Characterization of the Novel Leaderless Bacteriocin, Bawcin, from Bacillus wiedmannii
Abstract
:1. Introduction
2. Results and Discussion
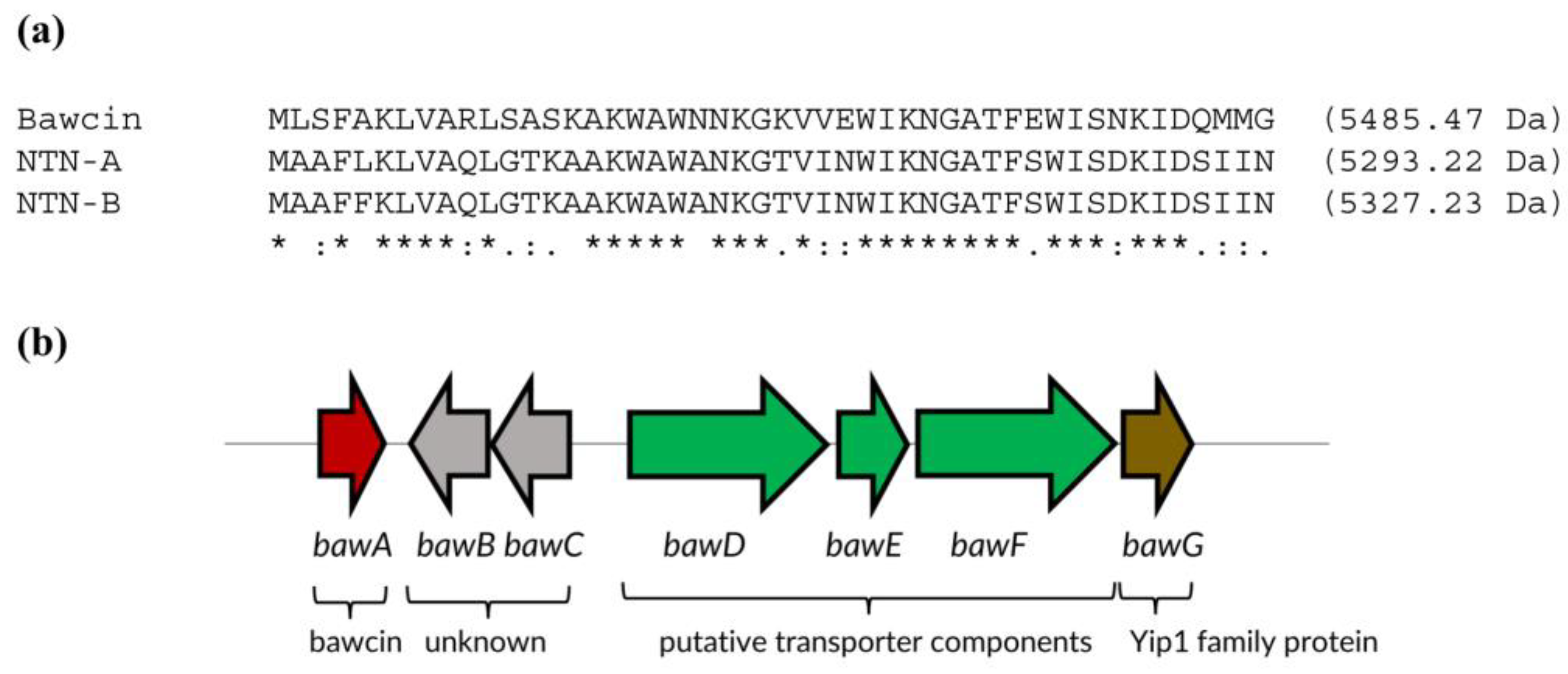
2.1. Bawcin Identification and Biosynthetic Gene Cluster
2.2. Antimicrobial Activity of Bawcin
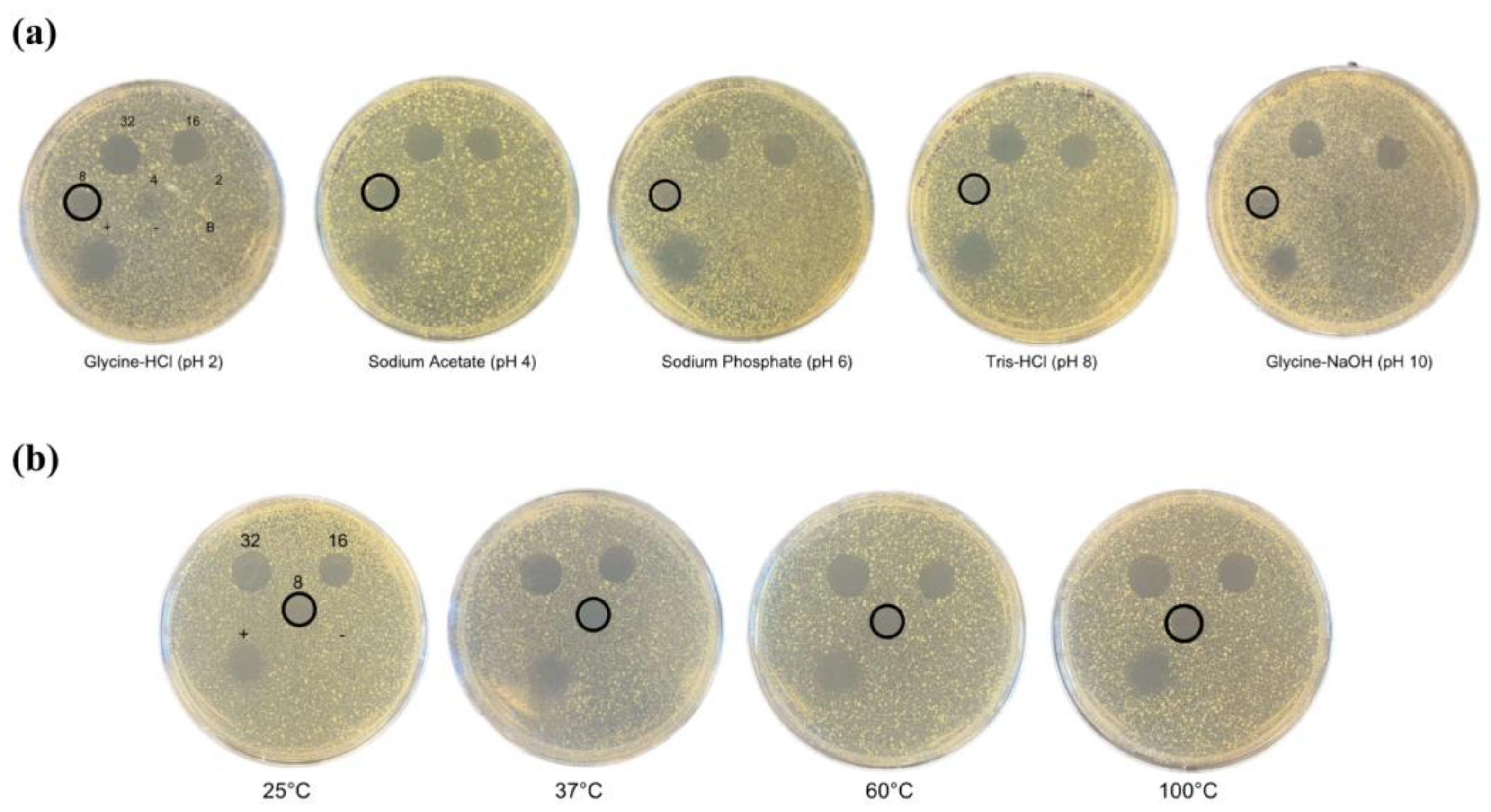
2.3. pH and Temperature Stability of Bawcin
2.4. Protease Stability of Bawcin
2.5. Structural Analysis of Bawcin
3. Materials and Methods
3.1. Bawcin and Bacterial Strains
3.2. Antimicrobial Activity Testing
3.3. pH Stability Testing
3.4. Temperature Stability Testing
3.5. Protease Sensitivity Testing
3.6. Sequence and Structural Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Okoye, C.O.; Okeke, E.S.; Ezeorba, T.P.C.; Chukwudozie, K.I.; Chiejina, C.O.; Fomena Temgoua, N.S. Microbial and Bio-Based Preservatives: Recent Advances in Antimicrobial Compounds; Springer: Singapore, 2022; pp. 53–74. [Google Scholar]
- Acedo, J.Z.; Chiorean, S.; Vederas, J.C.; van Belkum, M.J. The Expanding Structural Variety among Bacteriocins from Gram-Positive Bacteria. FEMS Microbiol. Rev. 2018, 42, 805–828. [Google Scholar] [CrossRef]
- Baindara, P.; Korpole, S.; Grover, V. Bacteriocins: Perspective for the Development of Novel Anticancer Drugs. Appl. Microbiol. Biotechnol. 2018, 102, 10393–10408. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Oman, T.J.; Van Der Donk, W.A. Follow the Leader: The Use of Leader Peptides to Guide Natural Product Biosynthesis. Nat. Chem. Biol. 2010, 6, 9–18. [Google Scholar] [CrossRef]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Circular and Leaderless Bacteriocins: Biosynthesis, Mode of Action, Applications, and Prospects. Front. Microbiol. 2018, 9, 2085. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Sivonen, K. Genome Mining Demonstrates the Widespread Occurrence of Gene Clusters Encoding Bacteriocins in Cyanobacteria. PLoS ONE 2011, 6, e22384. [Google Scholar] [CrossRef]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef]
- Mercado, V.; Olmos, J. Bacteriocin Production by Bacillus Species: Isolation, Characterization, and Application. Probiotics Antimicrob. Proteins 2022, 14, 1151–1169. [Google Scholar] [CrossRef]
- Miller, R.A.; Beno, S.M.; Kent, D.J.; Carroll, L.M.; Martin, N.H.; Boor, K.J.; Kovac, J. Bacillus wiedmannii sp. nov., a Psychrotolerant and Cytotoxic Bacillus cereus Group Species Isolated from Dairy Foods and Dairy Environments. Int. J. Syst. Evol. Microbiol. 2016, 66, 4744–4753. [Google Scholar] [CrossRef]
- Alkassab, D.; Sampang, J.; Budhwani, Z.; Buragina, J.; Lussier, L.; Acedo, J.Z. Genome Mining for Leaderless Bacteriocins and Discovery of Miticin. Can. J. Chem. 2023. submitted. [Google Scholar]
- Omachi, H.; Terahara, T.; Futami, K.; Kawato, S.; Imada, C.; Kamei, K.; Waku, T.; Kondo, A.; Naganuma, T.; Agustini, T.W.; et al. Distribution of Class IId Bacteriocin-Producing Virgibacillus salexigens in Various Environments. World J. Microbiol. Biotechnol. 2021, 37, 121. [Google Scholar] [CrossRef]
- Watterson, M.J.; Kent, D.J.; Boor, K.J.; Wiedmann, M.; Martin, N.H. Evaluation of Dairy Powder Products Implicates Thermophilic Sporeformers as the Primary Organisms of Interest. J. Dairy Sci. 2014, 97, 2487–2497. [Google Scholar] [CrossRef]
- Nucleotide [Internet]. Accession No. NZ_NUQK01000054.1, Bacillus Wiedmannii Strain AFS057349 AFS057349_106_H6_Contig54_140523, Whole Genome Shotgun Sequence; National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD, USA, 1988. Available online: Https://www.ncbi.nlm.nih.gov/nuccore/nz_nuqk01000054.1?from=6258&to=6404 (accessed on 10 August 2023).
- Nucleotide [Internet]. Accession No. NZ_NUEY01000011.1, Bacillus Wiedmannii Strain AFS001911 AFS001911_108_D10_Contig11_140523, Whole Genome Shotgun Sequence; National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD, USA, 1988. Available online: Https://www.ncbi.nlm.nih.gov/nuccore/nz_nuey01000011.1?from=11047&to=11193 (accessed on 10 August 2023).
- Major, D.; Flanzbaum, L.; Lussier, L.; Davies, C.; Caldo, K.M.P.; Acedo, J.Z. Transporter Protein-Guided Genome Mining for Head-to-Tail Cyclized Bacteriocins. Molecules 2021, 26, 7218. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, T.; Boon-Chieng, S.; Mitaku, S. SOSUI: Classification and Secondary Structure Prediction System for Membrane Proteins. Bioinformatics 1998, 14, 378–379. [Google Scholar] [CrossRef]
- Bhunia, A.K. Foodborne Microbial Pathogens; Springer: New York, NY, USA, 2008. [Google Scholar]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A Viable Alternative to Antibiotics? Nat. Rev. Microbiol. 2012, 11, 95–105. [Google Scholar] [CrossRef]
- Field, D.; Begley, M.; O’Connor, P.M.; Daly, K.M.; Hugenholtz, F.; Cotter, P.D.; Hill, C.; Ross, R.P. Bioengineered Nisin A Derivatives with Enhanced Activity against Both Gram Positive and Gram Negative Pathogens. PLoS ONE 2012, 7, e46884. [Google Scholar] [CrossRef]
- Cao-Hoang, L.; Marechal, P.A.; Le-Thanh, M.; Gervais, P. Synergistic Action of Rapid Chilling and Nisin on the Inactivation of Escherichia coli. Appl. Microbiol. Biotechnol. 2008, 79, 105–109. [Google Scholar] [CrossRef]
- Ovchinnikov, K.V.; Chi, H.; Mehmeti, I.; Holo, H.; Nes, I.F.; Diep, D.B. Novel Group of Leaderless Multipeptide Bacteriocins from Gram-Positive Bacteria. Appl. Environ. Microbiol. 2016, 82, 5216–5224. [Google Scholar] [CrossRef]
- Ovchinnikov, K.V.; Kristiansen, P.E.; Straume, D.; Jensen, M.S.; Aleksandrzak-Piekarczyk, T.; Nes, I.F.; Diep, D.B. The Leaderless Bacteriocin Enterocin K1 is Highly Potent against Enterococcus faecium: A Study on Structure, Target Spectrum and Receptor. Front. Microbiol. 2017, 8, 262767. [Google Scholar] [CrossRef]
- Iwatani, S.; Zendo, T.; Yoneyama, F.; Nakayama, J.; Sonomoto, K. Characterization and Structure Analysis of a Novel Bacteriocin, Lacticin Z, Produced by Lactococcus lactis QU 14. Biosci. Biotechnol. Biochem. 2007, 71, 1984–1992. [Google Scholar] [CrossRef]
- Band, V.I.; Weiss, D.S. Heteroresistance to Beta-Lactam Antibiotics May Often Be a Stage in the Progression to Antibiotic Resistance. PLoS Biol. 2021, 19, e3001346. [Google Scholar] [CrossRef]
- Pereira, C.; Larsson, J.; Hjort, K.; Elf, J.; Andersson, D.I. The Highly Dynamic Nature of Bacterial Heteroresistance Impairs Its Clinical Detection. Comm. Biol. 2021, 4, 521. [Google Scholar] [CrossRef]
- Dewachter, L.; Fauvart, M.; Michiels, J. Bacterial Heterogeneity and Antibiotic Survival: Understanding and Combatting Persistence and Heteroresistance. Mol. Cell 2019, 76, 255–267. [Google Scholar] [CrossRef]
- Band, V.I.; Hufnagel, D.A.; Jaggavarapu, S.; Sherman, E.X.; Wozniak, J.E.; Satola, S.W.; Farley, M.M.; Jacob, J.T.; Burd, E.M.; Weiss, D.S. Antibiotic Combinations That Exploit Heteroresistance to Multiple Drugs Effectively Control Infection. Nat. Microbiol. 2019, 4, 1627–1635. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, Q.; Wen, L.; Chen, J. Combined Effect of Polymyxin B and Tigecycline to Overcome Heteroresistance in Carbapenem-Resistant Klebsiella pneumoniae. Microbiol. Spectr. 2021, 9, e00152-21. [Google Scholar] [CrossRef]
- Deng, S.; Liu, S.; Li, X.; Liu, H.; Li, F.; Liu, K.; Zeng, H.; Zeng, X.; Xin, B. Thuricins: Novel Leaderless Bacteriocins with Potent Antimicrobial Activity Against Gram-Positive Foodborne Pathogens. J. Agric. Food Chem. 2022, 70, 9990–9999. [Google Scholar] [CrossRef]
- Liu, S.; Deng, S.; Liu, H.; Tang, L.; Wang, M.; Xin, B.; Li, F. Four Novel Leaderless Bacteriocins, Bacin A1, A2, A3, and A4 Exhibit Potent Antimicrobial and Antibiofilm Activities against Methicillin-Resistant Staphylococcus aureus. Microbiol. Spectr. 2022, 10, e0094522. [Google Scholar] [CrossRef]
- Wang, J.; Xu, H.; Liu, S.; Song, B.; Liu, H.; Li, F.; Deng, S.; Wang, G.; Zeng, H.; Zeng, X.; et al. Toyoncin, a Novel Leaderless Bacteriocin That Is Produced by Bacillus toyonensis XIN-YC13 and Specifically Targets B. cereus and Listeria monocytogenes. Appl. Environ. Microbiol. 2021, 87, e00185-21. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, F.; Imura, Y.; Ohno, K.; Zendo, T.; Nakayama, J.; Matsuzaki, K.; Sonomoto, K. Peptide-Lipid Huge Toroidal Pore, a New Antimicrobial Mechanism Mediated by a Lactococcal Bacteriocin, Lacticin Q. Antimicrob. Agents Chemother. 2009, 53, 3211–3217. [Google Scholar] [CrossRef]
- Netz, D.J.A.; Bastos, M.d.C.d.F.; Sahl, H.G. Mode of Action of the Antimicrobial Peptide Aureocin A53 from Staphylococcus aureus. Appl. Environ. Microbiol. 2002, 68, 5274–5280. [Google Scholar] [CrossRef]
- Uzelac, G.; Kojic, M.; Lozo, J.; Aleksandrzak-Piekarczyk, T.; Gabrielsen, C.; Kristensen, T.; Nes, I.F.; Diep, D.B.; Topisirovic, L. A Zn-Dependent Metallopeptidase Is Responsible for Sensitivity to LsbB, a Class II Leaderless Bacteriocin of Lactococcus lactis subsp. lactis BGMN1-5. J. Bacteriol. 2013, 195, 5614–5621. [Google Scholar] [CrossRef]
- Peng, Z.; Xiong, T.; Huang, T.; Xu, X.; Fan, P.; Qiao, B.; Xie, M. Factors Affecting Production and Effectiveness, Performance Improvement and Mechanisms of Action of Bacteriocins as Food Preservative. Crit. Rev. Food Sci. Nutr. 2022, 22, 1–14. [Google Scholar] [CrossRef]
- Daas, M.S.; Rosana, A.R.R.; Acedo, J.Z.; Douzane, M.; Nateche, F.; Kebbouche-Gana, S.; Vederas, J.C. Insights into the Draft Genome Sequence of Bioactives-Producing Bacillus thuringiensis DNG9 Isolated from Algerian Soil-Oil Slough. Stand. Genom. Sci. 2018, 13, 25. [Google Scholar] [CrossRef]
- Daas, M.S.; Acedo, J.Z.; Rosana, A.R.R.; Orata, F.D.; Reiz, B.; Zheng, J.; Nateche, F.; Case, R.J.; Kebbouche-Gana, S.; Vederas, J.C. Bacillus amyloliquefaciens ssp. plantarum F11 Isolated from Algerian Salty Lake as a Source of Biosurfactants and Bioactive Lipopeptides. FEMS Microbiol. Lett. 2018, 365, fnx248. [Google Scholar] [CrossRef]
- Banerjee, G.; Nandi, A.; Ray, A.K. Assessment of Hemolytic Activity, Enzyme Production and Bacteriocin Characterization of Bacillus subtilis LR1 Isolated from the Gastrointestinal Tract of Fish. Arch. Microbiol. 2017, 199, 115–124. [Google Scholar] [CrossRef]
- Lim, K.B.; Balolong, M.P.; Kim, S.H.; Oh, J.K.; Lee, J.Y.; Kang, D.-K. Isolation and Characterization of a Broad Spectrum Bacteriocin from Bacillus amyloliquefaciens RX7. Biomed. Res. Int. 2016, 2016, 8521476. [Google Scholar] [CrossRef]
- Senbagam, D.; Gurusamy, R.; Senthilkumar, B. Physical, Chemical and Biological Characterization of a New Bacteriocin Produced by Bacillus cereus NS02. Asian Pac. J. Trop. Med. 2013, 6, 934–941. [Google Scholar] [CrossRef]
- Delves-Broughton, J.; Blackburn, P.; Evans, R.J.; Hugenholtz, J. Applications of the Bacteriocin, Nisin. Antonie Van Leeuwenhoek 1996, 69, 193–202. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS); Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Dusemund, B.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gundert-Remy, U.; et al. Safety of Nisin (E 234) as a Food Additive in the Light of New Toxicological Data and the Proposed Extension of Use. EFSA J. 2017, 15, e05063. [Google Scholar]
- Hernández-González, J.C.; Martínez-Tapia, A.; Lazcano-Hernández, G.; García-Pérez, B.E.; Castrejón-Jiménez, N.S. Bacteriocins from Lactic Acid Bacteria. A Powerful Alternative as Antimicrobials, Probiotics, and Immunomodulators in Veterinary Medicine. Animals 2021, 11, 979. [Google Scholar] [CrossRef]
- Ibarra-Sánchez, L.A.; El-Haddad, N.; Mahmoud, D.; Miller, M.J.; Karam, L. Invited Review: Advances in Nisin Use for Preservation of Dairy Products. J. Dairy Sci. 2020, 103, 2041–2052. [Google Scholar] [CrossRef]
- Przemysław Zdziarski 1, K.S.J.M. Overuse of High Stability Antibiotics and Its Consequences in Public and Environmental Health. Acta Microbiol. Pol. 2003, 52, 5–13. [Google Scholar]
- Lahiri, D.; Nag, M.; Dutta, B.; Sarkar, T.; Pati, S.; Basu, D.; Abdul Kari, Z.; Wei, L.S.; Smaoui, S.; Wen Goh, K.; et al. Bacteriocin: A Natural Approach for Food Safety and Food Security. Front. Bioeng. Biotechnol. 2022, 10, 1005918. [Google Scholar] [CrossRef]
- Eghbal, N.; Chihib, N.E.; Gharsallaoui, A. Nisin. In Antimicrobials in Food; CRC Press: Boca Raton, FL, USA, 2020; p. 30. [Google Scholar]
- Mohd Azmi, S.I.; Kumar, P.; Sharma, N.; Sazili, A.Q.; Lee, S.-J.; Ismail-Fitry, M.R. Application of Plant Proteases in Meat Tenderization: Recent Trends and Future Prospects. Foods 2023, 12, 1336. [Google Scholar] [CrossRef]
- Gänzle, M.G.; Weber, S.; Hammes, W.P. Effect of Ecological Factors on the Inhibitory Spectrum and Activity of Bacteriocins. Int. J. Food Microbiol. 1999, 46, 207–217. [Google Scholar] [CrossRef]
- Mossallam, S.F.; Amer, E.I.; Diab, R.G. Potentiated Anti-Microsporidial Activity of Lactobacillus acidophilus CH1 Bacteriocin Using Gold Nanoparticles. Exp. Parasitol. 2014, 144, 14–21. [Google Scholar] [CrossRef]
- Oppegård, C.; Rogne, P.; Kristiansen, P.E.; Nissen-Meyer, J. Structure Analysis of the Two-Peptide Bacteriocin Lactococcin G by Introducing D-Amino Acid Residues. Microbiology 2010, 156, 1883–1889. [Google Scholar] [CrossRef]
- Powell, M.F.; Stewart, T.; Otvos, L.; Urge, L.; Gaeta, F.C.A.; Sette, A.; Arrhenius, T.; Thoon, D.; Soda, K.; Colon, S.M. Peptide Stability in Drug Development. II. Effect of Single Amino Acid Substitution and Glycosylation on Peptide Reactivity in Human Serum. Pharm. Res. 1993, 10, 1268–1273. [Google Scholar] [CrossRef]
- Tugyi, R.; Uray, K.; Iván, D.; Fellinger, E.; Perkins, A.; Hudecz, F. Partial D-Amino Acid Substitution: Improved Enzymatic Stability and Preserved Ab Recognition of a MUC2 Epitope Peptide. Proc. Natl. Acad. Sci. USA 2005, 102, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A Web-Based Environment for Protein Structure Homology Modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Delano, W.L. The PyMOL Molecular Graphics System. 2002. [Google Scholar]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An Automated Pipeline for the Setup of Poisson–Boltzmann Electrostatics Calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Acedo, J.Z.; Van Belkum, M.J.; Lohans, C.T.; Towle, K.M.; Miskolzie, M.; Vederas, J.C. Nuclear Magnetic Resonance Solution Structures of Lacticin Q and Aureocin A53 Reveal a Structural Motif Conserved among Leaderless Bacteriocins with Broad-Spectrum Activity. Biochemistry 2016, 55, 733–742. [Google Scholar] [CrossRef] [PubMed]
- de Planque, M.R.R.; Bonev, B.B.; Demmers, J.A.A.; Greathouse, D.V.; Koeppe, R.E., 2nd; Separovic, F.; Watts, A.; Killian, J.A. Interfacial Anchor Properties of Tryptophan Residues in Transmembrane Peptides Can Dominate over Hydrophobic Matching Effects in Peptide-Lipid Interactions. Biochemistry 2003, 42, 5341–5348. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Ichimasa, S.; Zendo, T.; Koga, S.; Yoneyama, F.; Nakayama, J.; Sonomoto, K. Structural Analysis and Characterization of Lacticin Q, a Novel Bacteriocin Belonging to a New Family of Unmodified Bacteriocins of Gram-Positive Bacteria. Appl. Environ. Microbiol. 2007, 73, 2871–2877. [Google Scholar] [CrossRef]
- Cladera-Olivera, F.; Caron, G.R.; Brandelli, A. Bacteriocin Production by Bacillus licheniformis Strain P40 in Cheese Whey Using Response Surface Methodology. Biochem. Eng. J. 2004, 21, 53–58. [Google Scholar] [CrossRef]
- Han, S.-K.; Shin, M.-S.; Park, H.-E.; Kim, S.-Y.; Lee, W.-K. Screening of Bacteriocin-Producing Enterococcus faecalis Strains for Antagonistic Activities against Clostridium perfringens. Korean J. Food Sci, Anim. Resour. 2014, 34, 614–621. [Google Scholar] [CrossRef]
- ProtParam, E. ExPASy-ProtParam Tool; SIB: Lausanne, Switzerland, 2005. [Google Scholar]


| Protein | Size (aa) | TM | Identity |
|---|---|---|---|
| BawA | 48 | 0 | Bawcin (bacteriocin) |
| BawB | 94 | 3 | Hypothetical protein |
| BawC | 98 | 0 | DUF2089 family protein |
| BawD | 390 | 1 | HlyD family efflux transporter |
| BawE | 226 | 0 | ABC transporter ATP-binding protein |
| BawF | 399 | 5 | ABC transporter permease |
| BawG | 223 | 5 | Yip1 family protein |
| Indicator Organism | MIC (µM) | Heteroresistance (µM) |
|---|---|---|
| Micrococcus luteus (Ward’s 85W 0966) | 4 | 4 |
| Lactococcus lactis ssp. lactis (Ward’s 85W 1774) | 4 | 4 |
| Lactobacillus delbruekii ssp. bulgaricus (VWR 470179-126) | 8 | 8 |
| Bacillus subtilis (Ward’s 85W 1650) | 8 | 8, 16 |
| Listeria monocytogenes ATCC 19115 | 8 | 8, 16 |
| Lactobacillus casei (Ward’s 85W 1682) | 8 | 8–32 |
| Bacillus cereus (Ward’s 85W 0200) | 16 | 16–64 |
| Staphylococcus aureus ATCC 6538 | 16 | 16–64 |
| Enterococcus faecalis (Ward’s 85W 1100) | 32 | 32–128 |
| Staphylococcus epidermidis (Ward’s 85W 1747) | - | - |
| Streptococcus pyogenes (Ward’s 85W 1180) | - | - |
| Geobacillus stearothermophilus 1010 | - | - |
| Pseudomonas aeruginosa (Ward’s 85W 1173) | - | - |
| Salmonella enterica (Ward’s 85W 1710) | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Budhwani, Z.; Buragina, J.T.; Lang, J.; Acedo, J.Z. Characterization of the Novel Leaderless Bacteriocin, Bawcin, from Bacillus wiedmannii. Int. J. Mol. Sci. 2023, 24, 16965. https://doi.org/10.3390/ijms242316965
Budhwani Z, Buragina JT, Lang J, Acedo JZ. Characterization of the Novel Leaderless Bacteriocin, Bawcin, from Bacillus wiedmannii. International Journal of Molecular Sciences. 2023; 24(23):16965. https://doi.org/10.3390/ijms242316965
Chicago/Turabian StyleBudhwani, Zafina, Jenna T. Buragina, Jen Lang, and Jeella Z. Acedo. 2023. "Characterization of the Novel Leaderless Bacteriocin, Bawcin, from Bacillus wiedmannii" International Journal of Molecular Sciences 24, no. 23: 16965. https://doi.org/10.3390/ijms242316965





