Comparative Transcriptome Sequencing and Endogenous Phytohormone Content of Annual Grafted Branches of Zelkova schneideriana and Its Dwarf Variety HenTianGao
Abstract
:1. Introduction
2. Results
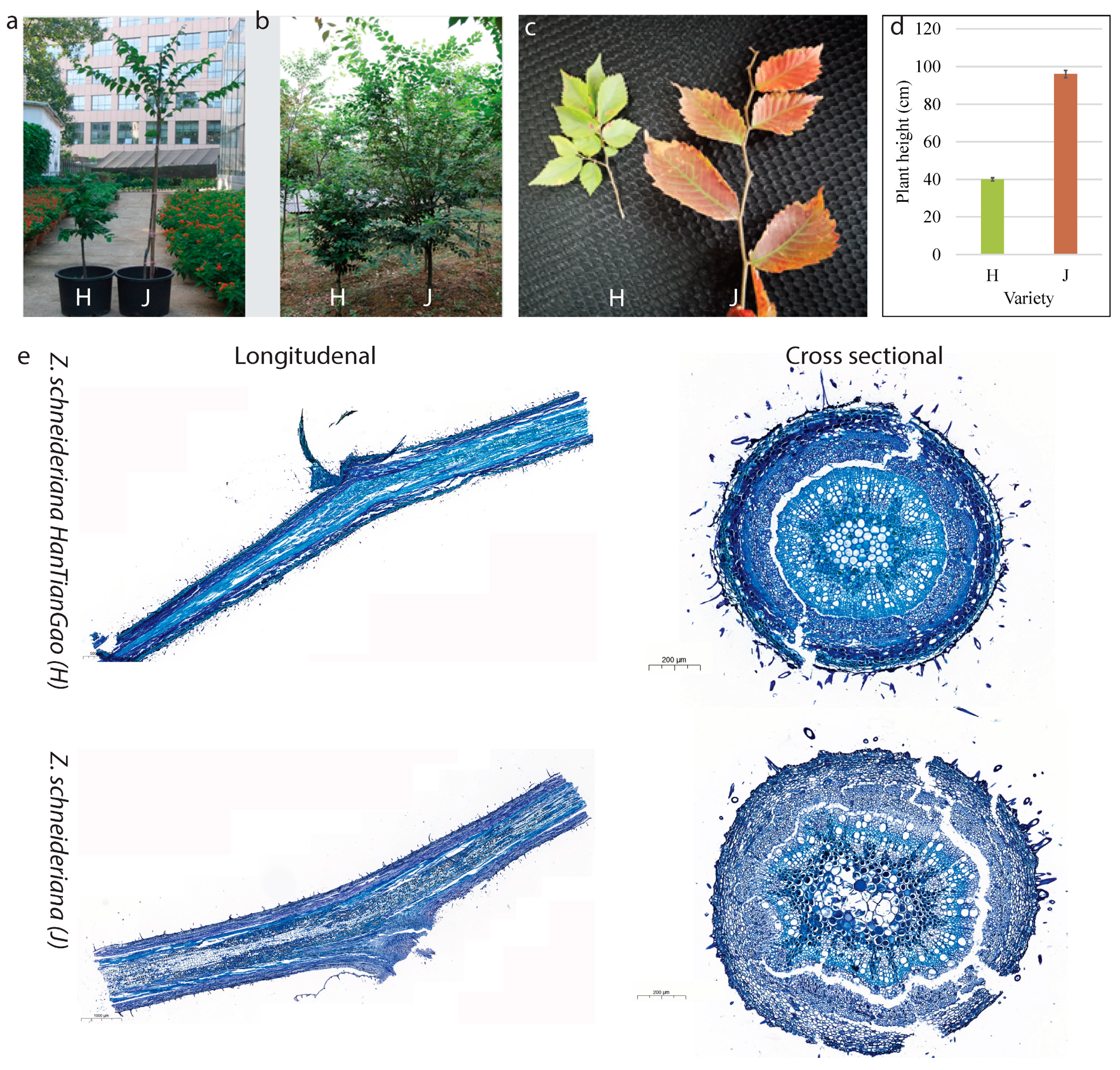
2.1. Growth Form and Histological Observations
2.2. Phytohormone Profiles of Z. schneideriana and HenTianGao
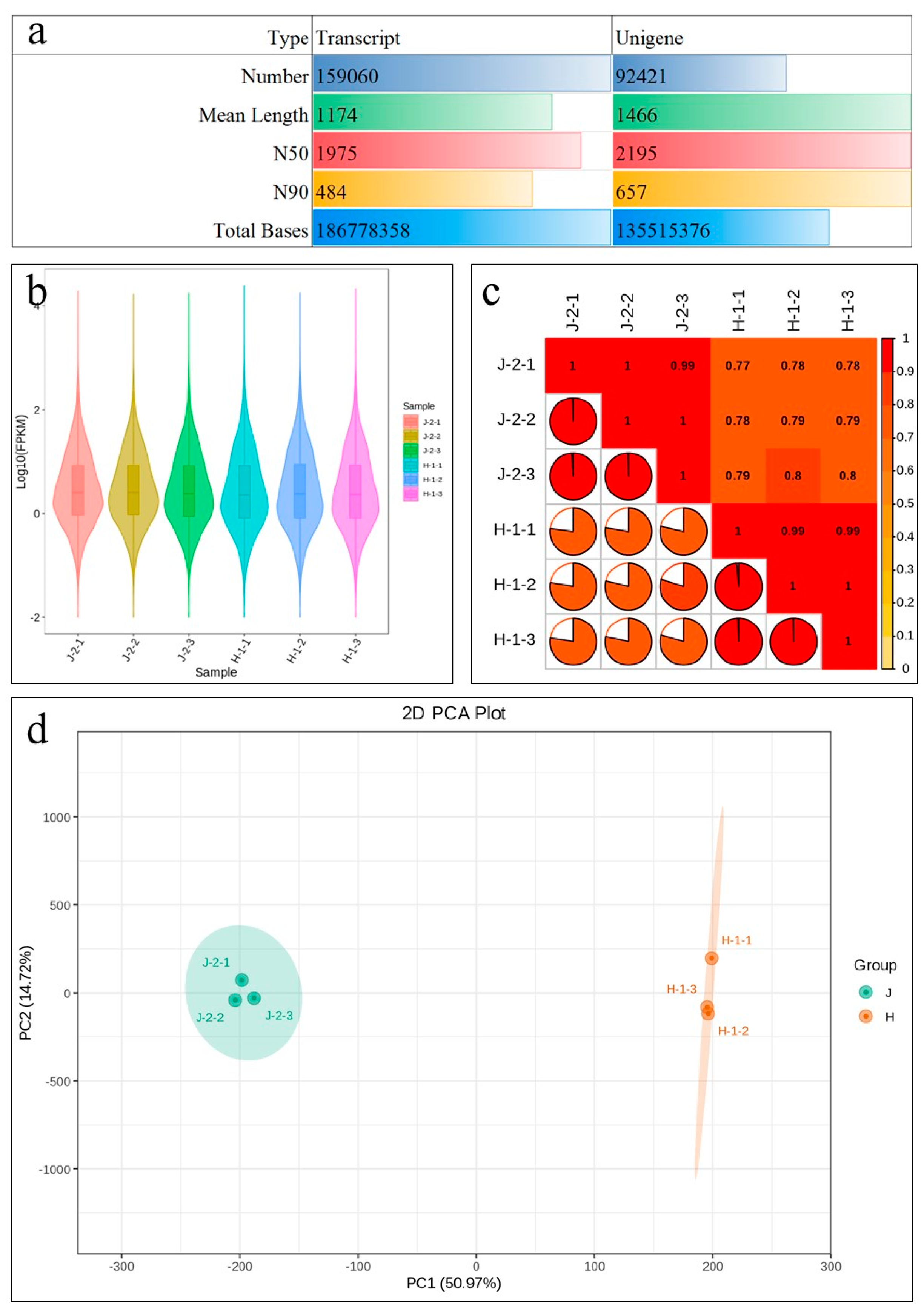
2.3. Transcriptome Sequencing of Z. schneideriana and HenTianGao
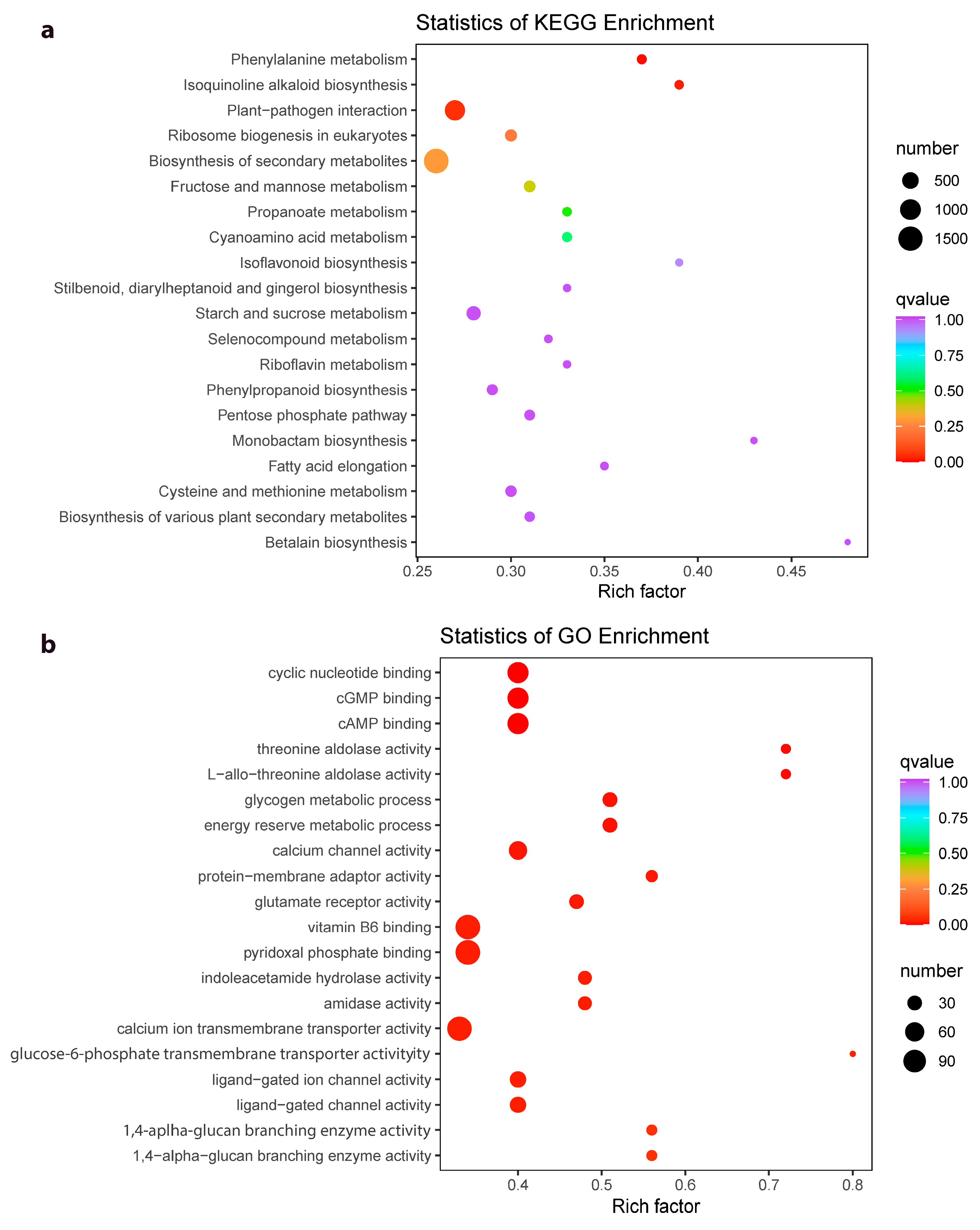
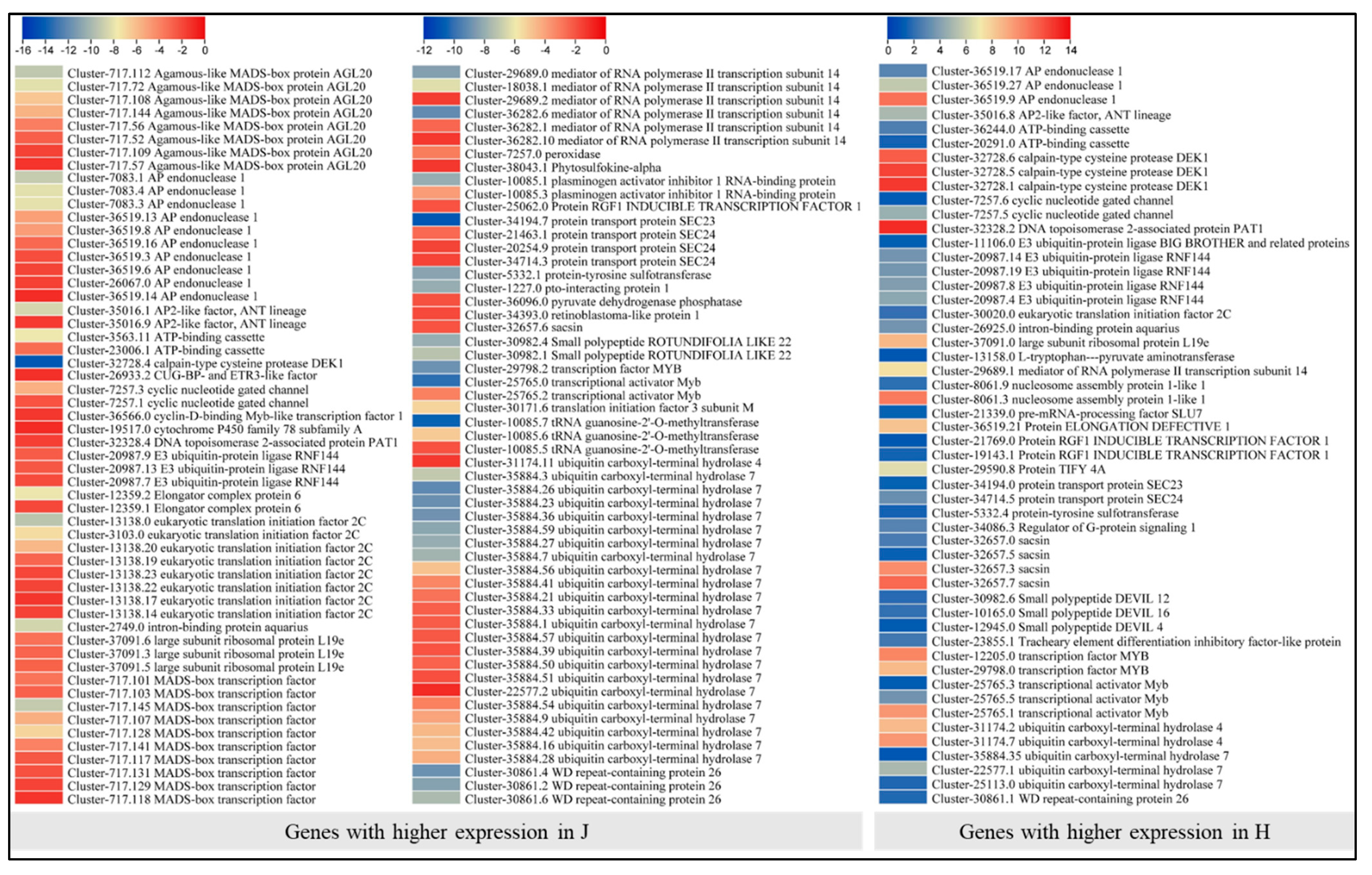
2.4. Differential Gene Expression between Z. schneideriana and HenTianGao
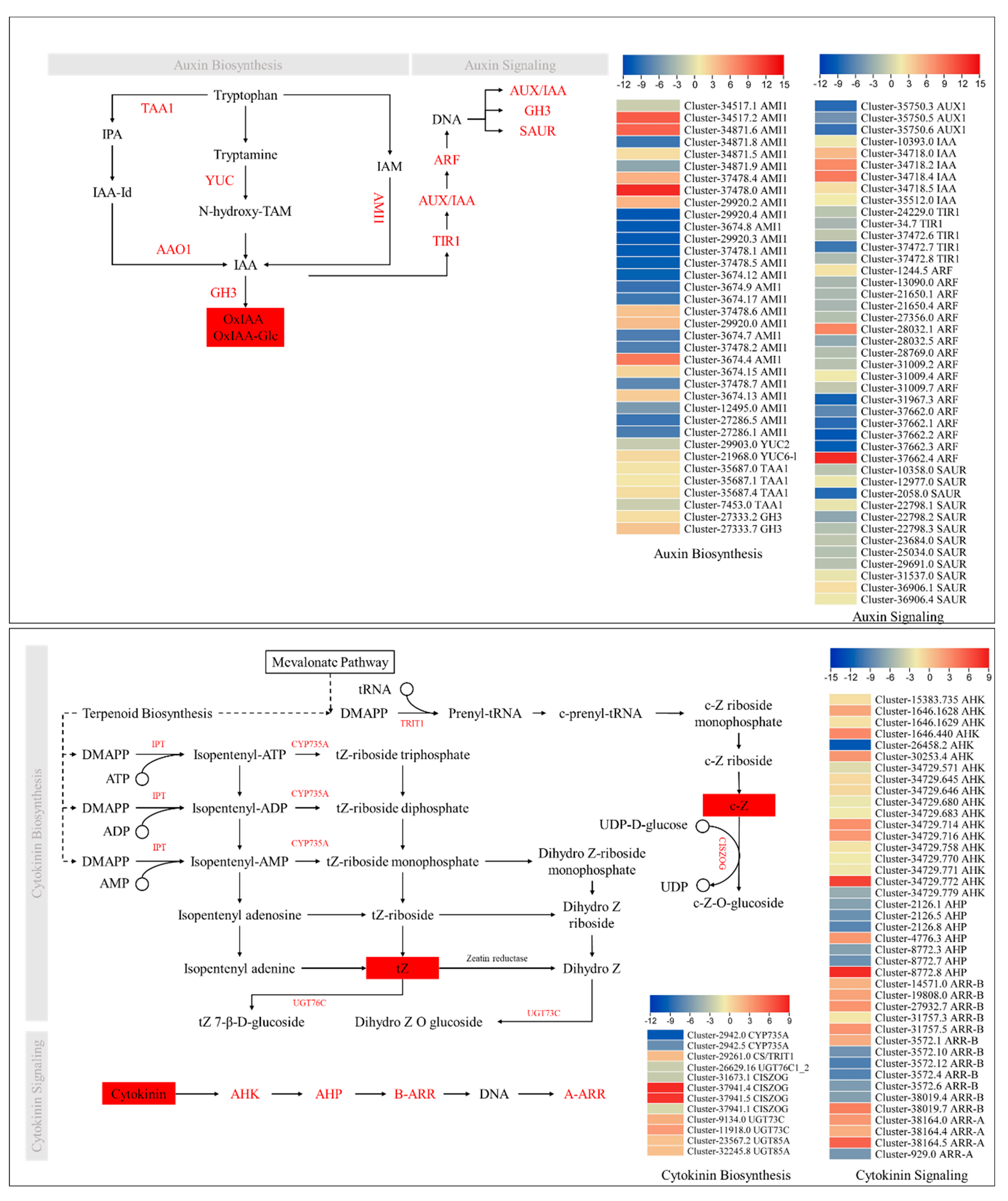
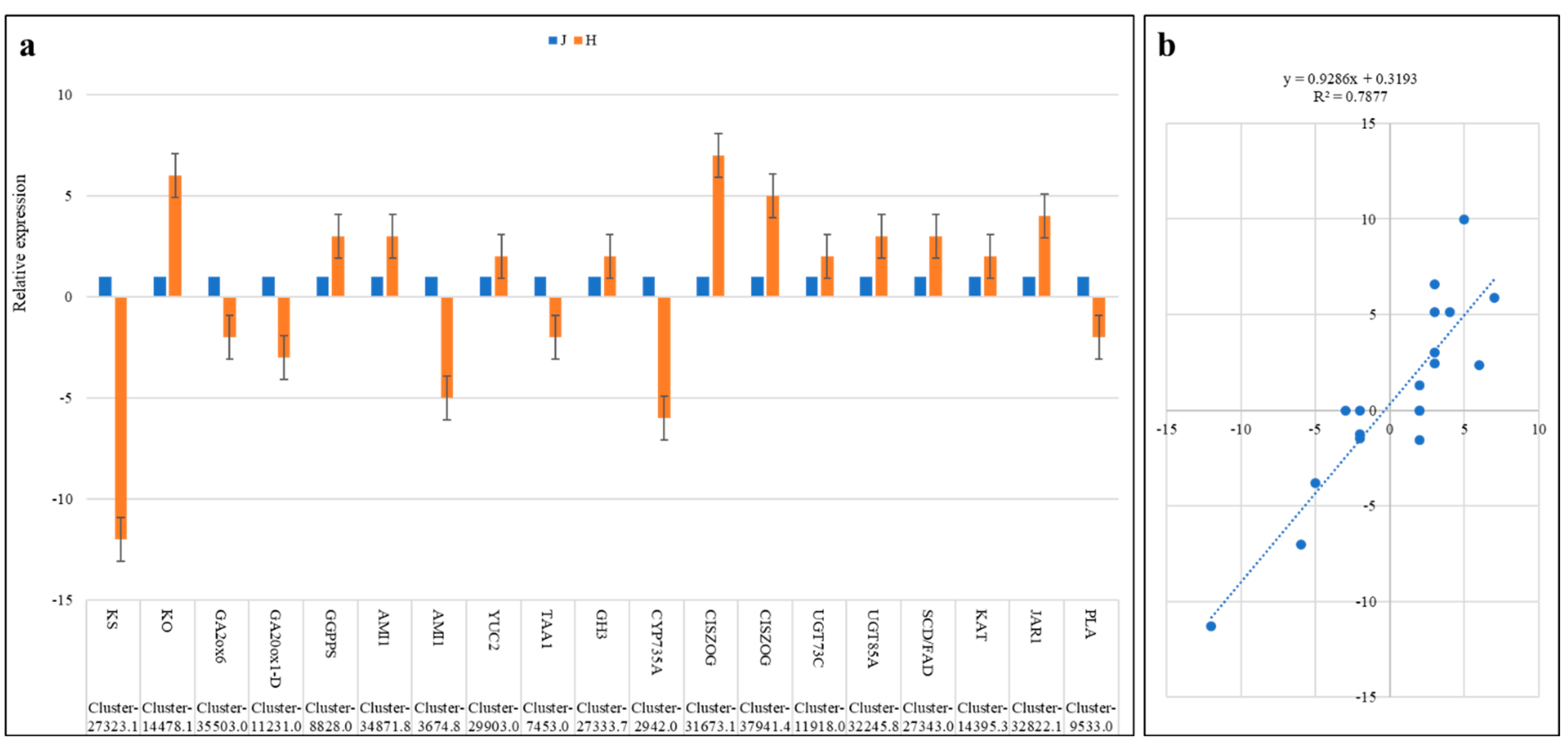
2.5. Transcriptome Sequencing Confirm the Phytohormone Accumulation Trends
2.6. Expression Changes in Genes Related to Stem Elongation, Cell Size, Number, and Proliferation
2.7. Differential Expression of Genes Related to Cell Wall
2.8. Differential Expression of Transcription Factors
2.9. Validation of Gene Expression by qRT-PCR Analysis
3. Discussion
4. Material and Methods
4.1. Plant Material and Growth Conditions
4.2. Histological Analysis
4.3. Quantification of Endogenous Phytohormones
4.4. Transcriptome Analysis
4.5. qRT-PCR Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Song, Y.; Bétrisey, S.; Kozlowski, G. Zelkova schneideriana. The IUCN Red List of Threatened Species 2018. In 2018 edn: IUCN Global Species Programme, the IUCN Species Survival Commission (SSC) and the IUCN Red List Partnership; WORLD HEADQUARTERS: Gland, Switzerland, 2018; p. 13. [Google Scholar]
- Sun, J.; Qiu, H.; Guo, J.; Xu, X.; Wu, D.; Zhong, L.; Jiang, B.; Jiao, J.; Yuan, W.; Huang, Y. Modeling the potential distribution of Zelkova schneideriana under different human activity intensities and climate change patterns in China. Glob. Ecol. Conserv. 2020, 21, e00840. [Google Scholar] [CrossRef]
- Jin, X.; Wang, X.; Liu, X.; Hu, X.; Liao, F. A New Variety of Zelkova schneideriana ‘HenTianGao’. Sci. Silvae Sin. 2016, 52, 1. [Google Scholar]
- Carra, A.; Catalano, C.; Badalamenti, O.; Carimi, F.; Pasta, S.; Motisi, A.; Abbate, L.; La Bella, F.; Fazan, L.; Kozlowski, G. Overcoming sexual sterility in conservation of endangered species: The prominent role of biotechnology in the multiplication of Zelkova sicula (Ulmaceae), a relict tree at the brink of extinction. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 137, 139–148. [Google Scholar] [CrossRef]
- Zhang, M.-L.; Wang, L.; Lei, Y.; Sanderson, S.C. Cenozoic evolutionary history of Zelkova (Ulmaceae), evidenced from ITS, trnL-trnF, psbA-trnH, and rbcL. Tree Genet. Genomes 2017, 13, 101. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, R.; Geng, M.; Qin, Y.; Liu, H.; Li, L.; Li, M. De novo transcriptome assembly and EST-SSR markers development for Zelkova schneideriana Hand.-Mazz.(Ulmaceae). 3 Biotech 2021, 11, 420. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, R.; Geng, M.; Zhu, J.; An, J.; Ma, J. Chloroplast analysis of Zelkova schneideriana (Ulmaceae): Genetic diversity, population structure, and conservation implications. Genet. Mol. Res 2016, 15, 1–9. [Google Scholar] [CrossRef]
- Dou, Q.; Zhang, M.; Huang, L.; Yu, S. Variation in growth of seedlings and saplings of plus tree progeny of Zelkova schneideriana. J. Jiangsu For. Sci. Technol. 2014, 41, 1–4. [Google Scholar]
- Wang, Y.; Zhao, J.; Lu, W.; Deng, D. Gibberellin in plant height control: Old player, new story. Plant Cell Rep. 2017, 36, 391–398. [Google Scholar] [CrossRef]
- Jiang, Z.-F.; Liu, D.-D.; Wang, T.-Q.; Liang, X.-L.; Cui, Y.-H.; Liu, Z.-H.; Li, W.-B. Concentration difference of auxin involved in stem development in soybean. J. Integr. Agric. 2020, 19, 953–964. [Google Scholar] [CrossRef]
- Yan, L.; Wang, W.-p.; Zhang, L.; Zhu, L.-f.; Zhang, X.-l.; Xin, H. The HD-Zip transcription factor GhHB12 represses plant height by regulating the auxin signaling in cotton. J. Integr. Agric. 2022, 22, 2015–2024. [Google Scholar]
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of jasmonic acid in plants: The molecular point of view. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Zhang, Z.; Li, S.; Zhang, S.; Li, Q.; Zhang, Z.-H.; Huang, S.; Yang, X. Genetic regulation of ethylene dosage for cucumber fruit elongation. Plant Cell 2019, 31, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Rupe, M.A.; Dieter, J.A.; Zou, J.; Spielbauer, D.; Duncan, K.E.; Howard, R.J.; Hou, Z.; Simmons, C.R. Cell Number Regulator1 affects plant and organ size in maize: Implications for crop yield enhancement and heterosis. Plant Cell 2010, 22, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.; Lenhard, M. Genetic control of plant organ growth. New Phytol. 2011, 191, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Carver, B.F.; Wang, S.; Zhang, F.; Yan, L. Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor. Appl. Genet. 2009, 118, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.-H. Characterization of a cinnamoyl-CoA reductase that is associated with stem development in wheat. J. Exp. Bot. 2007, 58, 2011–2021. [Google Scholar] [CrossRef]
- Hanzawa, Y.; Takahashi, T.; Michael, A.J.; Burtin, D.; Long, D.; Pineiro, M.; Coupland, G.; Komeda, Y. ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J. 2000, 19, 4248–4256. [Google Scholar] [CrossRef]
- Peaucelle, A.; Braybrook, S.; Höfte, H. Cell wall mechanics and growth control in plants: The role of pectins revisited. Front. Plant Sci. 2012, 3, 121. [Google Scholar] [CrossRef]
- Sun, T.-p. Gibberellin signal transduction in stem elongation & leaf growth. In Plant Hormones; Springer: Berlin/Heidelberg, Germany, 2010; pp. 308–328. [Google Scholar]
- Jan, A.; Yang, G.; Nakamura, H.; Ichikawa, H.; Kitano, H.; Matsuoka, M.; Matsumoto, H.; Komatsu, S. Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol. 2004, 136, 3670–3681. [Google Scholar] [CrossRef]
- Kou, E.; Huang, X.; Zhu, Y.; Su, W.; Liu, H.; Sun, G.; Chen, R.; Hao, Y.; Song, S. Crosstalk between auxin and gibberellin during stalk elongation in flowering Chinese cabbage. Sci. Rep. 2021, 11, 3976. [Google Scholar] [CrossRef]
- Keyes, G.; Sorrells, M.E.; Setter, T.L. Gibberellic acid regulates cell wall extensibility in wheat (Triticum aestivum L.). Plant Physiol. 1990, 92, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Derbyshire, P.; Findlay, K.; McCann, M.C.; Roberts, K. Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J. Exp. Bot. 2007, 58, 2079–2089. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid auxin-mediated cell expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Diffuse growth of plant cell walls. Plant Physiol. 2018, 176, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.A.; Rehman, H.M.; Imtiaz, M.; Baloch, F.S.; Lee, J.D.; Yang, S.H.; Lee, S.I.; Chung, G. Systems identification and characterization of cell wall reassembly and degradation related genes in Glycine max (L.) Merill, a bioenergy legume. Sci. Rep. 2017, 7, 10862. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.A.; Lin, X.; Chan, T.-F.; Imtiaz, M.; Rehman, H.M.; Ali, M.A.; Baloch, F.S.; Atif, R.M.; Yang, S.H.; Chung, G. Characterization of cellulose synthase A (CESA) gene family in eudicots. Biochem. Genet. 2019, 57, 248–272. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef]
- Pelletier, S.; Van Orden, J.; Wolf, S.; Vissenberg, K.; Delacourt, J.; Ndong, Y.A.; Pelloux, J.; Bischoff, V.; Urbain, A.; Mouille, G. A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol. 2010, 188, 726–739. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef]
- Park, J.; Lee, S.; Park, G.; Cho, H.; Choi, D.; Umeda, M.; Choi, Y.; Hwang, D.; Hwang, I. Cytokinin-responsive growth regulator regulates cell expansion and cytokinin-mediated cell cycle progression. Plant Physiol. 2021, 186, 1734–1746. [Google Scholar] [CrossRef]
- Robson, F.; Okamoto, H.; Patrick, E.; Harris, S.-R.; Wasternack, C.; Brearley, C.; Turner, J.G. Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 2010, 22, 1143–1160. [Google Scholar] [CrossRef]
- Mellor, N.; Band, L.R.; Pěnčík, A.; Novák, O.; Rashed, A.; Holman, T.; Wilson, M.H.; Voß, U.; Bishopp, A.; King, J.R. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 11022–11027. [Google Scholar] [CrossRef]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef]
- Hou, B.; Lim, E.-K.; Higgins, G.S.; Bowles, D.J. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 47822–47832. [Google Scholar] [CrossRef]
- Jarratt-Barnham, E.; Wang, L.; Ning, Y.; Davies, J.M. The complex story of plant cyclic nucleotide-gated channels. Int. J. Mol. Sci. 2021, 22, 874. [Google Scholar] [CrossRef]
- Do, T.H.T.; Martinoia, E.; Lee, Y.; Hwang, J.-U. 2021 update on ATP-binding cassette (ABC) transporters: How they meet the needs of plants. Plant Physiol. 2021, 187, 1876–1892. [Google Scholar] [CrossRef]
- Yang, L.; Liu, H.; Zhao, J.; Pan, Y.; Cheng, S.; Lietzow, C.D.; Wen, C.; Zhang, X.; Weng, Y. LITTLELEAF (LL) encodes a WD40 repeat domain-containing protein associated with organ size variation in cucumber. Plant J. 2018, 95, 834–847. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, Z.; Zhao, H. Preliminary investigation on the suitability of some deciduous broad-leaved trees in Beijing. J. Chin. Landsc. Arch. 2005, 8, 62–66. [Google Scholar]
- Gray, W.M. Hormonal regulation of plant growth and development. PLoS Biol. 2004, 2, e311. [Google Scholar] [CrossRef]
- Zhou, Y.; Underhill, S.J. Breadfruit (Artocarpus altilis) DELLA genes: Gibberellin-regulated stem elongation and response to high salinity and drought. Plant Growth Regul. 2017, 83, 375–383. [Google Scholar] [CrossRef]
- Sun, T.-P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arab. Book/Am. Soc. Plant Biol. 2008, 6, e0103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, W.; Huang, J.; Wang, Y.; Hu, L.; Yuan, Y.; Lyu, M.; Wu, B. Role of gibberellin and its three GID1 receptors in Jasminum sambac stem elongation and flowering. Planta 2022, 255, 17. [Google Scholar] [CrossRef] [PubMed]
- Pedmale, U.V.; Huang, S.-s.C.; Zander, M.; Cole, B.J.; Hetzel, J.; Ljung, K.; Reis, P.A.; Sridevi, P.; Nito, K.; Nery, J.R. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 2016, 164, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.J.; O’neill, D.P.; Wolbang, C.M.; Symons, G.M.; Reid, J.B. Auxin-gibberellin interactions and their role in plant growth. J. Plant Growth Regul. 2001, 20, 336–353. [Google Scholar] [CrossRef] [PubMed]
- Pěnčík, A.; Simonovik, B.; Petersson, S.V.; Henyková, E.; Simon, S.; Greenham, K.; Zhang, Y.; Kowalczyk, M.; Estelle, M.; Zažímalová, E. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell 2013, 25, 3858–3870. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.-i.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef]
- Reed, J.W.; Wu, M.-F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three auxin response factors promote hypocotyl elongation. Plant Physiol. 2018, 178, 864–875. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinin biosynthesis and regulation. Vitam. Horm. 2005, 72, 271–287. [Google Scholar]
- Schäfer, M.; Brütting, C.; Meza-Canales, I.D.; Großkinsky, D.K.; Vankova, R.; Baldwin, I.T.; Meldau, S. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef] [PubMed]
- Lacuesta, M.; Saiz-Fernández, I.; Podlešáková, K.; Miranda-Apodaca, J.; Novák, O.; Doležal, K.; De Diego, N. The trans and cis zeatin isomers play different roles in regulating growth inhibition induced by high nitrate concentrations in maize. Plant Growth Regul. 2018, 85, 199–209. [Google Scholar] [CrossRef]
- Kakimoto, T. Biosynthesis of cytokinins. J. Plant Res. 2003, 116, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, X.-M.; Kojima, M.; Sakakibara, H.; Hou, B.-K. N-glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 2200–2213. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Heinrich, M.; Hettenhausen, C.; Lange, T.; Wünsche, H.; Fang, J.; Baldwin, I.T.; Wu, J. High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of N icotiana attenuata stems. Plant J. 2013, 73, 591–606. [Google Scholar] [CrossRef]
- Wang, G.; Huang, W.; Li, M.; Xu, Z.; Wang, F.; Xiong, A. Expression profiles of genes involved in jasmonic acid biosynthesis and signaling during growth and development of carrot. Acta Biochim. Biophys. Sin. 2016, 48, 795–803. [Google Scholar] [CrossRef]
- Jang, G.; Chang, S.H.; Um, T.Y.; Lee, S.; Kim, J.-K.; Choi, Y.D. Antagonistic interaction between jasmonic acid and cytokinin in xylem development. Sci. Rep. 2017, 7, 10212. [Google Scholar] [CrossRef]
- Bowman, J.L.; Briginshaw, L.N.; Fisher, T.J.; Flores-Sandoval, E. Something ancient and something neofunctionalized—Evolution of land plant hormone signaling pathways. Curr. Opin. Plant Biol. 2019, 47, 64–72. [Google Scholar] [CrossRef]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef] [PubMed]
- Tapia-López, R.; García-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Pérez-Ruíz, R.V.; Kim, S.-H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.A.; Kohorn, B.D. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 2001, 13, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.; McDermott, H.I.; McAllister, T.; Cummins, M.; Silva, J.C.; Mollison, E.; Meikle, R.; Morris, J.; Hedley, P.E.; Waugh, R. APETALA2 control of barley internode elongation. Development 2019, 146, dev170373. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-Q.; Xu, K.-X.; Luo, B.; Wang, J.-W.; Chen, X.-Y. An ATP-binding cassette transporter GhWBC1 from elongating cotton fibers. Plant Physiol. 2003, 133, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Duszyn, M.; Świeżawska, B.; Szmidt-Jaworska, A.; Jaworski, K. Cyclic nucleotide gated channels (CNGCs) in plant signalling—Current knowledge and perspectives. J. Plant Physiol. 2019, 241, 153035. [Google Scholar] [CrossRef] [PubMed]
- Lertpiriyapong, K.; Sung, Z.R. The elongation defective1 mutant of Arabidopsis is impaired in the gene encoding a serine-rich secreted protein. Plant Mol. Biol. 2003, 53, 581–595. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.C.; Roberts, K. Changes in cell wall architecture during cell elongation. J. Exp. Bot. 1994, 45, 1683–1691. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; de Oliveira, D.M.; Andreotti, G.C.; de Souza, L.A.; Dos Santos, W.D.; Bonato, C.M.; Antunes, W.C. Increased gibberellins and light levels promotes cell wall thickness and enhance lignin deposition in xylem fibers. Front. Plant Sci. 2018, 9, 1391. [Google Scholar] [CrossRef]
- Sprangers, K.; Thys, S.; Van Dusschoten, D.; Beemster, G.T. Gibberellin enhances the anisotropy of cell expansion in the growth zone of the maize leaf. Front. Plant Sci. 2020, 11, 1163. [Google Scholar] [CrossRef] [PubMed]
- Buhrow, L.M.; Liu, Z.; Cram, D.; Sharma, T.; Foroud, N.A.; Pan, Y.; Loewen, M.C. Wheat transcriptome profiling reveals abscisic and gibberellic acid treatments regulate early-stage phytohormone defense signaling, cell wall fortification, and metabolic switches following Fusarium graminearum-challenge. BMC Genom. 2021, 22, 798. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Jiang, B.; Zhu, J.; Xiao, G. Auxin promotes fiber elongation by enhancing gibberellic acid biosynthesis in cotton. Plant Biotechnol. J. 2022, 20, 423. [Google Scholar] [CrossRef] [PubMed]
- Berry, H.M.; Argueso, C.T. More than growth: Phytohormone-regulated transcription factors controlling plant immunity, plant development and plant architecture. Curr. Opin. Plant Biol. 2022, 102309. [Google Scholar] [CrossRef]
- Shahnejat-Bushehri, S.; Tarkowska, D.; Sakuraba, Y.; Balazadeh, S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2016, 2, 16013. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.-H. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal. Behav. 2015, 10, e989746. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhou, C.; Nisa, Z.U.; El-Kassaby, Y.A.; Li, W. Exogenous 6-BA inhibited hypocotyl elongation under darkness in Picea crassifolia Kom revealed by transcriptome profiling. Front. Plant Sci. 2023, 14, 1086879. [Google Scholar] [CrossRef] [PubMed]
- Floková, K.; Tarkowská, D.; Miersch, O.; Strnad, M.; Wasternack, C.; Novák, O. UHPLC–MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 2014, 105, 147–157. [Google Scholar] [CrossRef]
- Niu, Q.; Zong, Y.; Qian, M.; Yang, F.; Teng, Y. Simultaneous quantitative determination of major plant hormones in pear flowers and fruit by UPLC/ESI-MS/MS. Anal. Methods 2014, 6, 1766–1773. [Google Scholar] [CrossRef]
- Cui, K.; Lin, Y.; Zhou, X.; Li, S.; Liu, H.; Zeng, F.; Zhu, F.; Ouyang, G.; Zeng, Z. Comparison of sample pretreatment methods for the determination of multiple phytohormones in plant samples by liquid chromatography–electrospray ionization-tandem mass spectrometry. Microchem. J. 2015, 121, 25–31. [Google Scholar] [CrossRef]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. The KEGG Database. In Novartis Foundation Symposium; Wiley Online Library: New York, NY, USA, 2002; pp. 91–100. [Google Scholar]
- Protocol for use with NEBNext Poly (A) mRNA Magnetic Isolation Module (NEB# E7490) and NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (E7760, E7765). Available online: https://www.neb.com/en/protocols/2014/12/01/protocol-for-use-with-nebnext-poly-a-mrna-magnetic-isolation-module-neb-e7490 (accessed on 11 November 2023).
- Lee, S.I.; Muthusamy, M.; Nawaz, M.A.; Hong, J.K.; Lim, M.-H.; Kim, J.A.; Jeong, M.-J. Genome-wide analysis of spatiotemporal gene expression patterns during floral organ development in Brassica rapa. Mol. Genet. Genom. 2019, 294, 1403–1420. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35 (Suppl. 1), D61–D65. [Google Scholar] [CrossRef]
- Apweiler, R. Functional information in SWISS-PROT: The basis for large-scale characterisation of protein sequences. Brief. Bioinform. 2001, 2, 9–18. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. 2), W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Suppl. 2), W316–W322. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Riano-Pachon, D.M.; Corrêa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38 (Suppl. 1), D822–D827. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Jin, X.; Lin, H.; He, J.; Chen, Y. Comparative Transcriptome Sequencing and Endogenous Phytohormone Content of Annual Grafted Branches of Zelkova schneideriana and Its Dwarf Variety HenTianGao. Int. J. Mol. Sci. 2023, 24, 16902. https://doi.org/10.3390/ijms242316902
Huang C, Jin X, Lin H, He J, Chen Y. Comparative Transcriptome Sequencing and Endogenous Phytohormone Content of Annual Grafted Branches of Zelkova schneideriana and Its Dwarf Variety HenTianGao. International Journal of Molecular Sciences. 2023; 24(23):16902. https://doi.org/10.3390/ijms242316902
Chicago/Turabian StyleHuang, Chenfei, Xiaoling Jin, Haiyan Lin, Jinsong He, and Yan Chen. 2023. "Comparative Transcriptome Sequencing and Endogenous Phytohormone Content of Annual Grafted Branches of Zelkova schneideriana and Its Dwarf Variety HenTianGao" International Journal of Molecular Sciences 24, no. 23: 16902. https://doi.org/10.3390/ijms242316902
APA StyleHuang, C., Jin, X., Lin, H., He, J., & Chen, Y. (2023). Comparative Transcriptome Sequencing and Endogenous Phytohormone Content of Annual Grafted Branches of Zelkova schneideriana and Its Dwarf Variety HenTianGao. International Journal of Molecular Sciences, 24(23), 16902. https://doi.org/10.3390/ijms242316902




