Deprivation of Sexual Reproduction during Garlic Domestication and Crop Evolution
Abstract
1. Introduction
2. Results and Discussion
2.1. Nucleotide Variation in Garlic Genotypes
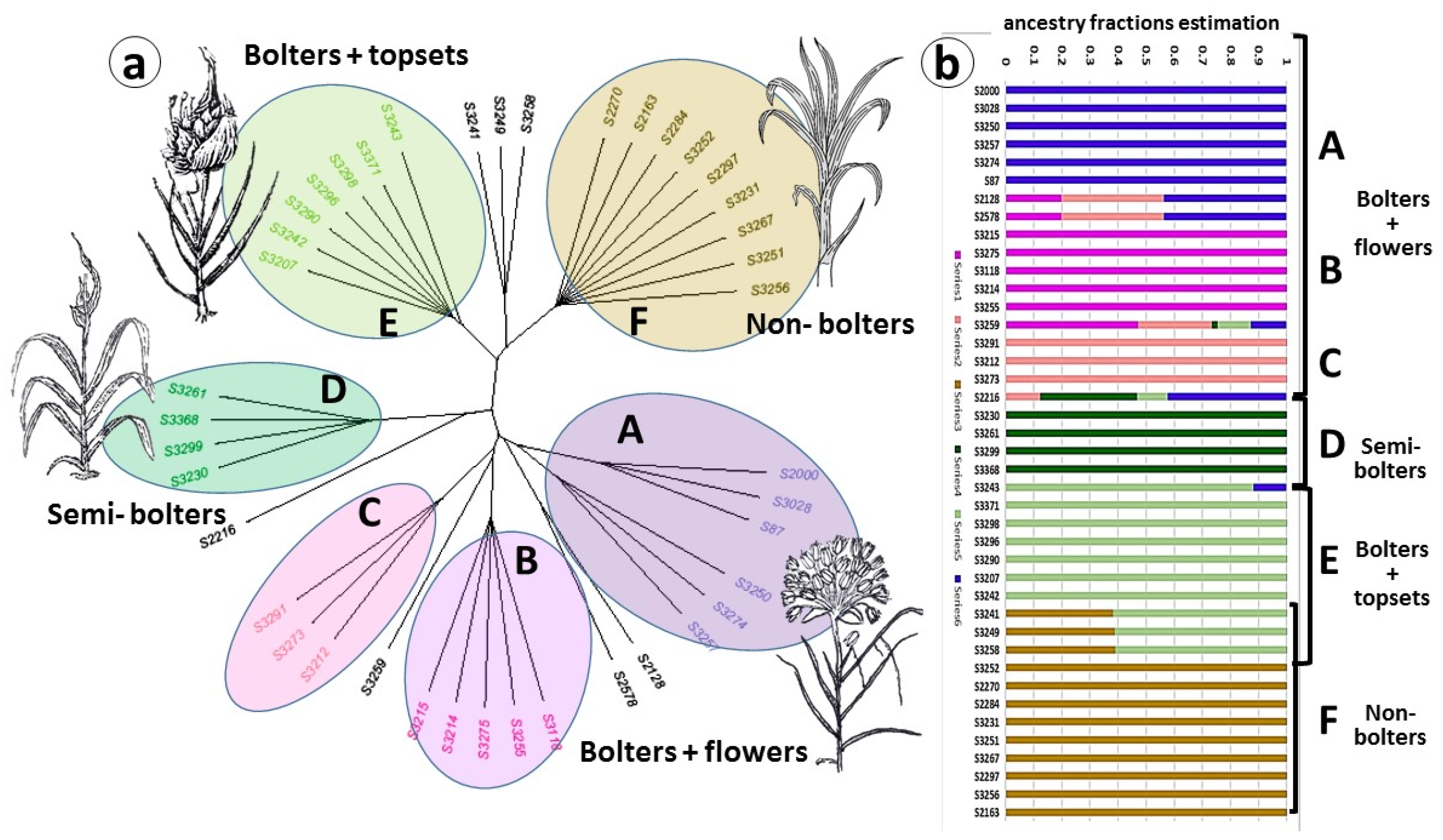
2.2. Population Structure and Diversity
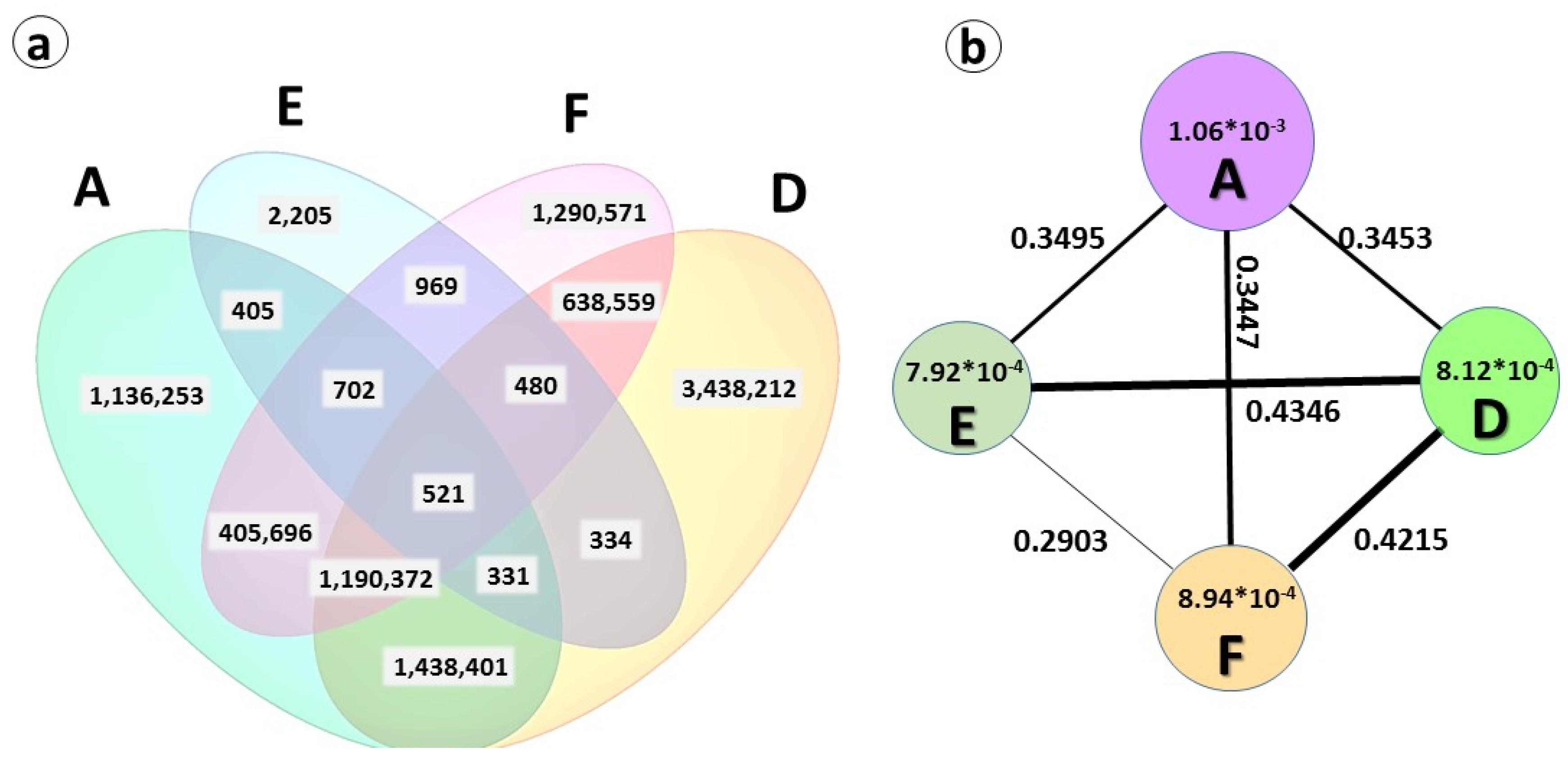
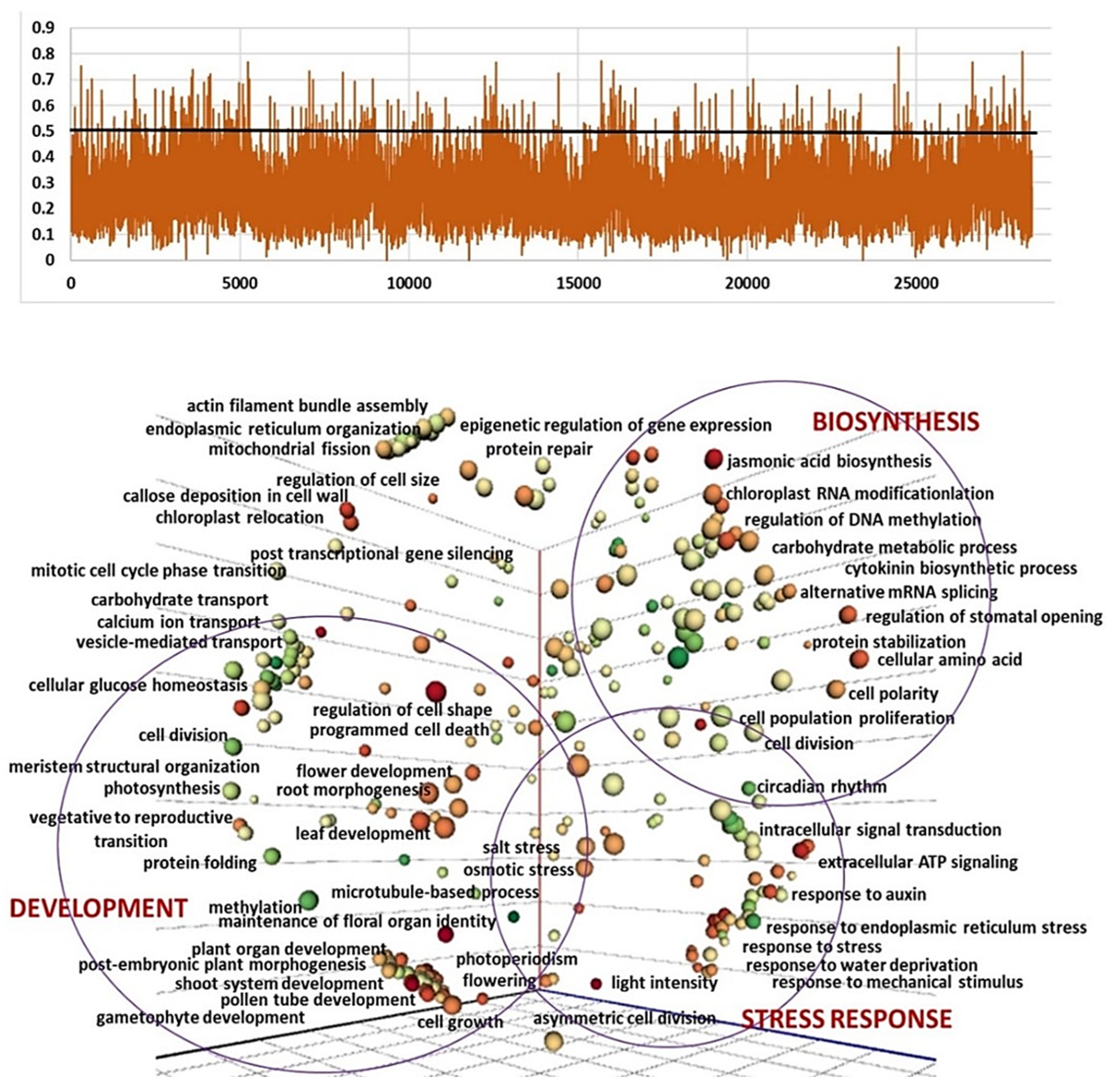
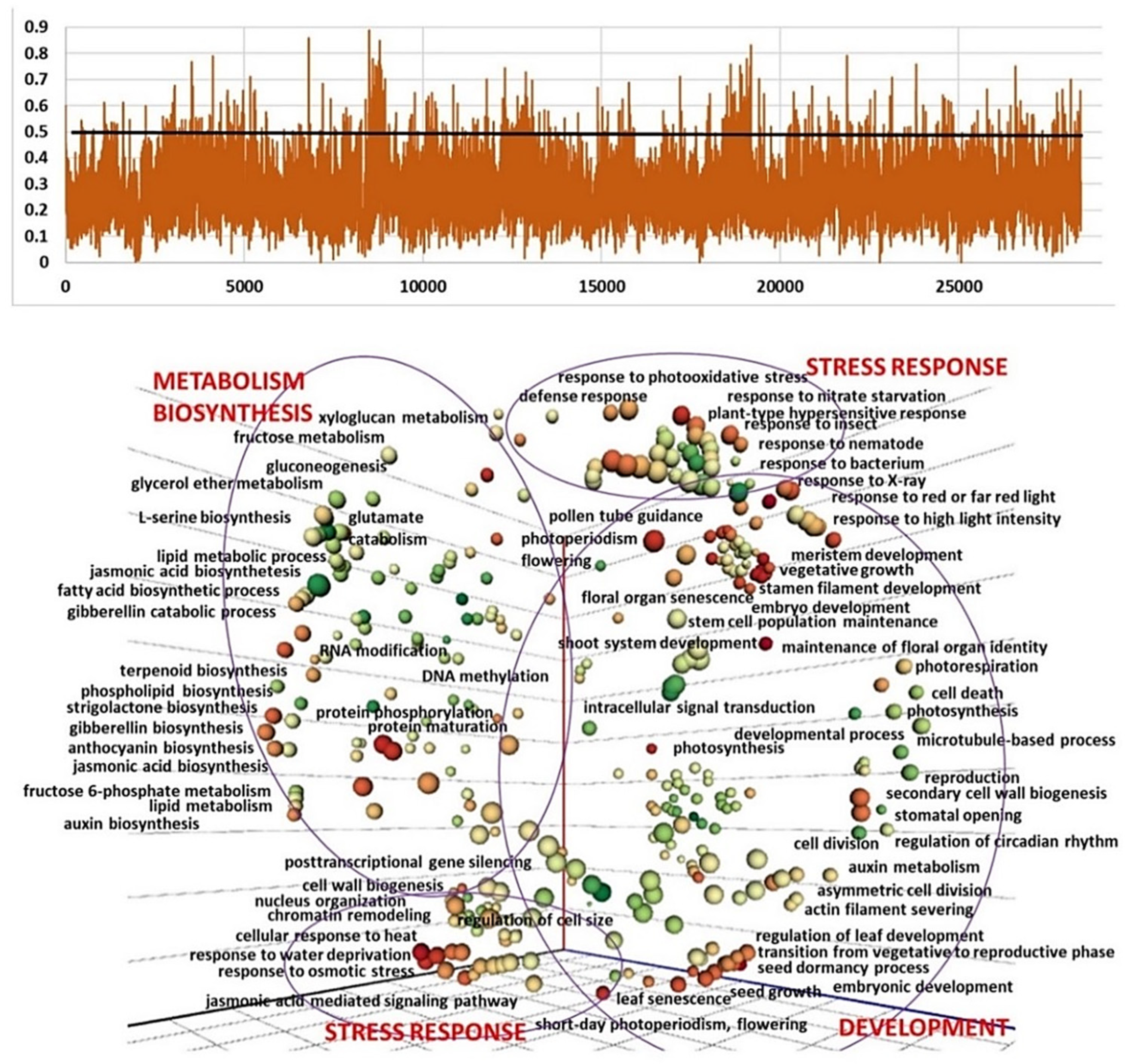
2.3. Genetic Diversity and Plant Architecture in Genomic Landscape
2.4. Genomic and Transcriptomic Analysis of Flowering-Related Gene Families
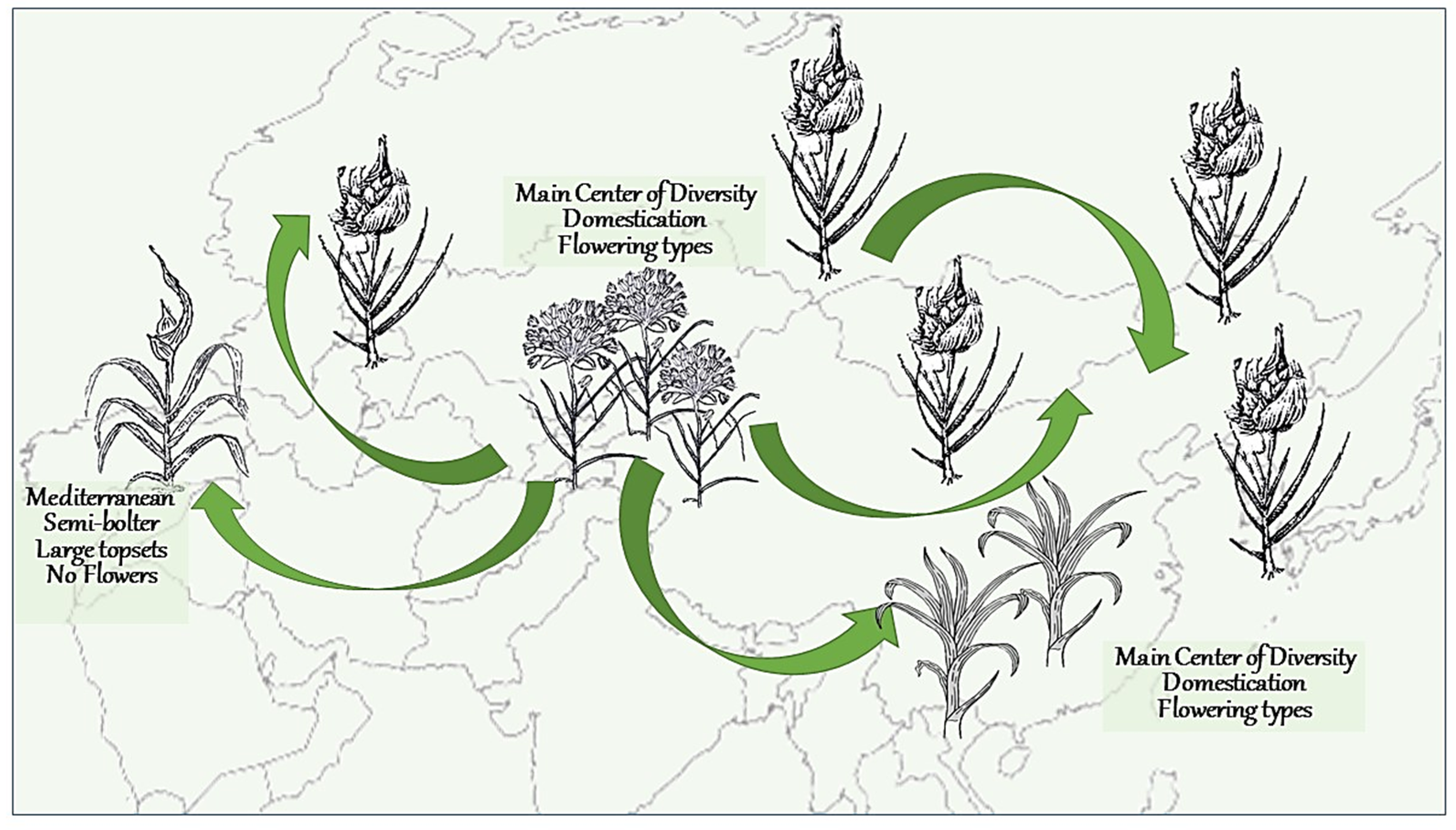
2.5. Crop Migration and Evolution Led to the Major Changes in Garlic Biology
3. Materials and Methods
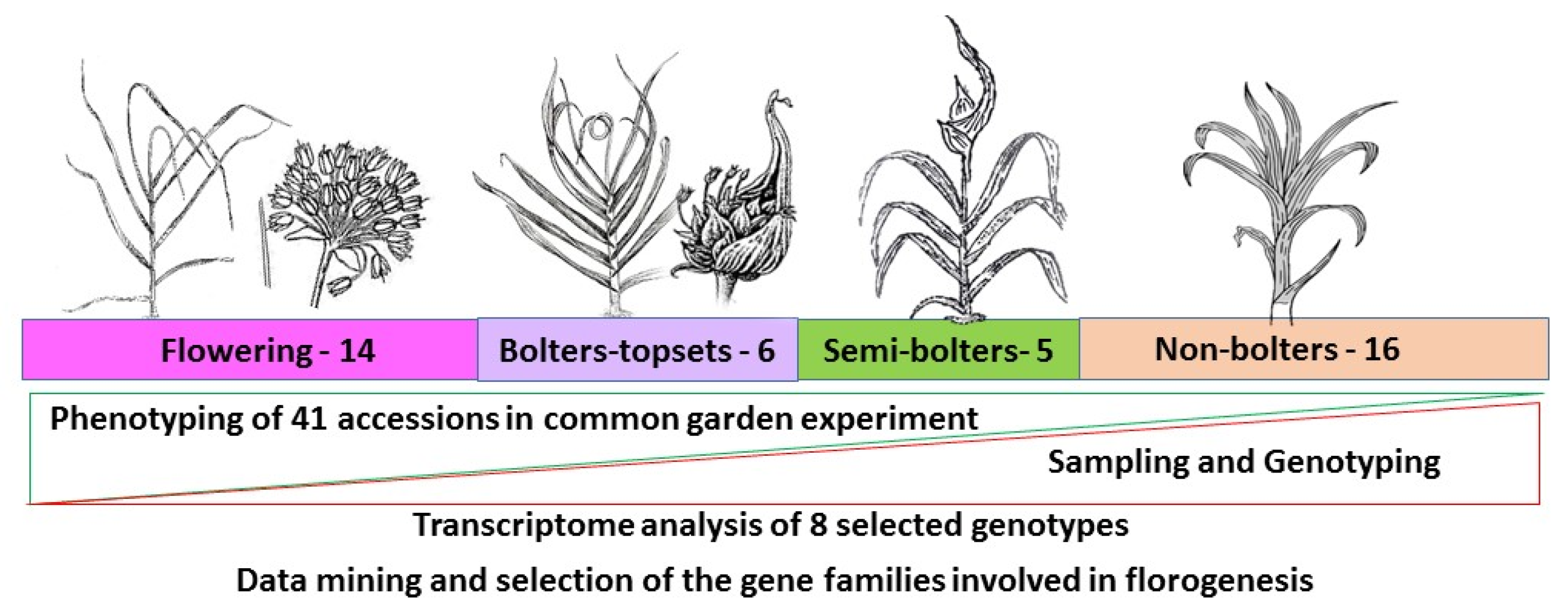
3.1. Plant Material and Phenology
3.2. Genome Sequencing
3.3. Variation Calling
3.4. Functional Annotation
3.5. Transcriptome Analysis
3.6. Data Mining for Flowering Genes
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yusupov, Z.; Deng, T.; Volis, S.; Khassanov, F.; Makhmudjanov, D.; Tojibaev, K.; Sun, H. Phylogenomics of Allium section Cepa (Amaryllidaceae) provides new insights on domestication of onion. Plant Divers. 2021, 43, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Tokenova, A.; Sitpayeva, G.; Gemejiyeva, N.; Suleimenova, S.; Friesen, N.; Batayeva, D. Wild Allium longicuspis Regel is a Feral Form of Allium sativum L. in Kazakhstan: A Comparative Molecular Genetic Analysis. Online J. Biol. Sci. 2023. [Google Scholar] [CrossRef]
- Etoh, T.; Simon, P.W. Diversity, fertility and seed production of garlic. In Allium Crop Sciences: Recent Advances; Rabinowitch, H.D., Currah, L., Eds.; CAB International: Wallingford, UK, 2002; pp. 101–117. [Google Scholar]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef]
- Takagi, H. Garlic (Allium sativum L.). In Onions and Allied Crops; Rabinowitch, H.D., Brewster, J.L., Eds.; CRC Press: Boca Raton, FL, USA, 1990; Volume III, pp. 109–157. [Google Scholar]
- Purugganan, M.D. Evolutionary insights into the nature of plant domestication. Curr. Biol. 2019, 29, R705–R714. [Google Scholar] [CrossRef] [PubMed]
- Denham, T.; Barton, H.; Castillo, C.; Crowther, A.; Dotte-Sarout, E.; Florin, S.A.; Pritchard, J.; Barron, A.; Zhang, Y.; Fuller, D.Q. The domestication syndrome in vegetatively propagated field crops. Ann. Bot. 2020, 125, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Maaß, H.I.; Klaas, M. Infraspecific differentiation of garlic (Allium sativum L.) by isozyme and RAPD markers. Theor. Appl. Genet. 1995, 91, 89–97. [Google Scholar] [CrossRef]
- Vavilov, N.I. The Origin, Variation, Immunity and Breeding of Cultivated Plants; Chester, S., Translator; The Chronica Botanica Co.: Waltham, MA, USA; Stechert-Hafner, Inc.: New York, NY, USA, 1951; Volume 72, No. 6. [Google Scholar]
- Ross-Ibarra, J.; Morrel, P.L.; Gaut, B.S. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. USA 2007, 104 (Suppl. S1), 8641–8648. [Google Scholar] [CrossRef]
- Shemesh-Mayer, E.; Kamenetsky-Goldstein, R. Traditional and novel approaches in garlic (Allium sativum L.) breeding. In Advances in Plant Breeding Strategies: Vegetable Crops; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2021; Volume 8, pp. 3–49. [Google Scholar]
- Tchórzewska, D.; Deryło, K.; Winiarczyk, K. Cytological and biophysical comparative analysis of cell structures at the microsporogenesis stage in sterile and fertile Allium species. Planta 2017, 245, 137–150. [Google Scholar] [CrossRef]
- Shemesh-Mayer, E.; Faigenboim, A.; Ben Michael, T.E.; Kamenetsky-Goldstein, R. Integrated genomic and transcriptomic elucidation of flowering in garlic. Int. J. Mol. Sci. 2022, 23, 13876. [Google Scholar] [CrossRef]
- Ricroch, A.; Yockteng, R.; Brown, S.C.; Nadot, S. Evolution of genome size across some cultivated Allium species. Genome 2005, 48, 511–520. [Google Scholar] [CrossRef]
- Fritsch, R.M.; Friesen, N. Evolution, domestication and taxonomy. In Allium Crop Sciences: Recent Advances; Rabinowitch, H.D., Currah, L., Eds.; CABI: Wallingford, UK, 2002; pp. 5–30. [Google Scholar]
- Sun, X.; Zhu, S.; Li, N.; Cheng, Y.; Zhao, J.; Qiao, X.; Lu, L.; Liu, S.; Wang, Y.; Liu, C.; et al. A chromosome-level genome assembly of garlic (Allium sativum) provides insights into genome evolution and allicin biosynthesis. Mol. Plant 2020, 13, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Kamenetsky, R.; Faigenboim, A.; Shemesh-Mayer, E.; Ben Michael, T.; Gershberg, C.; Kimhi, S.; Esquira, I.; Rohkin Shalom, S.; Eshel, D.; Rabinowitch, H.D. Integrated transcriptome catalogue and organ-specific profiling of gene expression in fertile garlic (Allium sativum L.). BMC Genom. 2015, 16, 1–16. [Google Scholar] [CrossRef]
- Liu, T.; Zeng, L.; Zhu, S.; Chen, X.; Tang, Q.; Mei, S.; Tang, S. Large-scale development of expressed sequence tag-derived simple sequence repeat markers by deep transcriptome sequencing in garlic (Allium sativum L.). Mol. Breed. 2015, 35, 204. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Zhang, H.; Liu, Z.; Wang, Y.; Xing, L.; He, Q.; Du, H. Plant pan-genomics: Recent advances, new challenges, and roads ahead. J. Genet. Genom. 2022, 49, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Shemesh-Mayer, E.; Winiarczyk, K.; Błaszczyk, L.; Kosmala, A.; Rabinowitch, H.D.; Kamenetsky, R. Male gametogenesis and sterility in garlic (Allium sativum L.): Barriers on the way to fertilization and seed production. Planta 2013, 237, 103–120. [Google Scholar] [CrossRef] [PubMed]
- Vavilov, N.I. Studies on the Origin of Cultivated Plants; Institut de Botanique Appliquée et d’Amélioration des Plantes: Leningrad, Russia, 1926. (In Russian) [Google Scholar]
- Kamenetsky, R.; London Shafir, I.; Khassanov, F.; Kik, C.; Van Heusden, A.W.; Vrielink-van Ginkel, M.; Burger-Meijer, K.; Auger, J.; Arnault, I.; Rabinowitch, H.D. Diversity in fertility potential and organo-sulphur compounds among garlics from Central Asia. Biodivers. Conserv. 2005, 14, 281–295. [Google Scholar] [CrossRef]
- Rohkin Shalom, S.; Gillett, D.; Zemach, H.; Kimhi, S.; Forer, I.; Zutahy, Y.; Tam, Y.; Teper-Bamnolker, P.; Kamenetsky, R.; Eshel, D. Storage temperature controls the timing of garlic bulb formation via shoot apical meristem termination. Planta 2015, 242, 951–962. [Google Scholar] [CrossRef]
- Spengler, R.N., III. Fruit from the Sands: The Silk Road Origins of the Foods We Eat; CORRECT: Oakland, CA, USA, 2019. [Google Scholar]
- Kazakova, A.A. Most common onion species, their origin and intraspecific classification. Tr. Prikl. Bot. Genet. I Sel. 1971, 45, 19–41. (In Russian) [Google Scholar]
- Engeland, R.L. Growing Great Garlic: The Definitive Guide for Organic Gardeners and Small Farmers; Filaree Productions: Okanogan, WA, USA., 1991. [Google Scholar]
- Shemesh, E.; Scholten, O.; Rabinowitch, H.D.; Kamenetsky, R. Unlocking variability: Inherent variation and developmental traits of garlic plants originated from sexual reproduction. Planta 2008, 227, 1013–1024. [Google Scholar] [CrossRef]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef]
- Holsinger, K.E.; Weir, B.S. Genetics in geographically structured populations: Defining, estimating and interpreting FST. Nat. Rev. Genet. 2009, 10, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Zhao, Q.; Song, J.; Zhang, X.; Yang, W.; Du, Z.; Zhu, Y.; Wang, H. Large-scale population structure and genetic architecture of agronomic traits of garlic. Hortic. Res. 2023, 10, uhad034. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.G.; Ai, X.Y.; Zhang, J.Z. Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip. Rev. RNA 2014, 5, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sung, S. Genetic and epigenetic mechanisms underlying vernalization. Arabidopsis. Book/Am. Soc. Plant Biol. 2014, 12, e0171. [Google Scholar] [CrossRef]
- Ream, T.S.; Woods, D.P.; Amasino, R.M. The molecular basis of vernalization in different plant groups. In Cold Spring Harbor Symposia on Quantitative Biology; Harbor Laboratory Press: Cold Spring, NY, USA, 2012; Volume 77, pp. 105–115. [Google Scholar]
- Kurokura, T.; Mimida, N.; Battey, N.H.; Hytönen, T. The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 2013, 64, 4131–4141. [Google Scholar] [CrossRef]
- Ben Michael, T.E.; Faigenboim, A.; Shemesh-Mayer, E.; Forer, I.; Gershberg, C.; Shafran, H.; Rabinowitch, H.D.; Kamenetsky-Goldstein, R. Crosstalk in the darkness: Bulb vernalization activates meristem transition via circadian rhythm and photoperiodic pathway. BMC Plant Biol. 2020, 20, 77. [Google Scholar] [CrossRef] [PubMed]
- Antoniou-Kourounioti, R.L.; Zhao, Y.; Dean, C.; Howard, M. Feeling every bit of winter–distributed temperature sensitivity in vernalization. Front. Plant Sci. 2021, 12, 628726. [Google Scholar] [CrossRef]
- Ben Michael, T.; Shemesh-Mayer, E.; Kimhi, S.; Gershberg, C.; Forer, I.; de Ávila, V.T.; Rabinowitch, H.D.; Kamenetsky-Goldstein, R. Temporal and spatial effect of low pre-planting temperatures on plant architecture and flowering in bolting garlic. Sci. Hortic. 2018, 242, 69–75. [Google Scholar] [CrossRef]
- Whittaker, C.; Dean, C. The FLC locus: A platform for discoveries in epigenetics and adaptation. Ann. Rev. Cell Dev. Biol. 2017, 33, 555–575. [Google Scholar] [CrossRef]
- Kennedy, A.; Geuten, K. The role of FLOWERING LOCUS C relatives in cereals. Front. Plant Sci. 2020, 11, 617340. [Google Scholar] [CrossRef]
- Maurya, J.P.; Bhalerao, R.P. Photoperiod-and temperature-mediated control of growth cessation and dormancy in trees: A molecular perspective. Ann. Bot. 2017, 120, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Aljaser, J.A.; Anderson, N.O.; Noyszewski, A. Discovery of UPSTREAM OF FLOWERING LOCUS C (UFC) and FLOWERING LOCUS C EXPRESSOR (FLX) in Gladiolus ×hybridus, G. dalenii. Ornam. Plant Res. 2022, 2, 13. [Google Scholar] [CrossRef]
- Finnegan, E.J.; Sheldon, C.C.; Jardinaud, F.; Peacock, W.J.; Dennis, E.S. A cluster of Arabidopsis genes with a coordinate response to an environmental stimulus. Curr. Biol. 2004, 14, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Ezer, D.; Jung, J.H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stöckle, D.; et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef]
- Junior, C.A.S.; D’Amico-Damião, V.; Carvalho, R.F. Phytochrome type B family: The abiotic stress responses signaller in plants. Ann. Appl. Biol. 2021, 178, 135–148. [Google Scholar] [CrossRef]
- Jin, S.; Nasim, Z.; Susila, H.; Ahn, J.H. Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin. Cell Dev. Boil. 2021, 109, 20–30. [Google Scholar] [CrossRef]
- Khosa, J.; Bellinazzo, F.; Kamenetsky Goldstein, R.; Macknight, R.; Immink, R.G. PHOSPHATIDYLETHANOLAMINE-BINDING PROTEINS: The conductors of dual reproduction in plants with vegetative storage organs. J. Exp. Bot. 2021, 72, 2845–2856. [Google Scholar] [CrossRef]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef]
- Etoh, T. Fertility of the garlic clones collected in Soviet Central Asia. J. Jpn. Soc. Hortic. Sci. 1986, 55, 312–319. [Google Scholar] [CrossRef]
- Kamenetsky, R.; Shafir London, I.; Baizerman, M.; Khassanov, F.; Kik, C.; Rabinowitch, H.D. Garlic (Allium sativum L.) and its wild relatives from central Asia: Evaluation for fertility potential. Acta Hortic. 2004, 637, 83–91. [Google Scholar] [CrossRef]
- Rivlin, R.S. Historical perspective on the use of garlic. J. Nutr. 2001, 131, 951S–954S. [Google Scholar] [CrossRef] [PubMed]
- Parreño, R.; Rodríguez-Alcocer, E.; Martínez-Guardiola, C.; Carrasco, L.; Castillo, P.; Arbona, V.; Jover-Gil, S.; Candela, H. Turning Garlic into a Modern Crop: State of the Art and Perspectives. Plants 2023, 12, 1212. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Gabor, M.; Goncalo, A.; Richard, D. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 1, 113–116. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Rotem, N.; Shemesh, E.; Peretz, Y.; Akad, F.; Edelbaum, O.; Rabinowitch, H.D.; Sela, I.; Kamenetsky, R. Reproductive development and phenotypic differences in garlic are associated with expression and splicing of LEAFY homologue gaLFY. J. Exp. Bot. 2007, 58, 1133–1141. [Google Scholar] [CrossRef][Green Version]
- Rotem, N.; David-Schwartz, R.; Peretz, Y.; Sela, I.; Rabinowitch, H.D.; Flaishman, M.; Kamenetsky, R. Flower development in garlic: The ups and downs of gaLFY expression. Planta 2011, 233, 1063–1072. [Google Scholar]
- Shemesh-Mayer, E.; Ben-Michael, T.; Rotem, N.; Rabinowitch, H.D.; Doron-Faigenboim, A.; Kosmala, A.; Perlikowski, D.; Sherman, A.; Kamenetsky, R. Garlic (Allium sativum L.) fertility: Transcriptome and proteome analyses provide insight into flower and pollen development. Front. Plant Sci. 2015, 6, 271. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shemesh-Mayer, E.; Faigenboim, A.; Sherman, A.; Gao, S.; Zeng, Z.; Liu, T.; Kamenetsky-Goldstein, R. Deprivation of Sexual Reproduction during Garlic Domestication and Crop Evolution. Int. J. Mol. Sci. 2023, 24, 16777. https://doi.org/10.3390/ijms242316777
Shemesh-Mayer E, Faigenboim A, Sherman A, Gao S, Zeng Z, Liu T, Kamenetsky-Goldstein R. Deprivation of Sexual Reproduction during Garlic Domestication and Crop Evolution. International Journal of Molecular Sciences. 2023; 24(23):16777. https://doi.org/10.3390/ijms242316777
Chicago/Turabian StyleShemesh-Mayer, Einat, Adi Faigenboim, Amir Sherman, Song Gao, Zheng Zeng, Touming Liu, and Rina Kamenetsky-Goldstein. 2023. "Deprivation of Sexual Reproduction during Garlic Domestication and Crop Evolution" International Journal of Molecular Sciences 24, no. 23: 16777. https://doi.org/10.3390/ijms242316777
APA StyleShemesh-Mayer, E., Faigenboim, A., Sherman, A., Gao, S., Zeng, Z., Liu, T., & Kamenetsky-Goldstein, R. (2023). Deprivation of Sexual Reproduction during Garlic Domestication and Crop Evolution. International Journal of Molecular Sciences, 24(23), 16777. https://doi.org/10.3390/ijms242316777








