Abstract
Polygalacturonase (PG) is one of the largest families of hydrolytic enzymes in plants. It is involved in the breakdown of pectin in the plant cell wall and even contributes to peel cracks. Here, we characterize PGs and outline their expression profiles using the available reference genome and transcriptome of Akebia trifoliata. The average length and exon number of the 47 identified AktPGs, unevenly assigned on 14 chromosomes and two unassembled contigs, were 5399 bp and 7, respectively. The phylogenetic tree of 191 PGs, including 47, 57, 51, and 36 from A. trifoliata, Durio zibethinus, Actinidia chinensis, and Vitis vinifera, respectively, showed that AktPGs were distributed in all groups except group G and that 10 AktPGs in group E were older, while the remaining 37 AktPGs were younger. Evolutionarily, all AktPGs generally experienced whole-genome duplication (WGD)/segmental repeats and purifying selection. Additionally, the origin of conserved domain III was possibly associated with a histidine residue (H) substitute in motif 8. The results of both the phylogenetic tree and expression profiling indicated that five AktPGs, especially AktPG25, could be associated with the cracking process. Detailed information and data on the PG family are beneficial for further study of the postharvest biology of A. trifoliata.
1. Introduction
Fruit ripening is a complex physiological and biochemical process, usually accompanied by changes in color, texture, and flavor, and in some fleshy fruits, even by cracking [1,2]. In many plants, fruit cracking has evolved as a sophisticated dispersal mechanism for spreading seeds and extending their range [3]. However, cracking can lead to low marketability, higher storage costs, and large economic losses in some fleshy fruits [4]. Understanding the details of cracking is very important for effectively solving the problem in commercial fruit production.
Cracking is common in legumes, with cracks developing along the ventral and dorsal sutures at maturity [5]. This cracking facilitates a range of production activities, such as threshing and seeding [6]. Cracking also occurs in a few fleshy species, such as Durio zibethinus [7], Prunus avium [8], and Citrullus lanatus [9]. Fleshy fruit cracking can be classified into two basic categories: noninherent and inherent. Noninherent cracking, such as that found in C. lanatus and Malus domestica, is occasionally caused by environmental factors. Inherent cracking, such as in D. zibethinus [10] and Akebia trifoliata [11], is one of the typical characteristics of a given species and is a programmed physiological process controlled by genetic regulation. In addition, the fruit of A. trifoliata also experiences “August cracking”, which are natural and regular crack along its ventral suture during its ripening season, so it is an ideal material for investigating the molecular mechanism responsible for inherent cracking.
Various factors, such as physiological status, genetic components, and environmental changes, can lead to cracking in the peel [12], which, as a mechanical barrier, plays an irreplaceable role in protecting the whole fruit against biotic or abiotic stress [2]. However, as the fruit grows and develops, the biochemical characteristics of the peel also change synchronously. When pressure inside the tissue is higher than the mechanical resistance of the peel cell wall, cracks occur [13,14]. Since pectin is a common and major component of the primary cell wall and intercellular layer, respectively, and plays an important role in intercellular adhesion [15,16], this may indicate that pectin hydrolysis could be the essential process underlying fruit cracking. Therefore, identifying the genetic component associated with pectin disassembly is very important in order to further understand the molecular mechanism of fruit cracking.
Among hydrolases, polygalacturonase (PG) is one of the largest families and is also the most widely studied class of pectin hydrolases [17,18]. PG plays a central role in pectin degradation by catalyzing the breakdown of α-(1,4)-galacturonic bonds in the pectin molecule [19] and is classified into endo-PGs, exo-PGs, and rhamnose-PGs according to its mode of action on pectin [18]. To date, the PG gene family has been identified in several species, including Zea mays [20], C. lanatus [21], Pyrus bretschneideri [22], Actinidia chinensis [23], and Vitis vinifera [24]. Various studies have suggested that PG genes are widely involved in plant growth and development processes such as seed germination [25], organ abscission [26], pod cracking [27], anther dehiscence [28], and fruit softening [22].
A. trifoliata, commonly known as augmelon and wild banana [11], is a perennial woody vine mainly distributed in East Asian countries such as China, Japan, and Korea [29] and is becoming a new fruit crop in various regions of China due to its high nutritional and medicinal value [30,31]. Previous studies have suggested that the fruit of A. trifoliata physiologically belongs to a typical respiratory climacteric fruit and naturally cracks along the ventral suture line in the late stage of fruit development [11,32]. In addition, high temperatures and rain are usually accompanied by the harvesting of A. trifoliata fruits. They contribute to the fruit’s deterioration through contamination by biotic and abiotic factors, largely increasing the difficulty of storage and significantly shortening its shelf life. Clearly, cracking would be a major limiting factor for the commercial exploitation of A. trifoliata as a fresh fruit crop.
In the past, only a few studies have explored the cracking issue of A. trifoliata fruit. The results of a comprehensive transcriptomic and proteomic analysis suggested that the cracking of A. trifoliata fruit was associated with cell wall structural changes and rearrangements [33], and this view was subsequently supported by the results of a metabolic analysis [34]. At the same time, another report showed that the degradation process of the A. trifoliata cell wall was highly associated with PG activity [35], which indicated that PG could largely regulate fruit cracking. However, the molecular mechanism by which PGs regulate fruit cracking is still unknown.
During the ripening of A. trifoliata, PGs may play a significant role in fruit cracking. However, we still lack knowledge on PGs in A. trifoliata. To provide basic information to further elucidate the detailed regulatory mechanism, we planned to systemically outline the structural profile, such as the component, number, size, cis-acting element, chromosomal position and conserved domain of putative proteins of PGs, to investigate the evolutionary events experienced by PGs, and to determine their expression level using available genomic and transcriptomic data. The results would be helpful to study the cracking trait of A. trifoliata in the future.
2. Result
2.1. Systemic Identification of PGs in the A. trifoliata Genome
The results of both BLAST and BLASTP analyses consistently showed that there were 47 AktPGs in the A. trifoliata genome. They were named AktPG1 to AktPG47 according to their chromosomal locations. Physically, only AktPG46 and AktPG47 were assigned to unassembled contig00874 and contig00909, respectively (Table 1), while the other 45 AktPGs were unevenly distributed on the 16 chromosomes, except for chromosomes 6 and 15. In addition, they were mainly distributed in the end region of the chromosomes. The number of AktPGs on chromosomes 5, 10, and 16 was the highest, at six, while that on chromosomes 2, 7, 12, and 13 was only one (Figure 1).

Table 1.
Detailed characteristics of both AktPGs and putative AktPGs.
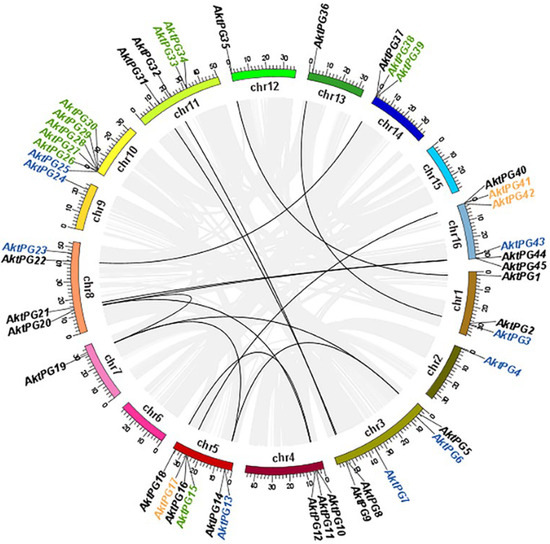
Figure 1.
Chromosomal distribution of PGs in Akebia trifoliata. Different segments represent different chromosomes. Gray and black lines represent segmental duplication pairs in the whole A. trifoliata and between AktPG pairs, respectively. Genes marked in black, blue, green, and yellow were putatively produced by whole-genome duplication (WGD) or segmental, dispersed, tandem, and proximal duplication, respectively.
In terms of genetic structure, the average length and exon number of all 47 AktPGs were 5398.7 bp, varying from 1650 to 28,733 bp, and 7, varying from 4 to 10, respectively. The average length, isoelectric point (PI), molecular weight (MW), and instability index of the putative proteins were 440.42, 7.54, 47.75 kDa, and 35.02, respectively (Table 1). Evolutionarily, the results of intraspecies collinearity analysis showed that twenty-three (48.94%), eleven (23.40%), ten (21.28%), and three (6.38%) AktPGs were produced by whole-genome duplication (WGD) or segmental duplication, dispersed, tandem, and proximal duplication, respectively.
2.2. Phylogenetic Tree of AktPGs
The phylogenetic tree, consisting of 47 AktPGs for A. trifoliata, 57 DzPGs for D. zibethinus, 51 AcPGs for A. chinensis, and 36 VvPGs for V. vinifera, showed that all 191 PGs were clustered into seven branches from A to G (Figure 2). In groups A, B, D, and E, there were similar PG numbers or similar percentages of PGs per species. In group C, the number of DzPGs was only three; moreover, there were no VvPGs in group F. Group G consisted of only two genes (AcPG15 and DzPG49), neither being AktPG or VvPG.
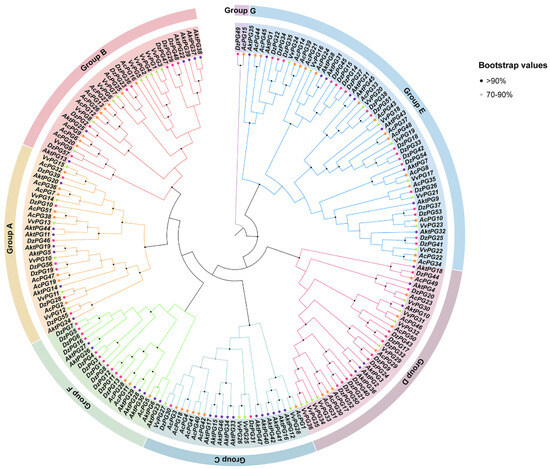
Figure 2.
Phylogenetic tree of PGs in A. trifoliata, Durio zibethinus, Actinidia chinensis, and Vitis vinifera. Different shapes represent different species. According to the tree, all AktPGs were divided into six groups.
2.3. Interspecific Collinearity
Through interspecific collinearity, we detected a total of 156 homogenous gene pairs between A. trifoliata and the other three species (Figure 3), which consisted of 66 pairs between 26 (55.32%) AktPGs and 36 (63.16%) DzPGs, 59 pairs between 26 (55.32%) AktPGs and 36 (70.59%) AcPGs, and 31 pairs between 24 (51.06%) AktPGs and 18 (50%) VvPGs.
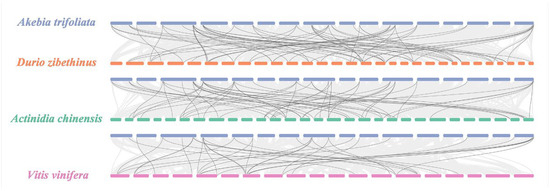
Figure 3.
PGs collinearity analysis among A. trifoliata and D. zibethinus, A. chinensis, and V. vinifera. Black lines represent collinearity among different PGs.
2.4. Conserved Structures and Motifs of Putative AktPGs
The results of multiple sequence ratios and conserved structural domain analyses revealed that most AktPGs contained four conserved structural domains (I, II, III, and IV) associated with protein catalytic and binding sites (Figure 4). The conservation degree of structural domain III was obviously lower than those of structures I, II, and IV. In addition, 10 AktPGs (AktPG1, AktPG7-9, AktPG21, AktPG31, AktPG32, AktPG35, AktPG43, and AktPG45) were usually absent in the conserved structural domain III. In contrast, five AAs, including the fourth and sixth in conserved structural domain I, the first and third AAs in conserved domain II, and the third AA in conserved structure domain IV, did not show any variation in any of the 47 AktPGs.
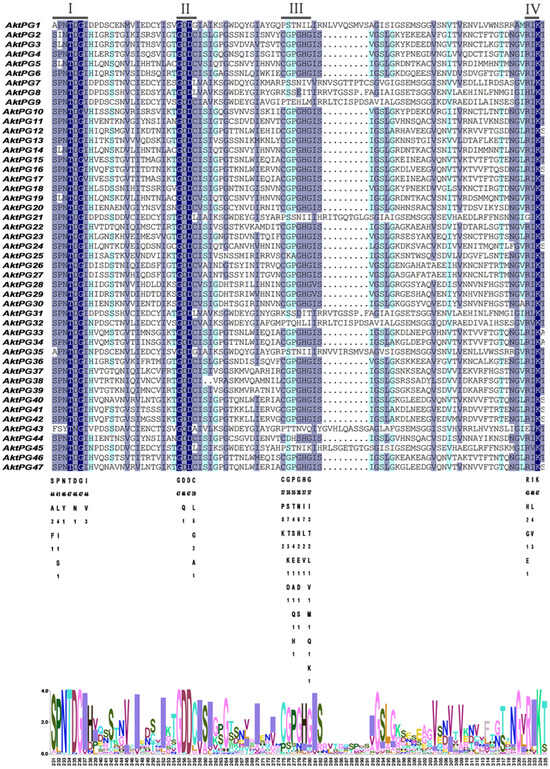
Figure 4.
Multiple sequence alignment of AktPGs. Underlining indicates the conserved domains of AktPGs: I (SPNTDGI), II (GDDC), III (CGPGHG), and IV (RIK). The shade of the color represents the degree of sequence similarity.
The results of conserved motif analyses showed that the number of conserved motifs ranged from six (only AktPG25) to nine. Conserved motif 1 covered conserved structural domains I and II, and motif 3 and motif 4 covered conserved structural domains IV and III, respectively (Table 2). The numbers of AktPGs containing seven, eight, and nine motifs were three, nineteen, and twenty-four, respectively (Figure 5). However, conserved motifs 1, 2, 3, and 6 widely existed in all 47 AktPGs. The rates of motifs 4, 5, 7, 9, and 10 reached 0.79, 0.64, 0.94, 0.91, and 0.89, respectively. Motif 8 only existed in all 10 (21.3%) AktPGs of group E. Moreover, AktPGs in groups A, C, and D had identical motif compositions, while those in groups B, E, and F had different motif compositions.

Table 2.
Information on the conserved motifs of 47 AktPGs identified.
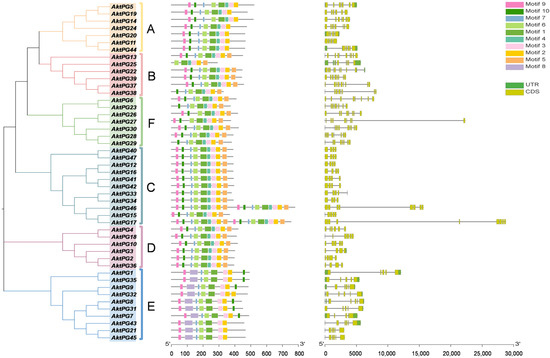
Figure 5.
Gene structures and conserved motifs of AktPGs. The left side indicates the phylogenetic tree of 47 AktPGs. The middle shows the distribution of conserved motifs of 47 AktPGs. The right side shows the exon/intron structures of AktPGs. UTR, CDS, and black lines represent noncoding regions, coding regions, and introns, respectively.
2.5. Ka/Ks Value of Homologous AktPG Pairs
Ka/Ks value is the ratio of the nonsynonymous substitution rate (Ka) to the synonymous substitution rate (Ks) of two protein-coding genes, which is often used to determine whether there is selective pressure acting on protein-coding genes, thus reflecting the evolutionary selection of the species. A total of 467 homologous AktPG pairs were detected, in which only one gene pair (AktPG15 and AktPG17) had a Ka/Ks value (1.122) larger than 1, while all values of the other homologous AktPG pairs were less than 0.5 (Table S1), which indicated that AktPGs mainly experienced purifying selection.
2.6. Cis-Acting Elements of the AktPGs
A total of 745 cis-acting elements were identified within the 2000 bp upstream region of the AktPG promoter (Figure 6), and they could be classified into two types with 12 subtypes: hormone-responsive with five subtypes (abscisic acid (ABA)-, auxin-, gibberellin (GA)-, methyl jasmonate (MeJA)-, and salicylic acid (SA)-responsive elements) and environment-responsive with seven subtypes (anaerobic induction, defense and stress response, light response, physiological rhythm, drought induction, low-temperature response, and damage response). MeJA-responsive elements and light-responsive elements were the most hormone- and environment-responsive elements and accounted for 39% and 54% of the total, respectively. In addition, 40 (85%) and 45 (96%) AktPGs had ABA-responsive cis-acting and light-responsive cis-acting elements, respectively.
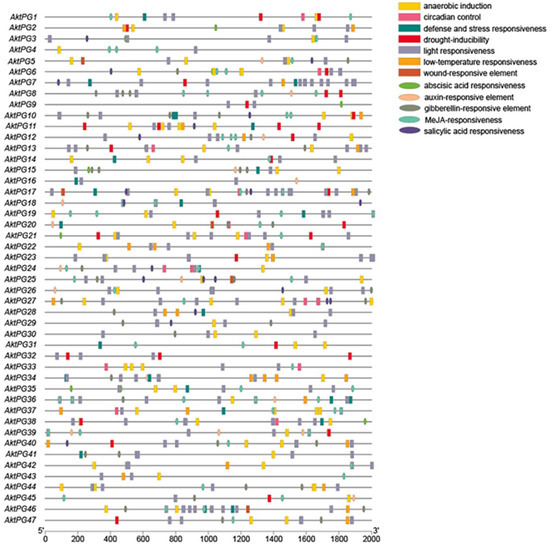
Figure 6.
Distribution of cis-acting elements within the 2000 bp upstream region of AktPGs. Different icons represent different types of promoter cis-acting elements.
The number of cis-acting elements each AktPG exhibited showed a large variation, from four (AktPG16) to thirty-three (AktPG17); similarly, the number of subtypes that each AktPG contained also showed a large variation, from three (AktPG16 and AktPG12) to eleven (AktPG17). In addition, although the numbers and types of cis-acting elements of AktPGs exhibited large variation, all AktPGs simultaneously contained at least one hormone-responsive and one environment-responsive subtype.
2.7. Expression Profiles of AktPGs in Two Independent Transcriptome Datasets
The expression level of more than 30 AktPGs was either undetectable or very low in the two transcriptome datasets, and there were only 16 and 15 AktPGs with an FPKM value detectable in one sample in the tissue- and development-related transcriptome (Figure 7a) and disease- and development-related transcriptome (Figure 7b) of A. trifoliata, respectively. In addition, 14 AktPGs were simultaneously expressed in the two transcriptome datasets, although their expression profiles were not completely the same. Both AktPG22 and AktPG33 exhibited a high expression level in tissue- and development-related transcriptome data, while only AktPG42 exhibited high expression in disease- and development-related transcriptome data. Moreover, AktPG22 and AktPG33 exhibited tissue- and developmental-stage-specific expression, while AktPG42 also exhibited differential expression among samples with different disease resistance levels. We further found that AktPG22 and AktPG33 only exhibited high expression levels in the later stages of both flesh and seed.
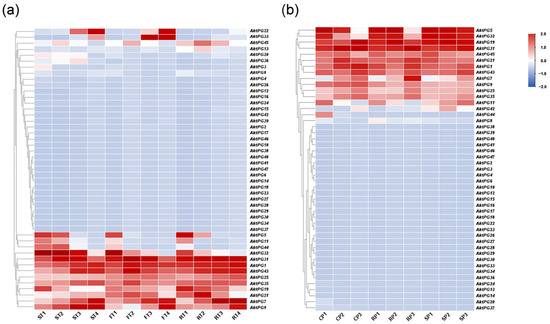
Figure 7.
Expression profiles of AktPGs in different stages. The degree of change from blue to red indicates the intensity of the expression level. (a) Expression profiles of AktPGs in tissue- and development-related transcriptomes. ST, FT, RT and 1, 2, 3, 4 represent three different tissues (seed, flesh, and peel tissues) and four different developmental stages (immature, enlargement, coloring, and mature stages) of A. trifoliata, respectively. (b) Expression profiles of AktPGs in disease- and development-related transcriptomes. CP, RP, SP and 1, 2, 3 represent three different treatment objects (control peel group, disease-resistant peel group, and disease resistant–susceptible mixed pool) and developmental stages (May, June, and July) of A. trifoliata, respectively.
In fact, we also would like to note the AktPGs that exhibited developmental-stage-specific expression in two transcriptome datasets, namely AktPG5, AktPG7, AktPG11, AktPG32, and AktPG44. Among the five genes, the expression levels of AktPG5, AktPG32, and AktPG44 generally showed a gradual decrease, while that of AktPG7 showed a gradual increase, with developmental progress in the two transcriptome datasets. However, although the expression level of AktPG11 also showed a gradual decrease with developmental progress in tissue- and development-related transcriptome data, the disease- and development-related expression profiles showed obvious differences among different samples with different disease resistance levels.
3. Discussion
Previous publications have suggested that PGs, mainly involved in pectin degradation by hydrolysis, are a very ancestral gene family and exist in almost all land plants, especially flowering plants [19,36]. However, PGs of different species possibly experienced different events in evolutionary processes that contributed to their current structural and functional characteristics. Therefore, we are mainly concerned with the possible evolutionary events, structural characteristics, and functional divergences of PGs in A. trifoliata.
3.1. Putative Evolutionary Events Experienced by AktPGs
At present, PGs have been widely identified in various species from algae to angiosperms [37], such as Chlamydomonas reinhardtii [36], Z. mays [20], and Populus [38]. However, reports examining basal eudicot PGs as an evolutionarily important branch because of their role as a bridge between basal angiosperms and core eudicots (comprising 80% extant land plants) have been very few [39]. Recently, omics data have supported, especially through the various versions of the genome and different tissue transcriptomes of A. trifoliata, their being typical representatives of basal eudicots [40], which provided opportunities to systemically investigate the possible evolutionary history of AktPGs.
In the present study, the A. trifoliata genome referred to is that reported by [41] in 2022 because the long terminal repeat (LTR) assembly index of this version is as high as 11.9, and it could be truly called the reference genome according to the assembly quality standard suggested by [42]. Moreover, only the corresponding files of that version were available. A total of 47 AktPGs were identified (Table 1), which was generally fewer than that in many core eudicots [37,38]. Both the selection style and duplication type of members are the two most important factors when we investigate the evolutionary history of a given gene family. Here, we only found one homogenous gene pair (AktPG15 and AktPG17) with a Ka/Ks value (1.122) larger than 1 among a total of 467 homologous gene pairs, while all the others were less than 1 or even 0.5 (Supplementary Table S1), which suggested that AktPGs as well as AktNBSs [43], AktMADS-boxs [44], AktWRKYs [45], AktNACs [46], AktSODs [47], and AktDofs [48] experienced strong purifying selection in the evolutionary process.
However, AktPGs, similar to AktNBSs [43], were produced by more duplication types compared with the other reported gene families in A. trifoliata (Table 1, Table S2), which indirectly supported the view that AktPGs could be an ancestral gene family. In addition, this interpretation also agreed with the results of both interspecific collinearity (Figure 3) and phylogenetic tree analysis (Figure 2) because genes in the E group have been confirmed to be essential and indispensable in almost all plants of different species [20,36]. Comprehensively, AktPGs could be of ancestral origin, and they may have experienced both various duplication types and strong purifying selection.
3.2. A Histidine Residue (H) Substitute Could Influence the Structural Characteristics of AktPGs
The structural domains of proteins are closely related to their functions [49]. Significant differences in amino acid sequences among members of PGs typically contain four conserved domains [50]. The core amino acid sequences of domains I and II are SPNTDG and GDDC, respectively, in which three aspartic acid residues (D) form the catalytic site [51]. The core amino acid sequence of domain III is CGPGHG, and histidine residues (H) are thought to be involved in the catalytic reaction [52]. Domain IV, consisting of RIK, interacts with the carboxylic acid groups in the substrate and binds to the substrate [53].
Here, sequence comparison found that domains I, II, and IV widely existed in each AktPG, while domain III was absent in all 10 AktPGs of the E group (Figure 4). Moreover, there was at least one completely identical AA in domains I, II, and IV among all 47 AktPGs (Figure 4). Obviously, domains I, II, and IV were highly conserved, while the conservation of domain III was obviously low. By further comparison, we found a significant difference in the fifth histidine residue (H) involved in the catalytic reaction between the 10 AktPGs in the E group and the other 37 AktPGs in the domain III sequence [52], and among all 37 AktPGs in domain III, the fifth AA had a consistent H, while the 10 AktPGs in group E outside of domain III did not have H, so H could be the hallmark of domain III.
Similarly, the other interesting result of sequence analysis was that motif 8 was also only distributed in all 10 AktPGs in the E group, while motif 4, covering domain III specifically, existed in the other 37 AktPGs. The next problem is determining which motif is older. Various studies have confirmed that PGs in group E are widely distributed in plant species and are the oldest [36,54,55], so motif 8 could be older than motif 4. In addition, the significantly lower exon number (5~7) of the 10 AktPGs in the E group compared with that (6~10) of most AktPGs also further reinforces this view because gene family evolution usually results in an increase in exon number [56]. Finally, the high expression ratio (90%) of the 10 AktPGs and the low ratio (18.9% and 16.2% in the tissue- and development-related transcriptome and disease- and development-related transcriptome, respectively) of the 37 AktPGs also provide persuasive evidence for this view because the last member of the same gene family is usually absent in the corresponding function due to a short evolutionary history [57,58].
To further identify the possible ancestor of the 37 younger AktPGs among the 10 older AktPGs in group E, we compared the Ka/Ks values between each member of the 10 AktPGs in group E and the other 37 AktPGs one by one. The results showed that among the 10 older AktPGs, AktPG9 had the smallest average Ka/Ks value (0.23) among the other 37 AktPGs, and the Ka/Ks value ranged from 0.03 (AktPG9 and AktPG12) to 0.39 (AktPG9 and AktPG16). Likewise, among the 37 AktPGs, AktPG12 also had the smallest average Ka/Ks value (0.21) among the 10 AktPGs in the E group. Considering this together, it is reasonable to assume that a histidine residue (H) substitute in motif 8 of a given member among the 10 AktPGs in group E further produced the 37 AktPGs within domain III, and that the process putatively occurred in AktPG9 and consequently produced AktPG12 as the first AktPG within domain III.
3.3. Several Putative Candidate AktPGs Possibly Associated with Fruit Cracking in A. trifoliata
The results of cis-acting element analysis revealed that each AktPG contained at least one hormone and one abiotic stress-related response element at the same time (Figure 6), which indicated that AktPGs would be differentially expressed during plant development, especially in various environmental conditions. In highly expressed AktPGs, nine AktPGs out of the ten AktPGs in group E exhibited high expression levels (Figure 7), but we would like to exclude them as candidate genes associated with fruit cracking because genes in group E, as the oldest PGs, would have been responsible for common biological processes rather than species-specific physiological behavior [21]. In addition, A. trifoliata still retains both asexual and sexual reproduction, yet the effectiveness of sexual reproduction, mainly by seeds, is enhanced, which is generally facilitated by fruit cracking behavior [59]. Newly produced physiological behavior usually accompanies newly functionalized members rather than older members of the same gene family.
Among the remaining highly expressed AktPGs, only the five simultaneously highly expressed genes (AktPG5, AktPG11, AktPG19, and AktPG44 in group A and AktPG25 in group B) could be associated with fruit cracking in A. trifoliata because developmental change was the common factor of the two transcriptome datasets (Figure 7). Previous studies have suggested that genes in groups A and B have species-specific functions [60]. For example, ADPG1 (At3g57510), ADPG2 (At2g41850), QRT2 (At3g07970) [27], and DzPG25 [61], which are responsible for anther dehiscence in Arabidopsis and fruit cracking in D. zibethinus, respectively, belong to group B. Therefore, we would like to recognize AktPG25 in group B as the candidate gene associated with fruit cracking in A. trifoliata, which also agreed well with the expression change in AktPG25 (Figure 7). Confirming the functional relationship between the putative candidate genes, especially AktPG25, and fruit cracking in A. trifoliata is our next objective in the future.
4. Materials and Methods
4.1. Identification and Characterization Analysis of AktPGs
Genome sequence annotation files of A. trifoliata (accession IDs: GWHBISH00000000) were downloaded from the National Genomics Data Centre (https://ngdc.cncb.ac.cn/; accessed on 21 March 2023) [45]. The hidden Markov model of conserved PG sequences, glycosyl hydrolase family 28 (PF00295), was downloaded from the Pfam database (http://pfam-legacy.xfam.org/; accessed on 21 March 2023) to identify AktPGs, and then the sequences of all the proteins of A. trifoliata were scanned by HMMER 3.0 using the HMM with an E value of 1 × 10−5. At the same time, amino acid sequences of Arabidopsis PG were downloaded from the TAIR website (https://www.arabidopsis.org/; accessed on 21 March 2023) and used to identify AktPGs. A local BLASTP was conducted to find each putative candidate with a similarity > 50%, identity > 30%, query coverage > 95%, and E-value < 1 × 10−10. The physicochemical properties of putative AktPG were calculated using the Expasy Protoparam website (https://web.expasy.org/protparam/; accessed on 28 March 2023). The gene replication events among A. trifoliata PGs were analyzed using multiple collinear scanning toolkits (MCScanX) with the default parameters.
4.2. Phylogenetic Analysis, Selective Pressure, and Collinearity of AktPGs
The reference genome sequences of D. zibethinus (accession IDs: PRJNA407962) and A. chinensis (accession IDs: ASM966300v1) as well as of V. vinifera (genome version 2.1) were downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/; accessed on 3 April 2023) and the Ensembl Plants website (https://plants.ensembl.org/index.html; accessed on 3 April 2023), respectively. The phylogenetic tree of all AktPGs and 144 reference PGs, including 57 DzPGs of respiratory climacteric and cracking D. zibethinus, 51 AcPGs of respiratory climacteric and non-cracking A. chinensis, and 36 VvPGs of neither climacteric nor cracking V. vinifera, was constructed using MEGA 11 software (v11.0.10, Auckland, New Zealand) according to a previously reported program [62]. To display the evolutionary selection pressure between AktPG pairs, the Ka/Ks ratio was calculated by TBtools-II software (v2.012, Chengjie Chen, China). Similarly, we also performed a collinearity analysis using TBtools software.
4.3. Gene Structure and Conserved Motif Analysis
Multiple sequence comparisons of PGs were performed using DNAMAN software (version8, San Ramon, CA, USA) and the conserved AktPG motifs were identified using the MEME website (https://memesuite.org/me-me/tools/meme; accessed on 6 April 2023), in which the maximum motif parameter was set to 10, while the remaining parameters were default values. The final schematic diagram of the gene structure and conserved motifs of AktPGs was drawn using TBtools software.
4.4. Analysis of the Cis-Acting Elements of AktPGs
The possible cis-acting elements within the 2000 bp upstream region of the start codon of the AktPGs were predicted using the PlantCARE website (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/; accessed on 13 April 2023), and the predicted results were then visualized using TBtools software.
4.5. Expression Analysis of the AktPGs
Growth and development transcriptome data (accession IDs: PRJNA671772) of A. trifoliata were downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/; accessed on 10 April 2023) [48]. These data included four different developmental stages (immature, enlargement, coloring, and mature stages) in three different tissues (flesh, seeds, and peels). Disease transcriptome data (accession IDs: PRJCA014987) of A. trifoliata were downloaded from the National Genomics Data Centre (https://ngdc.cncb.ac.cn/; accessed on 11 July 2023). Transcriptome data included three different treatment objects (control peel group, disease-resistant peel group, and disease-susceptible group) at three developmental stages (May, June, and July). FPKM values were extracted from the transcriptome data using Hisat2 software (v2.1.0, Mihaela Pertea, USA) and DESeq2 (v1.36.0, Michael I Love, Germany) to estimate gene expression levels. TBtools software was used to construct a heatmap of AktPG expression.
5. Conclusions
A total of 47 AktPGs were identified in the A. trifoliata reference genome, of which 45 (95.7%) were unevenly assigned to 14 high-quality assembled pseudochromosomes. Evolutionarily, they were mainly produced by WGD/segmental repeats and purifying selection. Further phylogenetic analysis classified them into six groups (A–F), and 10 AktPGs belonging to group E were the oldest group. We found a mutually exclusive relationship between motif 4 and motif 8 among 47 AktPGs, and after carefully comparing the sequences, we would like to suggest that a histidine residue substitute in motif 8 could be highly associated with the origin of conserved structural domain III of AktPGs in motif 4. Together, we speculated that five AktPGs (AktPG5, AktPG11, AktPG19, AktPG44, and AktPG25), especially AktPG25, could be associated with the cracking process. The results, including data, gene resources, and conclusions, will be helpful to further genetically improve the traits of interest in A. trifoliata in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms242316973/s1.
Author Contributions
X.Y. and P.L. designed the experiment and drafted the manuscript; W.C., J.G., J.Z., Q.Z., H.Y. (Huai Yang) and H.Y. (Hao Yang) collected the materials and organized the data; S.Z., C.C., F.T. and T.R. provided the analysis tools and technical guidance. All the authors contributed to the revisions and comments on the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Science and Technology Department of Sichuan Province, grant number 2022ZHXC0002, 2022ZHXC0028; Ya’an Science and Technology Bureau, grant number 22SXHZ0071; and Sichuan Provincial Administration of Traditional Chinese Medicine, grant number 2023zd026.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available in this article and the Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Barry, C.S.; Giovannoni, J.J. Ethylene and fruit ripening. J. Plant Growth Regul. 2007, 26, 143–159. [Google Scholar] [CrossRef]
- Santos, M.; Egea-Cortines, M.; Gonçalves, B.; Matos, M. Molecular mechanisms involved in fruit cracking: A review. Front. Plant Sci. 2023, 14, 561. [Google Scholar] [CrossRef] [PubMed]
- Galstyan, A.; Hay, A. Snap, crack and pop of explosive fruit. Curr. Opin. Genet. Dev. 2018, 51, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Khadivi-Khub, A. Physiological and genetic factors influencing fruit cracking. Acta Physiol. Plant. 2015, 37, 1718. [Google Scholar] [CrossRef]
- Christiansen, L.; Dal Degan, F.; Ulvskov, P.; Borkhardt, B. Examination of the dehiscence zone in soybean pods and isolation of a dehiscence-related endopolygalacturonase gene. Plant Cell Environ. 2002, 25, 479–490. [Google Scholar] [CrossRef]
- Esgici, R.; Pekitkan, F.G.; Sessiz, A. Correlation between rice stem cutting resistance and cracking force of rice kernel. Fresenius Environ. Bull. 2019, 28, 3014–3021. [Google Scholar]
- Khurnpoon, L.; Siriphanich, J.; Labavitch, J. Cell wall metabolism during durian fruit dehiscence. Postharvest Biol. Technol. 2008, 48, 391–401. [Google Scholar] [CrossRef]
- Correia, S.; Schouten, R.; Silva, A.P.; Gonçalves, B. Sweet cherry fruit cracking mechanisms and prevention strategies: A review. Sci. Hortic. 2018, 240, 369–377. [Google Scholar] [CrossRef]
- Liao, N.; Hu, Z.; Li, Y.; Hao, J.; Chen, S.; Xue, Q.; Ma, Y.; Zhang, K.; Mahmoud, A.; Ali, A. Ethylene-responsive factor 4 is associated with the desirable rind hardness trait conferring cracking resistance in fresh fruits of watermelon. Plant Biotechnol. J. 2020, 18, 1066–1077. [Google Scholar] [CrossRef]
- Palapol, Y.; Kunyamee, S.; Thongkhum, M.; Ketsa, S.; Ferguson, I.B.; van Doorn, W.G. Expression of expansin genes in the pulp and the dehiscence zone of ripening durian (Durio zibethinus) fruit. J. Plant Physiol. 2015, 182, 33–39. [Google Scholar] [CrossRef]
- Yang, H.; Chen, W.; Fu, P.; Zhong, S.; Guan, J.; Luo, P. Developmental stages of Akebia trifoliata fruit based on volume. Hortic. Sci. Technol. 2021, 39, 823–831. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, L.; Zhao, X.; Zhao, Y.; Hao, Z.; Luo, H.; Yuan, Z. Advances in mechanisms and omics pertaining to fruit cracking in horticultural plants. Agronomy 2021, 11, 1045. [Google Scholar] [CrossRef]
- Brüggenwirth, M.; Knoche, M. Cell wall swelling, fracture mode, and the mechanical properties of cherry fruit skins are closely related. Planta 2017, 245, 765–777. [Google Scholar] [CrossRef]
- Coen, E.; Cosgrove, D.J. The mechanics of plant morphogenesis. Science 2023, 379, e8055. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, M.; Briggs, S.; Knox, J. Intercellular adhesion and cell separation in plants. Plant Cell Environ. 2003, 26, 977–989. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef]
- Markovič, O.; Janeček, Š. Pectin degrading glycoside hydrolases of family 28: Sequence-structural features, specificities and evolution. Protein Eng. 2001, 14, 615–631. [Google Scholar] [CrossRef]
- Fischer, R.L.; Bennett, A.B. Role of cell wall hydrolases in fruit ripening. Annu. Rev. Plant Biol. 1991, 42, 675–703. [Google Scholar] [CrossRef]
- Lu, L.; Hou, Q.; Wang, L.; Zhang, T.; Zhao, W.; Yan, T.; Zhao, L.; Li, J.; Wan, X. Genome-Wide Identification and Characterization of Polygalacturonase Gene Family in Maize (Zea mays L.). Int. J. Mol. Sci. 2021, 22, 10722. [Google Scholar] [CrossRef]
- Yu, Y.; Liang, Y.; Lv, M.; Wu, J.; Lu, G.; Cao, J. Genome-wide identification and characterization of polygalacturonase genes in Cucumis sativus and Citrullus lanatus. Plant Physiol. Biochem. 2014, 74, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, M.; Zhang, H.; Zhang, S.; Qian, M.; Zhang, Z.; Luo, W.; Fan, J.; Liu, Z.; Wang, L. Genome-wide analysis of polygalacturonase gene family from pear genome and identification of the member involved in pear softening. BMC Plant Biol. 2019, 19, 587–598. [Google Scholar] [CrossRef]
- Huang, W.; Chen, M.; Zhao, T.; Han, F.; Zhang, Q.; Liu, X.; Jiang, C.; Zhong, C. Genome-wide identification and expression analysis of polygalacturonase gene family in Kiwifruit (Actinidia chinensis) during fruit softening. Plants 2020, 9, 327. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Fatima, F.; Haider, M.S.; Shazadee, H.; Liu, Z.; Zheng, T.; Fang, J. Genome-wide identification and expression profiling of the polygalacturonase (PG) and pectin methylesterase (PME) genes in grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2019, 20, 3180. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Fernández, R.; Matilla, A.; Rodríguez-Gacio, M.; Fernández-Otero, C.; de La Torre, F. The polygalacturonase gene PdPG1 is developmentally regulated in reproductive organs of Prunus domestica L. subsp. insititia. Plant Sci. 2007, 172, 763–772. [Google Scholar] [CrossRef]
- Peng, G.; Wu, J.; Lu, W.; Li, J. A polygalacturonase gene clustered into clade E involved in lychee fruitlet abscission. Sci. Hortic. 2013, 150, 244–250. [Google Scholar] [CrossRef]
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef]
- Chen, D.; Wei, X.; Xu, L.; Xu, J.; Zeng, L. Genome-wide identification and expression analysis of PG gene family in wax apple [Syzygium samarangense (Bl.) Merr. et Perry]. J. Fruit Sci. 2022, 39, 548–563. [Google Scholar]
- Li, L.; Yao, X.; Zhong, C.; Chen, X.; Huang, H. Akebia: A potential new fruit crop in China. HortScience 2010, 45, 4–10. [Google Scholar] [CrossRef]
- Zhou, Y.; Jie, B.; Wu, G.; Cao, Y.; Wu, W.; Meng, H. Research progress of Akebia trifoliata (Thunb.) Koidz. Agric. Biotechnol. 2017, 6, 1–8. [Google Scholar]
- Zou, S.; Gao, P.; Jia, T.; Huang, H. Physicochemical characteristics and nutritional composition during fruit ripening of Akebia trifoliata (Lardizabalaceae). Horticulturae 2022, 8, 326. [Google Scholar] [CrossRef]
- Cao, Y.; Xiong, D.; Zhu, J.; Yu, H.; Li, G. Study on the respiration physiology of Akebia trifoliate fruit and the suitable storage conditions. J. Fruit Sci. 2003, 20, 512–514. [Google Scholar]
- Niu, J.; Shi, Y.; Huang, K.; Zhong, Y.; Chen, J.; Sun, Z.; Luan, M.; Chen, J. Integrative transcriptome and proteome analyses provide new insights into different stages of Akebia trifoliata fruit cracking during ripening. Biotechnol. Biofuels 2020, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yin, H.; Wang, D.; Zhong, Y.; Deng, Y. Exploring the mechanism of Akebia trifoliata fruit cracking based on cell-wall metabolism. Food Res. Int. 2022, 157, 111219. [Google Scholar] [CrossRef]
- Jiang, Y.; Yin, H.; Wang, D.; Zhong, Y.; Deng, Y. Combination of chitosan coating and heat shock treatments to maintain postharvest quality and alleviate cracking of Akebia trifoliate fruit during cold storage. Food Chem. 2022, 394, 133330. [Google Scholar] [CrossRef] [PubMed]
- Park, K.C.; Kwon, S.J.; Kim, N.S. Intron loss mediated structural dynamics and functional differentiation of the polygalacturonase gene family in land plants. Genes Genom. 2010, 32, 570–577. [Google Scholar] [CrossRef]
- Mahmood, U.; Fan, Y.; Wei, S.; Niu, Y.; Li, Y.; Huang, H.; Chen, Y.; Tang, Z.; Liu, L.; Qu, C. Comprehensive analysis of polygalacturonase genes offers new insights into their origin and functional evolution in land plants. Genomics 2021, 113, 1096–1108. [Google Scholar] [CrossRef]
- Yang, Z.L.; Liu, H.J.; Wang, X.R.; Zeng, Q.Y. Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol. 2013, 197, 1353–1365. [Google Scholar] [CrossRef]
- Sun, G.; Dilcher, D.L.; Wang, H.; Chen, Z. A eudicot from the Early Cretaceous of China. Nature 2011, 471, 625–628. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, J.; Zhang, N.; Shan, H.; Su, K.; Zhang, J.; Meng, Z.; Kong, H.; Chen, Z. Interactions among proteins of floral MADS-box genes in basal eudicots: Implications for evolution of the regulatory network for flower development. Mol. Biol. Evol. 2010, 27, 1598–1611. [Google Scholar] [CrossRef]
- Zhong, S.; Li, B.; Chen, W.; Wang, L.; Guan, J.; Wang, Q.; Yang, Z.; Yang, H.; Wang, X.; Yu, X. The chromosome-level genome of Akebia trifoliata as an important resource to study plant evolution and environmental adaptation in the Cretaceous. Plant J. 2022, 112, 1316–1330. [Google Scholar] [CrossRef]
- Ou, S.; Chen, J.; Jiang, N. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 2018, 46, e126. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhong, S.; Yang, H.; Chen, C.; Chen, W.; Yang, H.; Guan, J.; Fu, P.; Tan, F.; Ren, T. Identification and characterization of NBS resistance genes in Akebia trifoliata. Front. Plant Sci. 2021, 12, 758559. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Yang, H.; Guan, J.; Shen, J.; Ren, T.; Li, Z.; Tan, F.; Li, Q.; Luo, P. Characterization of the MADS-Box Gene Family in Akebia trifoliata and Their Evolutionary Events in Angiosperms. Genes 2022, 13, 1777. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhong, S.; Guan, J.; Chen, W.; Yang, H.; Yang, H.; Chen, C.; Tan, F.; Ren, T.; Li, Z. Genome-Wide Identification and Expression Analysis of WRKY Transcription Factors in Akebia trifoliata: A Bioinformatics Study. Genes 2022, 13, 1540. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, S.; Wu, X.; Li, J.; Xu, C.; Huang, M.; Wang, H.; Liu, H.; Zhao, Z. Identification of the NAC Transcription Factor Family during Early Seed Development in Akebia trifoliata (Thunb.) Koidz. Plants 2023, 12, 1518. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, Q.; Zhong, S.; Yang, H.; Ren, T.; Chen, C.; Tan, F.; Cao, G.; Liu, J.; Luo, P. Genome-Wide Identification of Superoxide Dismutase and Expression in Response to Fruit Development and Biological Stress in Akebia trifoliata: A Bioinformatics Study. Antioxidants 2023, 12, 726. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhong, S.; Dong, Q.; Yang, H.; Yang, H.; Tan, F.; Chen, C.; Ren, T.; Shen, J.; Cao, G. Identification of Photoperiod-and Phytohormone-Responsive DNA-Binding One Zinc Finger (Dof) Transcription Factors in Akebia trifoliata via Genome-Wide Expression Analysis. Int. J. Mol. Sci. 2023, 24, 4973. [Google Scholar] [CrossRef]
- Lyu, M.; Iftikhar, J.; Guo, R.; Wu, B.; Cao, J. Patterns of expansion and expression divergence of the polygalacturonase gene family in Brassica oleracea. Int. J. Mol. Sci. 2020, 21, 5706. [Google Scholar] [CrossRef]
- Tebbutt, S.J.; Rogers, H.J.; Lonsdale, D.M. Characterization of a tobacco gene encoding a pollen-specific polygalacturonase. Plant Mol. Biol. 1994, 25, 283–297. [Google Scholar] [CrossRef]
- Rexová-Benková, L. Evidence for the role of carboxyl groups in activity of endopolygalacturonase of Aspergillus niger. Chemical modification by carbodiimide reagent. Collect. Czechoslov. Chem. Commun. 1990, 55, 1389–1395. [Google Scholar] [CrossRef]
- Rao, M.N.; Kembhavi, A.A.; Pant, A. Implication of tryptophan and histidine in the active site of endo-polygalacturonase from Aspergillus ustus: Elucidation of the reaction mechanism. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 1996, 1296, 167–173. [Google Scholar]
- Bussink, H.J.; Buxton, F.P.; Visser, J. Expression and sequence comparison of the Aspergillus niger and Aspergillus tubigensis genes encoding polygalacturonase II. Curr. Genet. 1991, 19, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Son, J.H.; Park, K.C.; Oh, H.Y.; Kim, P.H.; Byeon, W.H.; Kim, N.S. Structural dynamics and divergence of the polygalacturonase gene family in land plants. Nat. Preced. 2008. [Google Scholar] [CrossRef]
- Mahmood, U.; Li, X.; Qian, M.; Fan, Y.; Yu, M.; Li, S.; Shahzad, A.; Qu, C.; Li, J.; Liu, L. Comparative transcriptome and co-expression network analysis revealed the genes associated with senescence and polygalacturonase activity involved in pod shattering of rapeseed. Biotechnol. Biofuels Bioprod. 2023, 16, 20–36. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Li, W.H.; Yang, J.; Gu, X. Expression divergence between duplicate genes. TRENDS Genet. 2005, 21, 602–607. [Google Scholar] [CrossRef]
- Kim, J.; Shiu, S.H.; Thoma, S.; Li, W.H.; Patterson, S.E. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006, 7, 1–14. [Google Scholar]
- Shcherbakov, V.P. Biological species is the only possible form of existence for higher organisms: The evolutionary meaning of sexual reproduction. Biol. Direct 2010, 5, 1–22. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, Y.; Cui, J.; Lyu, M.; Xu, L.; Cao, J. A comparative analysis of the evolution, expression, and cis-regulatory element of polygalacturonase genes in grasses and dicots. Funct. Integr. Genom. 2016, 16, 641–656. [Google Scholar] [CrossRef]
- Liu, L. Identification of PG Gene Family in Durian and Pectin Changes during Fruit Dehiscence. Master’s Thesis, Anhui Agricultural University, Hefei, China, 2022. [Google Scholar]
- Russo, C.A.; Selvatti, A.P. Bootstrap and rogue identification tests for phylogenetic analyses. Mol. Biol. Evol. 2018, 35, 2327–2333. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).