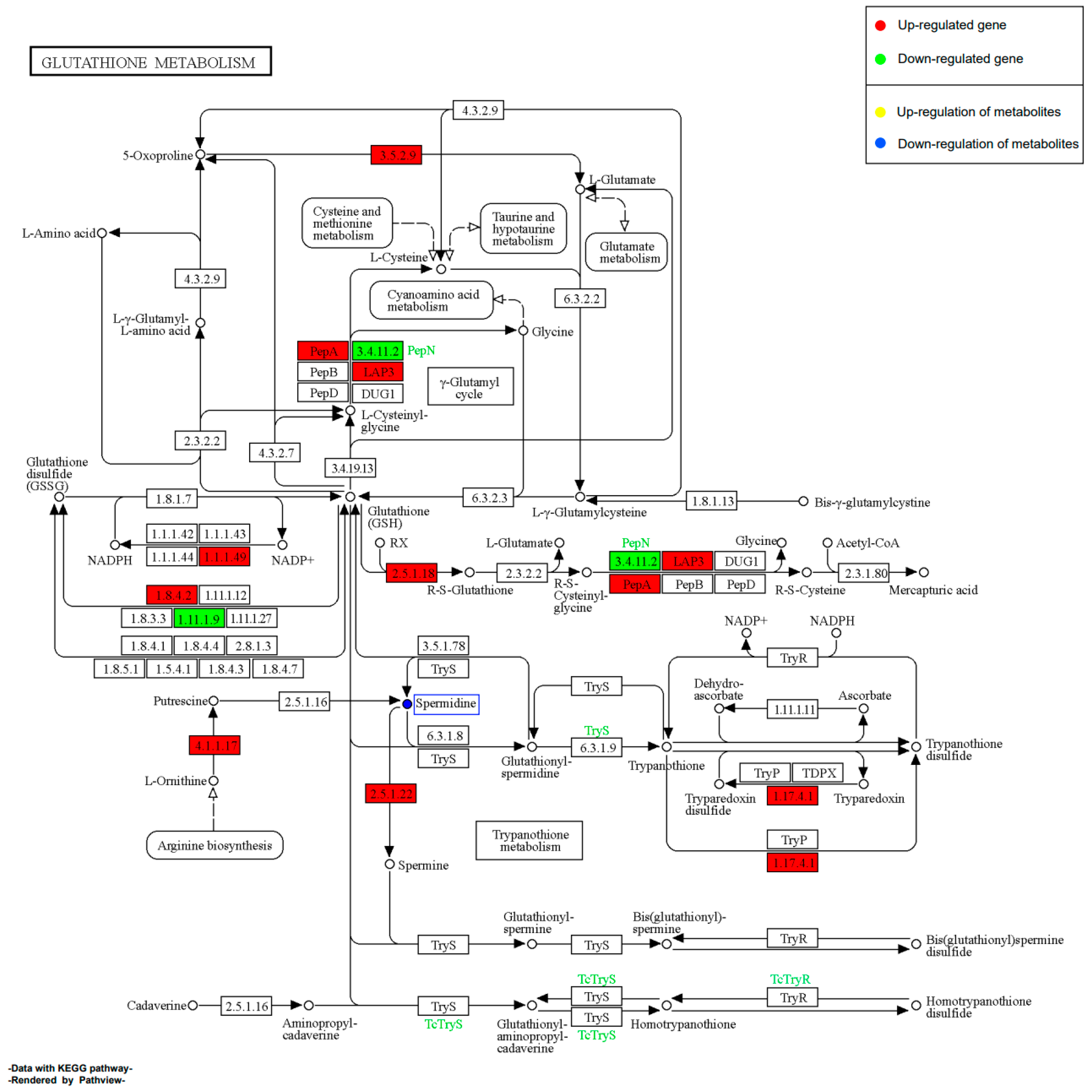
2.11.2. Effect of EHP on Glutathione Metabolism during Early Infection
2.11.3. Effect of EHP on the Metabolism of Glycerophospholipids and Sphingolipids during Late Infection
2.11.4. Effect of EHP on Glutathione Metabolism during Late Infection
3. Discussion
4. Materials and Methods
4.1. Pacific White Shrimp Samples
4.2. Multiple Pathogen Screening and Quantitative PCR Test of EHP
4.3. RNA Extraction and Quality Control
4.4. Assembly of Transcriptomic Data
4.5. Processing and Annotation of Transcriptome Data
4.6. Analysis of Differentially Expressed Genes
4.7. Quantitative RT-PCR Analysis
4.8. Metabolite Extraction and Liquid LC-MS Analysis
4.9. Differential Metabolites Identification
4.10. Correlation Analysis of the Transcriptome and Metabolome
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, E.; Xu, C.; Wang, X.; Wang, S.; Zhao, Q.; Zhang, M.; Qin, J.G.; Chen, L. Gut microbiota and its modulation for healthy farming of Pacific white shrimp Litopenaeus vannamei. Rev. Fish. Sci. Aquac. 2018, 26, 381–399. [Google Scholar] [CrossRef]
- Newman, S.G. Microsporidian Impacts Shrimp Production; Global Seafood Alliance: Portsmouth, NH, USA, 2018. [Google Scholar]
- Shen, H.; Qiao, Y.; Wan, X.; Jiang, G.; Fan, X.; Li, H.; Shi, W.; Wang, L.; Zhen, X. Prevalence of shrimp microsporidian parasite Enterocytozoon hepatopenaei in Jiangsu Province, China. Aquacult Int. 2019, 27, 675–683. [Google Scholar] [CrossRef]
- Chaijarasphong, T.; Munkongwongsiri, N.; Stentiford, G.D.; Aldama-Cano, D.J.; Thansa, K.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. The shrimp microsporidian Enterocytozoon hepatopenaei (EHP): Biology, pathology, diagnostics and control. J. Invertebr. Pathol. 2021, 186, 107458. [Google Scholar] [CrossRef]
- Chayaburakul, K.; Nash, G.; Pratanpipat, P.; Sriurairatana, S.; Withyachumnarnkul, B. Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in Thailand. Dis. Aquat. Organ. 2004, 60, 89–96. [Google Scholar] [CrossRef]
- Tourtip, S.; Wongtripop, S.; Stentiford, G.D.; Bateman, K.S.; Sriurairatana, S.; Chavadej, J.; Sritunyalucksana, K.; Withyachumnarnkul, B. Enterocytozoon hepatopenaei sp. nov.(Microsporida: Enterocytozoonidae), a parasite of the black tiger shrimp Penaeus monodon (Decapoda: Penaeidae): Fine structure and phylogenetic relationships. J. Invertebr. Pathol. 2009, 102, 21–29. [Google Scholar] [CrossRef]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P.; Salachan, P.V.; Sritunyalucksana, K.; Flegel, T.W.; Itsathitphaisarn, O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 2016, 452, 69–87. [Google Scholar] [CrossRef]
- Tangprasittipap, A.; Srisala, J.; Chouwdee, S.; Somboon, M.; Chuchird, N.; Limsuwan, C.; Srisuvan, T.; Flegel, T.W.; Sritunyalucksana, K. The microsporidian Enterocytozoon hepatopenaei is not the cause of white feces syndrome in whiteleg shrimp Penaeus (Litopenaeus) vannamei. BMC Vet. Res. 2013, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Vávra, J.; Lukeš, J. Microsporidia and ‘the art of living together’. Adv. Parasitol. 2013, 82, 253–319. [Google Scholar] [CrossRef] [PubMed]
- Stentiford, G.D.; Feist, S.W.; Stone, D.M.; Bateman, K.S.; Dunn, A.M. Microsporidia: Diverse, dynamic, and emergent pathogens in aquatic systems. Trends Parasitol. 2013, 29, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Stentiford, G.D.; Bass, D.; Williams, B.A. Ultimate opportunists—The emergent Enterocytozoon group microsporidia. PLoS Pathog. 2019, 15, e1007668. [Google Scholar] [CrossRef]
- Trentmann, O.; Horn, M.; van Scheltinga, A.C.T.; Neuhaus, H.E.; Haferkamp, I. Enlightening energy parasitism by analysis of an ATP/ADP transporter from chlamydiae. PLoS Biol. 2007, 5, e231. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, J.; Liao, G.; Hu, M.; Zhang, Q.; Meng, X.; Li, T.; Long, M.; Fan, X.; Yu, Q. Down-Regulation of lipid metabolism in the hepatopancreas of shrimp Litopenaeus vannamei upon light and heavy infection of Enterocytozoon hepatopenaei: A comparative proteomic study. Int. J. Mol. Sci. 2022, 23, 11574. [Google Scholar] [CrossRef]
- Kumar, T.S.; Makesh, M.; Alavandi, S.; Vijayan, K. Clinical manifestations of White feces syndrome (WFS), and its association with Enterocytozoon hepatopenaei in Penaeus vannamei grow-out farms: A pathobiological investigation. Aquaculture 2022, 547, 737463. [Google Scholar] [CrossRef]
- Shen, H.; Dou, Y.; Li, H.; Qiao, Y.; Jiang, G.; Wan, X.; Cheng, J.; Fan, X.; Li, H.; Wang, L. Changes in the intestinal microbiota of Pacific white shrimp (Litopenaeus vannamei) with different severities of Enterocytozoon hepatopenaei infection. J. Invertebr. Pathol. 2022, 191, 107763. [Google Scholar] [CrossRef]
- Ye, S.; Xingqiang, W.; Mei, C.; Nianhao, Z.; Baiyao, C.; Chuanxin, Q. Transcriptome analysis of Exopalaemon carinicauda under low salinity stress. South China Fish. Sci. 2020, 16, 19–32. (In Chinese) [Google Scholar] [CrossRef]
- Mercuro, G.; Bassareo, P.P.; Deidda, M.; Cadeddu, C.; Barberini, L.; Atzori, L. Metabolomics: A new era in cardiology? J. Cardiovasc. Med. 2011, 12, 800–805. [Google Scholar] [CrossRef]
- Mutz, K.-O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.-G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotech. 2013, 24, 22–30. [Google Scholar] [CrossRef]
- Duan, Y.; Chen, H.; Wang, J.; Zeng, S.; Wang, Y.; Mo, Z.; Dan, X.; Li, Y. Response signatures of Litopenaeus vannamei to natural Enterocytozoon hepatopenaei infection revealed by the integration of the microbiome and transcriptome. Aquaculture 2021, 542, 736885. [Google Scholar] [CrossRef]
- Ning, M.; Wei, P.; Shen, H.; Wan, X.; Jin, M.; Li, X.; Shi, H.; Qiao, Y.; Jiang, G.; Gu, W. Proteomic and metabolomic responses in hepatopancreas of whiteleg shrimp Litopenaeus vannamei infected by microsporidian Enterocytozoon hepatopenaei. Fish. Shellfish. Immun. 2019, 87, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Szumowski, S.C.; Troemel, E.R. Microsporidia–host interactions. Curr. Opin. Microbiol. 2015, 26, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Wiredu Boakye, D.; Jaroenlak, P.; Prachumwat, A.; Williams, T.A.; Bateman, K.S.; Itsathitphaisarn, O.; Sritunyalucksana, K.; Paszkiewicz, K.H.; Moore, K.A.; Stentiford, G.D. Decay of the glycolytic pathway and adaptation to intranuclear parasitism within Enterocytozoonidae microsporidia. Environ. Microbiol. 2017, 19, 2077–2089. [Google Scholar] [CrossRef]
- Antúnez, K.; Martín-Hernández, R.; Prieto, L.; Meana, A.; Zunino, P.; Higes, M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 2009, 11, 2284–2290. [Google Scholar] [CrossRef]
- Weng, K.; Zuo, H.; Zhu, Z.; Guo, Z.; Weng, S.; He, J.; Xu, X. Ftz-F1H promotes white spot syndrome virus infection in shrimp by suppressing the Dorsal pathway. Aquaculture 2022, 548, 737708. [Google Scholar] [CrossRef]
- Zuo, H.; Weng, K.; Zhu, Z.; Guo, Z.; Weng, S.; He, J.; Xu, X. Suppression of a JAK-STAT pathway target gene Ftz-F1H enhances the resistance of shrimp to Vibrio parahaemolyticus. Aquaculture 2021, 543, 737022. [Google Scholar] [CrossRef]
- Diao, M.-Q.; Li, C.; Xu, J.-D.; Zhao, X.-F.; Wang, J.-X. RPS27, a sORF-encoded polypeptide, functions antivirally by activating the NF-κB pathway and interacting with viral envelope proteins in shrimp. Front. Immunol. 2019, 10, 2763. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, M.; Ponniah, A. Recent insights into host–pathogen interaction in white spot syndrome virus infected penaeid shrimp. J. Fish Dis. 2015, 38, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yao, X.; Ding, Y.; Xu, Z.; Liang, R.; Zhang, Y.; Wu, Y.; Li, B.; Guan, B. PI3K signaling pathways modulated white spot syndrome virus (WSSV) replication in Procambarus clarkii. Fish. Shellfish. Immun. 2018, 76, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Padayatty, S.J.; Katz, A.; Wang, Y.; Eck, P.; Kwon, O.; Lee, J.-H.; Chen, S.; Corpe, C.; Dutta, A.; Dutta, S.K. Vitamin C as an antioxidant: Evaluation of its role in disease prevention. J. Am. Coll. Nutr. 2003, 22, 18–35. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Kaur, V.I.; Baruah, K.; Nguyen, V.D.; Bossier, P. High doses of sodium ascorbate act as a prooxidant and protect gnotobiotic brine shrimp larvae (Artemia franciscana) against Vibrio harveyi infection coinciding with heat shock protein 70 activation. Dev. Comp. Immunol. 2019, 92, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Formentini, L.; Pereira, M.P.; Sánchez-Cenizo, L.; Santacatterina, F.; Lucas, J.J.; Navarro, C.; Martínez-Serrano, A.; Cuezva, J.M. In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 2014, 33, 762–778. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.-J.; Lu, P.-Y.; Li, D.-X.; Shi, X.-Z. The sORF-Encoded Peptides, ATP Synthase Subunits, Facilitate WSSV Duplication in Shrimp. Viruses 2022, 14, 2449. [Google Scholar] [CrossRef]
- Dean, P.; Hirt, R.P.; Embley, T.M. Microsporidia: Why make nucleotides if you can steal them? PLoS Pathog. 2016, 12, e1005870. [Google Scholar] [CrossRef]
- Heinz, E.; Hacker, C.; Dean, P.; Mifsud, J.; Goldberg, A.V.; Williams, T.A.; Nakjang, S.; Gregory, A.; Hirt, R.P.; Lucocq, J.M. Plasma membrane-located purine nucleotide transport proteins are key components for host exploitation by microsporidian intracellular parasites. PLoS Pathog. 2014, 10, e1004547. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Ma, Y.; Tu, V.; Tomita, T.; Mayoral, J.; Williams, T.; Horta, A.; Huang, H.; Weiss, L.M. Microsporidia interact with host cell mitochondria via voltage-dependent anion channels using sporoplasm surface protein 1. MBio 2019, 10, 10–128. [Google Scholar] [CrossRef]
- Jimenez-Gutierrez, L.; Uribe-Carvajal, S.; Sanchez-Paz, A.; Chimeo, C.; Muhlia-Almazan, A. The cytochrome c oxidase and its mitochondrial function in the whiteleg shrimp Litopenaeus vannamei during hypoxia. J. Bioenerg. Biomembr. 2014, 46, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Dolgikh, V.V.; Sokolova, J.J.; Issi, I.V. Activities of enzymes of carbohydrate and energy metabolism of the spores of the microsporidian, Nosema grylli. J. Eukaryot. Microbiol. 1997, 44, 246–249. [Google Scholar] [CrossRef]
- Sun, S.; Xuan, F.; Fu, H.; Zhu, J.; Ge, X. Molecular cloning and functional characterization of a hexokinase from the oriental river prawn Macrobrachium nipponense in response to hypoxia. Int. J. Mol. Sci. 2017, 18, 1256. [Google Scholar] [CrossRef]
- Cuomo, C.A.; Desjardins, C.A.; Bakowski, M.A.; Goldberg, J.; Ma, A.T.; Becnel, J.J.; Didier, E.S.; Fan, L.; Heiman, D.I.; Levin, J.Z. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 2012, 22, 2478–2488. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Qi, X.; Tao, M.; Liu, X.; Kang, D.; Qin, G.; Pan, G.; Zhou, Z. Comparative Genomic Analysis of Secreted Proteins in Four Microsporidian Species. J. Seric. Sci. 2013, 39, 527–536. (In Chinese) [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, S.; Mei, X.; Yu, B.; Sun, B.; Li, B.; Wei, J.; Chen, J.; Li, T.; Pan, G. A secretory hexokinase plays an active role in the proliferation of Nosema bombycis. Peerj 2018, 6, e5658. [Google Scholar] [CrossRef]
- Dolgikh, V.V.; Tsarev, A.A.; Timofeev, S.A.; Zhuravlyov, V.S. Heterologous overexpression of active hexokinases from microsporidia Nosema bombycis and Nosema ceranae confirms their ability to phosphorylate host glucose. Parasitol. Res. 2019, 118, 1511–1518. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; He, Q.; Xu, J.-Z.; Xu, C.; Han, Y.-Z.; Gao, H.-L.; Meng, X.-Z.; Pan, G.-Q.; Li, T.; Zhou, Z.-Y. Microsporidia infection upregulates host energy metabolism but maintains ATP homeostasis. J. Invertebr. Pathol. 2021, 186, 107596. [Google Scholar] [CrossRef]
- Ding, Z. Lipid metabolism disorders contribute to the pathogenesis of Hepatospora eriocheir in the crab Eriocheir sinensis. J. Fish Dis. 2021, 44, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Alaoui, H.E.; Bata, J.; Bauchart, D.; Doré, J.-C.; Vivares, C.P. Lipids of three microsporidian species and multivariate analysis of the host–parasite relationship. J. Parasitol. 2001, 87, 554–559. [Google Scholar] [CrossRef]
- El Alaoui, H.; Bata, J.; Peyret, P.; Vivarès, C.P. Encephalitozoon cuniculi (Microspora): Characterization of a phospholipid metabolic pathway potentially linked to therapeutics. Exp. Parasitol. 2001, 98, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Franchet, A.; Niehus, S.; Caravello, G.; Ferrandon, D. Phosphatidic acid as a limiting host metabolite for the proliferation of the microsporidium Tubulinosema ratisbonensis in Drosophila flies. Nat. Microbiol. 2019, 4, 645–655. [Google Scholar] [CrossRef]
- Han, B.; Takvorian, P.M.; Weiss, L.M. Invasion of host cells by microsporidia. Front. Microbiol. 2020, 11, 172. [Google Scholar] [CrossRef]
- Jeon, J. Characterizing the Role of Host Lipid Metabolism on Microsporidia Infection in Caenorhabditis elegans; University of Toronto: Toronto, ON, Canada, 2021. [Google Scholar]
- Lang, H.; Wang, H.; Wang, H.; Zhong, Z.; Xie, X.; Zhang, W.; Guo, J.; Meng, L.; Hu, X.; Zhang, X. Engineered symbiotic bacteria interfering Nosema redox system inhibit microsporidia parasitism in honeybees. Nat. Commun. 2023, 14, 2778. [Google Scholar] [CrossRef]
- El Bissati, K.; Redel, H.; Ting, L.-M.; Lykins, J.D.; McPhillie, M.J.; Upadhya, R.; Woster, P.M.; Yarlett, N.; Kim, K.; Weiss, L.M. Novel synthetic polyamines have potent antimalarial activities in vitro and in vivo by decreasing intracellular spermidine and spermine concentrations. Front. Cell. Infect. Microbiol. 2019, 9, 9. [Google Scholar] [CrossRef]
- Bacchi, C.; Yarlett, N.; Weiss, L. Polyamine Metabolism in the Microsporidia; Portland Press Ltd.: London, UK, 2003. [Google Scholar] [CrossRef]
- Han, B.; Weiss, L.M. Therapeutic targets for the treatment of microsporidiosis in humans. Expert Opin. Ther. Targets 2018, 22, 903–915. [Google Scholar] [CrossRef]
- Millamena, O.M.; Teruel, M.; Kanazawa, A.; Teshima, S. Quantitative dietary requirements of postlarval tiger shrimp, Penaeus monodon, for histidine, isoleucine, leucine, phenylalanine and tryptophan. Aquaculture 1999, 179, 169–179. [Google Scholar] [CrossRef]
- Li, A.; Yue, Y.; Zhang, Y.; Zhang, L.; Zhang, K.; Jie, S.; Tan, S.; Shi, W. Effects of Paranosema locustae (Microsporidia) on the development and morphological phase transformation of Locusta migratoria (Orthoptera: Acrididae) through modulation of the neurotransmitter taurine. J. Integr. Agric. 2020, 19, 204–210. [Google Scholar] [CrossRef]
- Wang, L.; Lv, Q.; Meng, X.; Chen, J.; Wang, Y.; Pan, G.; Long, M.; Zhou, Z. Identification and characterization polar tube protein 2 (PTP2) from Enterocytozoon hepatopenaei and its potential effect on shrimp microsporidian germination activity evaluation. Aquaculture 2021, 544, 737062. [Google Scholar] [CrossRef]
- Liu, Y.-M.; Qiu, L.; Sheng, A.-Z.; Wan, X.-Y.; Cheng, D.-Y.; Huang, J. Quantitative detection method of Enterocytozoon hepatopenaei using TaqMan probe real-time PCR. J. Invertebr. Pathol. 2018, 151, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L. Integrated nr database in protein annotation system and its localization. Nat. Commun. 2010, 6, 1–8. [Google Scholar]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32 (Suppl. 1), D115–D119. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32 (Suppl. 1), D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]












| Sample | Raw Reads | Clean Reads | Error Rate (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|
| Asn1 | 60,517,026 | 58,658,178 | 0.0254 | 93.89 | 47.89 |
| Asn2 | 50,392,962 | 48,311,954 | 0.0252 | 93.99 | 48.49 |
| Asn3 | 46,760,842 | 45,547,650 | 0.0251 | 94.14 | 48.49 |
| Asp1 | 73,211,792 | 71,633,060 | 0.0249 | 94.31 | 49.49 |
| Asp2 | 51,861,550 | 50,571,484 | 0.0251 | 94.17 | 48.49 |
| Asp3 | 44,573,798 | 43,024,636 | 0.0255 | 93.78 | 48.93 |
| Jsn1 | 42,488,742 | 40,402,838 | 0.0257 | 93.48 | 43.21 |
| Jsn2 | 44,758,988 | 43,049,092 | 0.0253 | 93.94 | 48.68 |
| Jsn3 | 42,868,272 | 40,989,324 | 0.0255 | 93.72 | 44.95 |
| Jsp1 | 42,872,192 | 41,092,782 | 0.0255 | 93.69 | 42.85 |
| Jsp2 | 43,511,964 | 41,948,760 | 0.0256 | 93.62 | 45.78 |
| Jsp3 | 42,276,170 | 41,297,560 | 0.0256 | 93.61 | 44.83 |
| Expre Gene Number | Percentage (%) | |
|---|---|---|
| GO | 13,406 | 61.37 |
| KEGG | 11,612 | 53.16 |
| COG | 15,150 | 69.36 |
| NR | 21,135 | 96.75 |
| Swiss-Prot | 14,008 | 64.13 |
| Pfam | 14,978 | 68.57 |
| Total | 21,844 | 100 |
| Gene Id | Primer Name | Sequence (5′–3′) |
|---|---|---|
| LOC113812219 | CLECG4-R | TTGTCGCTGACGCCTTCC |
| CLECG4-F | GATGCTGGAGGACTTGAGGAG | |
| LOC113817723 | AMY1-R | CTCGGGCTGATGATGTTGAAG |
| AMY1-F | CGTGATGTCGTCGTACTACTG | |
| LOC113817801 | RPN2-R | GGCTTGGTCAGGCTTAATTGG |
| RPN2-F | GATGAAGTTGATGGGCGGATG | |
| LOC113818949 | MAN-R | GGTCGCTTCGGTCCTTCAG |
| MAN-F | GCCTACGAGCACGGATACAG | |
| LOC113805243 | LvPLP-R | CTCCGCTGTGGGTCTTGC |
| LvPLP-F | TGTCGCCAGTGTTCGTTCG | |
| LOC113826472 | CLCA1-R | ATGACGATGCCGACGATGG |
| CLCA1-F | ATGAAGTGAGGGAGATGGTGTC |
| Gene Id | Primer Name | Sequence (5′–3′) |
|---|---|---|
| LOC113809619 | cenE4-R | TTGGTCAGCGAAGTCGTAGAG |
| cenE4-F | CACCCAGGCACGGAGTTG | |
| LOC113813736 | RPL13a-R | TTGAAGGAGCACGCTGATGG |
| RPL13a-F | CCTTGTGTTCTCGGGCATTTTC | |
| LOC113829084 | MGST3-R | ACCATATAGCACCACCAACGG |
| MGST3-F | CCACCAGAACACACTTGAGAAC | |
| LOC113818949 | MAN-R | GGTCGCTTCGGTCCTTCAG |
| MAN-F | GCCTACGAGCACGGATACAG | |
| LOC113823620 | HcC-R | GGGAGCAGGAATCGGTTAGG |
| HcC-F | TCAGAATCAGCAGTCACAGTCC | |
| LOC113812602 | mtATPase-R | AATGATGGCTGGGTCAACTTTC |
| mtATPase-F | GCTGCTGTAAGAGGAGAGGTC |
| Gene Id | Primer Name | Sequence (5′–3′) |
|---|---|---|
| LOC113817185 | CNDP-R | AGCCAGTAGTTGTCGGAGATG |
| CNDP-F | TCTTTGAGGGAATGGAGGAGTC | |
| LOC113806448 | HBPs-R | TTGAATGGTGCGTCGTATGC |
| HBPs-F | GAGGAAGTTCGGTGGATATGC | |
| LOC113808797 | CP4-R | TCTCCGTCTCGCCGTAGC |
| CP4-F | CCAGCCGTCCTTCCAGTTC | |
| LOC113829084 | MGST3-R | ACCATATAGCACCACCAACGG |
| MGST3-F | CCACCAGAACACACTTGAGAAC | |
| LOC113817723 | AMY1-R | CTCGGGCTGATGATGTTGAAG |
| AMY1-F | CGTGATGTCGTCGTACTACTG | |
| LOC113827622 | DPP1-R | GCAACTGGCGTCCGTATCG |
| DPP1-F | GTCGTGCTCCGTGCTGTC |
| Gene Id | Primer Name | Sequence (5′–3′) |
|---|---|---|
| LOC113829084 | MGST3-R | ACCATATAGCACCACCAACGG |
| MGST3-F | CCACCAGAACACACTTGAGAAC | |
| LOC113817723 | AMY1-R | CTCGGGCTGATGATGTTGAAG |
| AMY1-F | CGTGATGTCGTCGTACTACTG | |
| LOC113827551 | NEP1-R | GCCACCATAGCATACTTGAAGG |
| NEP1-F | ACCACAAACCAGCGGACAC | |
| LOC113827755 | TECR-R | TTGCCCACACTGCCATCTG |
| TECR-F | CTCACGCTGCTCTTCAATGC | |
| LOC113803442 | NCX1-R | CGCCTTCTGAACCTGGATTAAC |
| NCX1-F | TGGACCATGAAGACGAGCAC | |
| LOC113810457 | Inx2-R | CGCTCCTGTAGTGGCTCTTC |
| Inx2-F | TGGCTGGTGCTGCTAATGAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhang, S.; Qiao, Y.; Cao, X.; Cheng, J.; Meng, Q.; Shen, H. Dynamic Interplay of Metabolic and Transcriptional Responses in Shrimp during Early and Late Infection Stages of Enterocytozoon hepatopenaei (EHP). Int. J. Mol. Sci. 2023, 24, 16738. https://doi.org/10.3390/ijms242316738
Zhang L, Zhang S, Qiao Y, Cao X, Cheng J, Meng Q, Shen H. Dynamic Interplay of Metabolic and Transcriptional Responses in Shrimp during Early and Late Infection Stages of Enterocytozoon hepatopenaei (EHP). International Journal of Molecular Sciences. 2023; 24(23):16738. https://doi.org/10.3390/ijms242316738
Chicago/Turabian StyleZhang, Leiting, Sheng Zhang, Yi Qiao, Xiaohui Cao, Jie Cheng, Qingguo Meng, and Hui Shen. 2023. "Dynamic Interplay of Metabolic and Transcriptional Responses in Shrimp during Early and Late Infection Stages of Enterocytozoon hepatopenaei (EHP)" International Journal of Molecular Sciences 24, no. 23: 16738. https://doi.org/10.3390/ijms242316738
APA StyleZhang, L., Zhang, S., Qiao, Y., Cao, X., Cheng, J., Meng, Q., & Shen, H. (2023). Dynamic Interplay of Metabolic and Transcriptional Responses in Shrimp during Early and Late Infection Stages of Enterocytozoon hepatopenaei (EHP). International Journal of Molecular Sciences, 24(23), 16738. https://doi.org/10.3390/ijms242316738





