Adhesion G Protein-Coupled Receptor G2 Promotes Hepatocellular Carcinoma Progression and Serves as a Neutrophil-Related Prognostic Biomarker
Abstract
:1. Introduction
2. Results
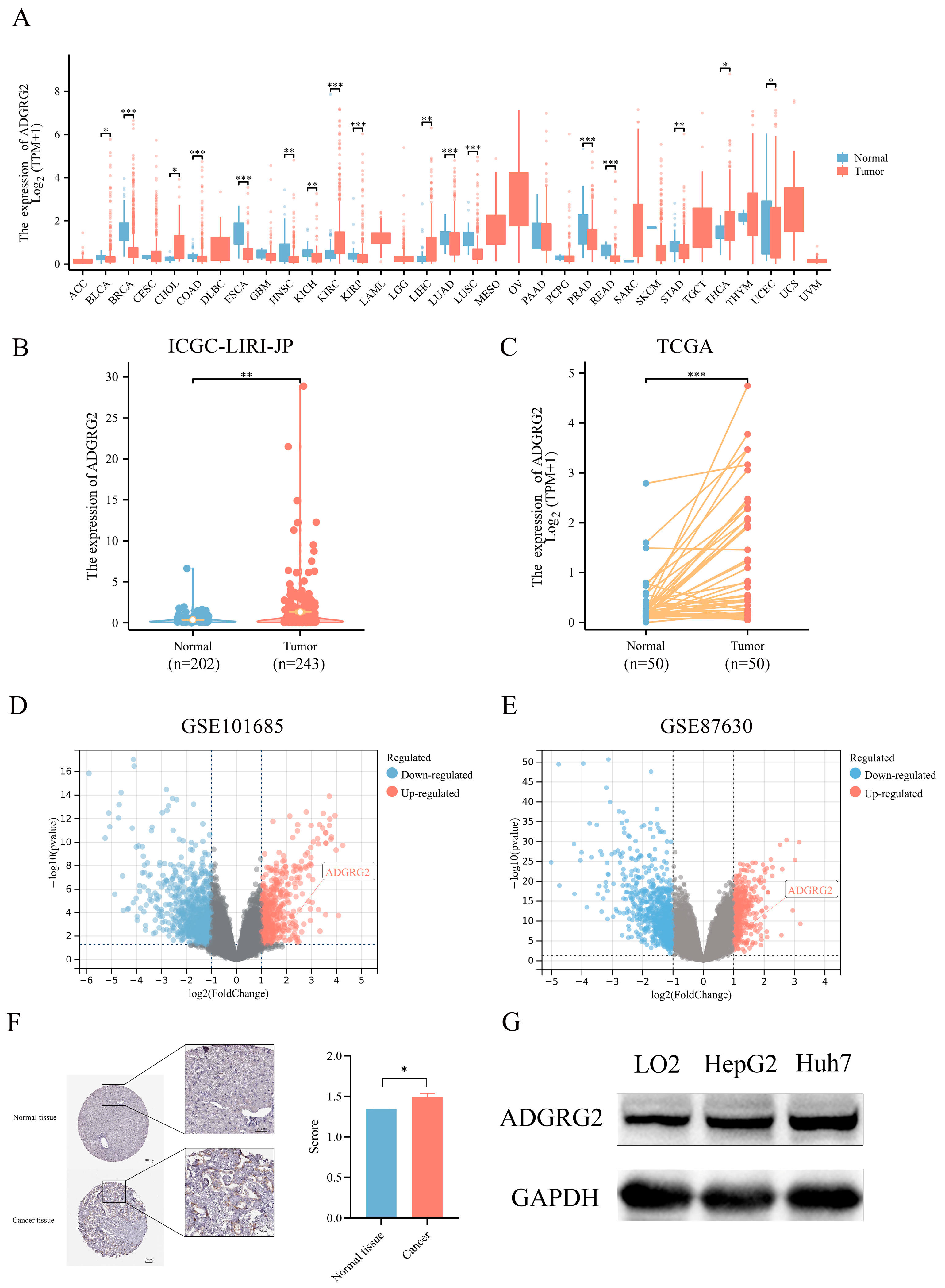
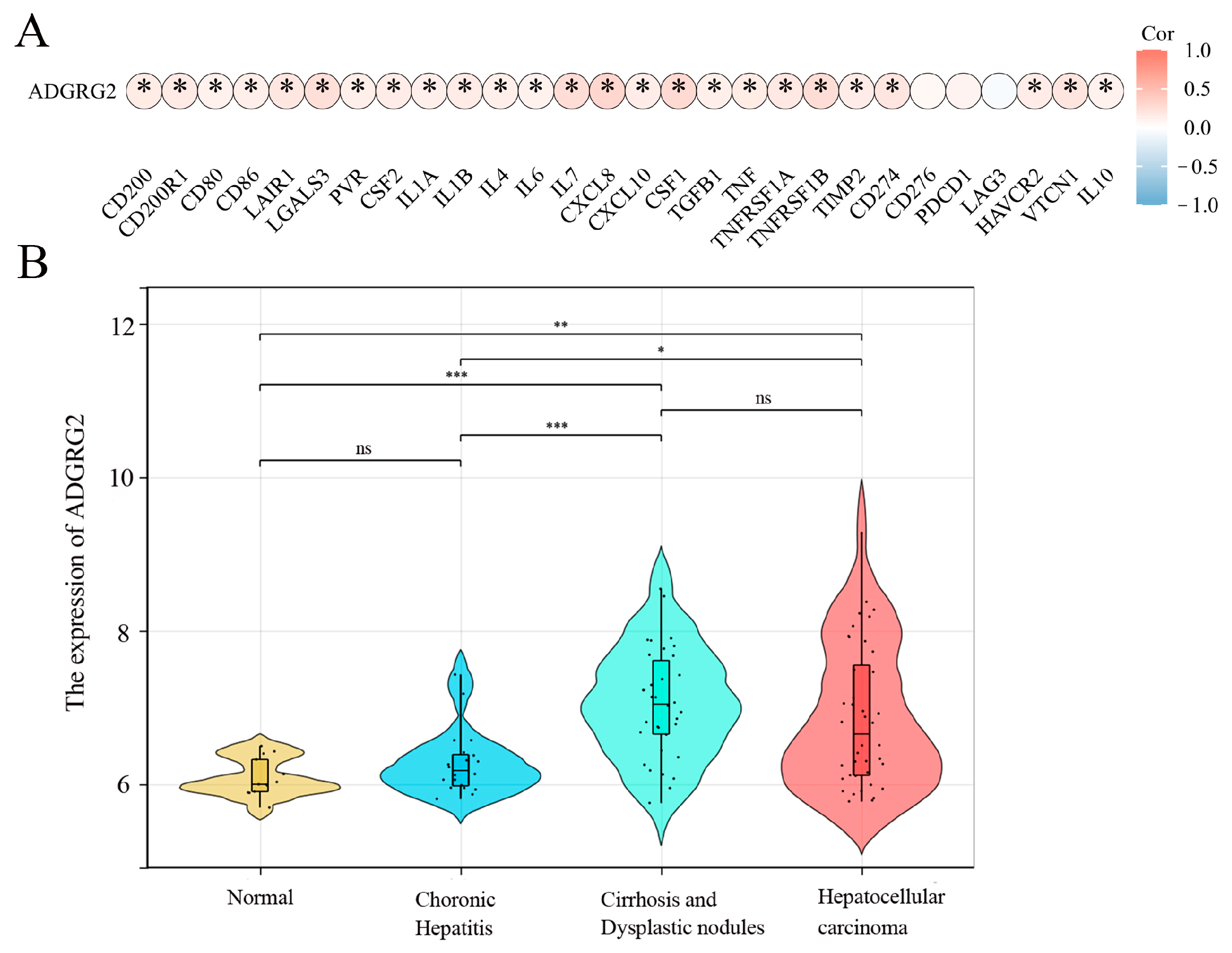
2.1. High Expression of ADGRG2 in HCC
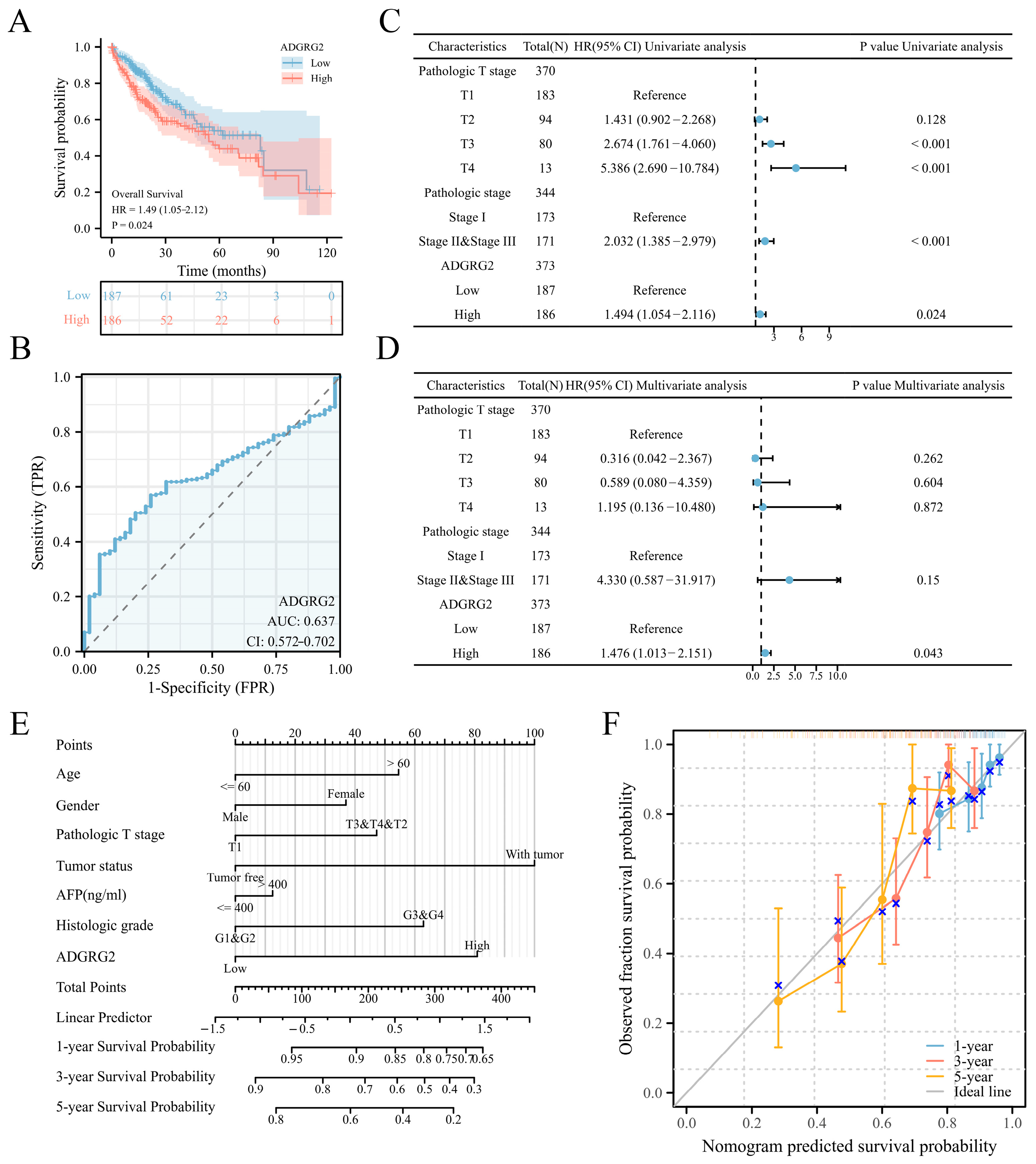
2.2. High Expression of ADGRG2 Was Associated with Adverse Clinicopathological Factors and Worse Prognosis in HCC
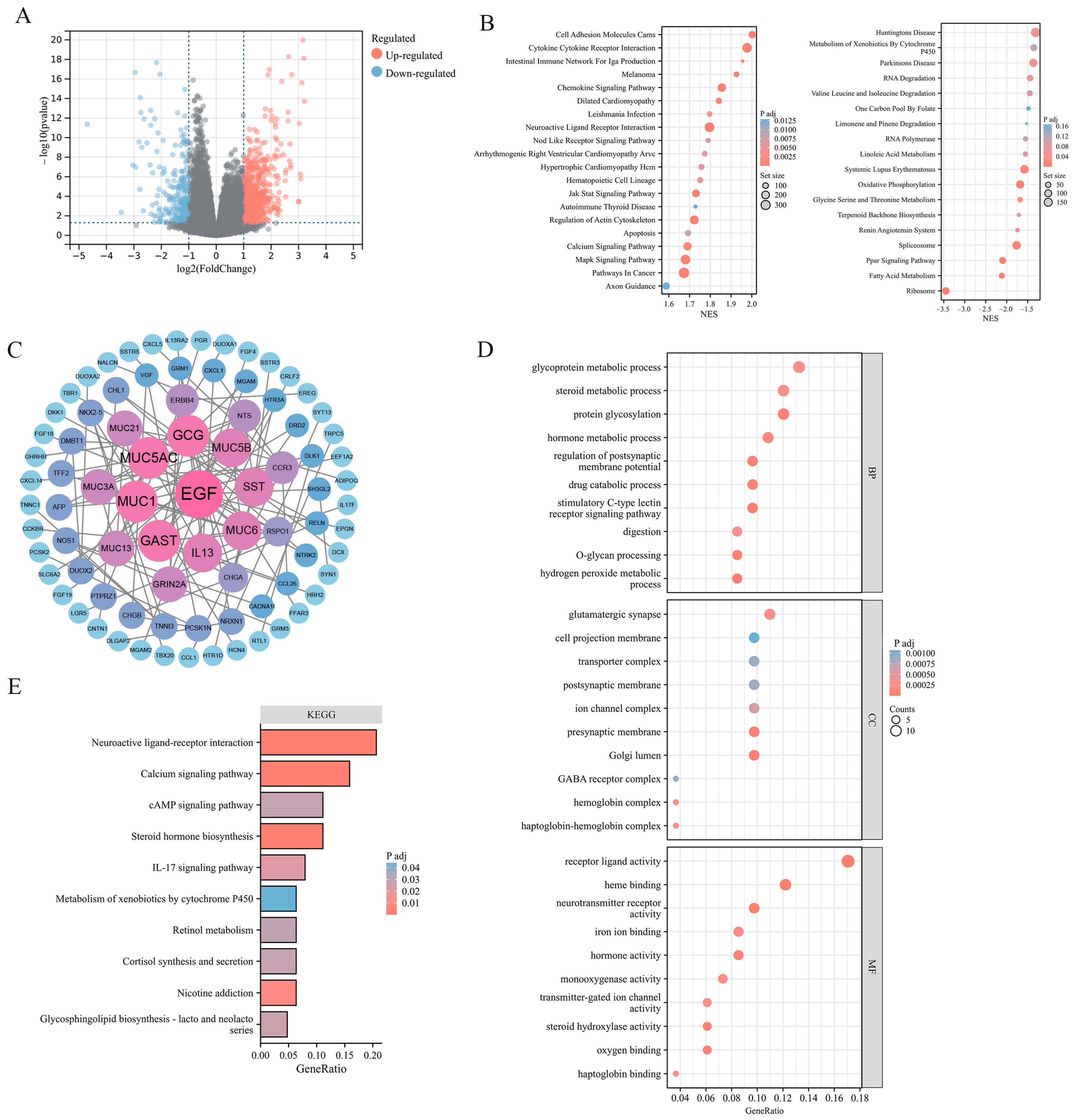
2.3. Enrichment Analysis of ADGRG2-Related DEGs in HCC
2.4. Knockdown of ADGRG2 Inhibited the Proliferation and Migration of HCC Cells
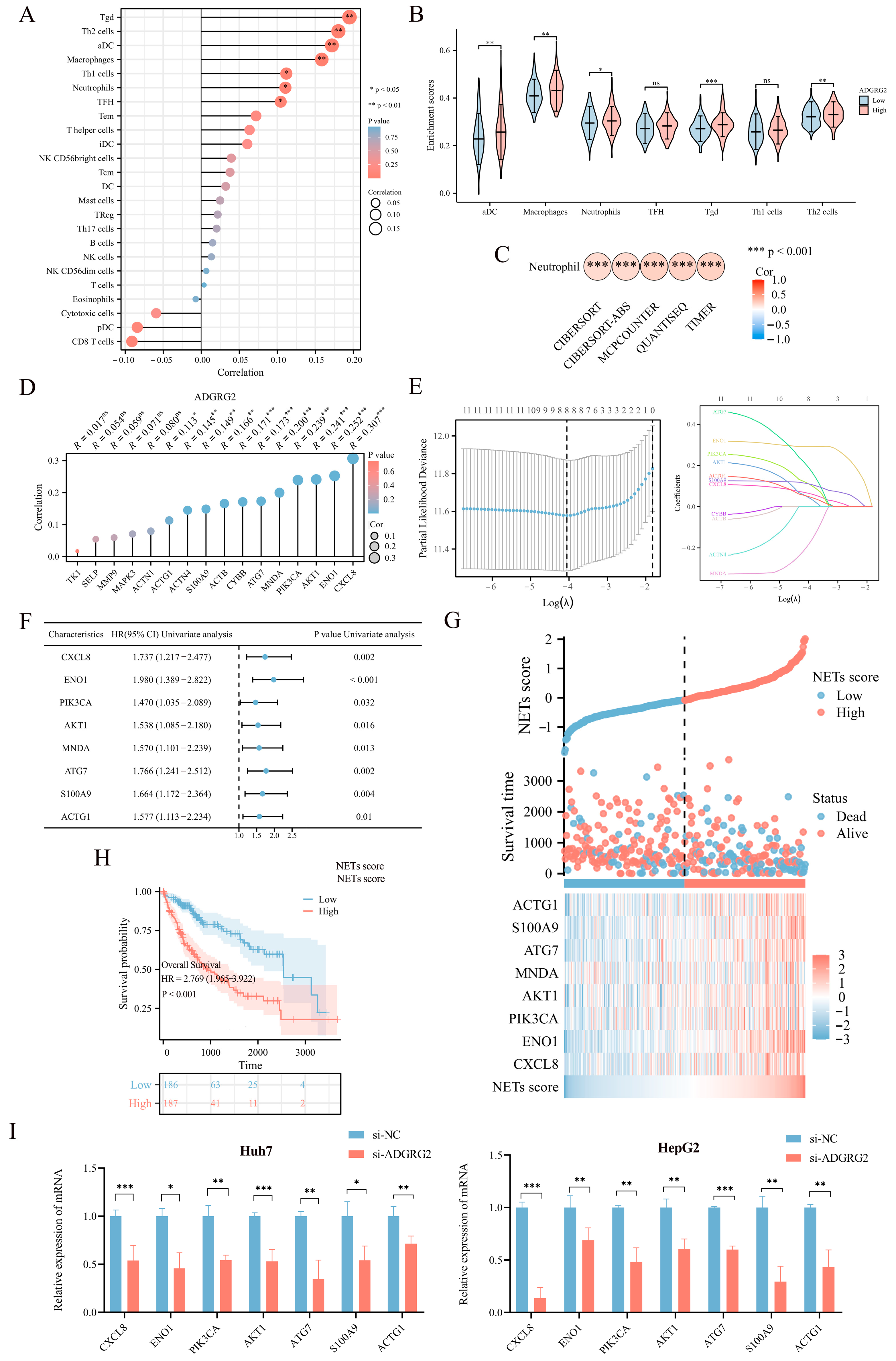
2.5. Associations between ADGRG2 and Neutrophil Infiltration in HCC
2.6. The Potential Role of ADGRG2 in Inflammation
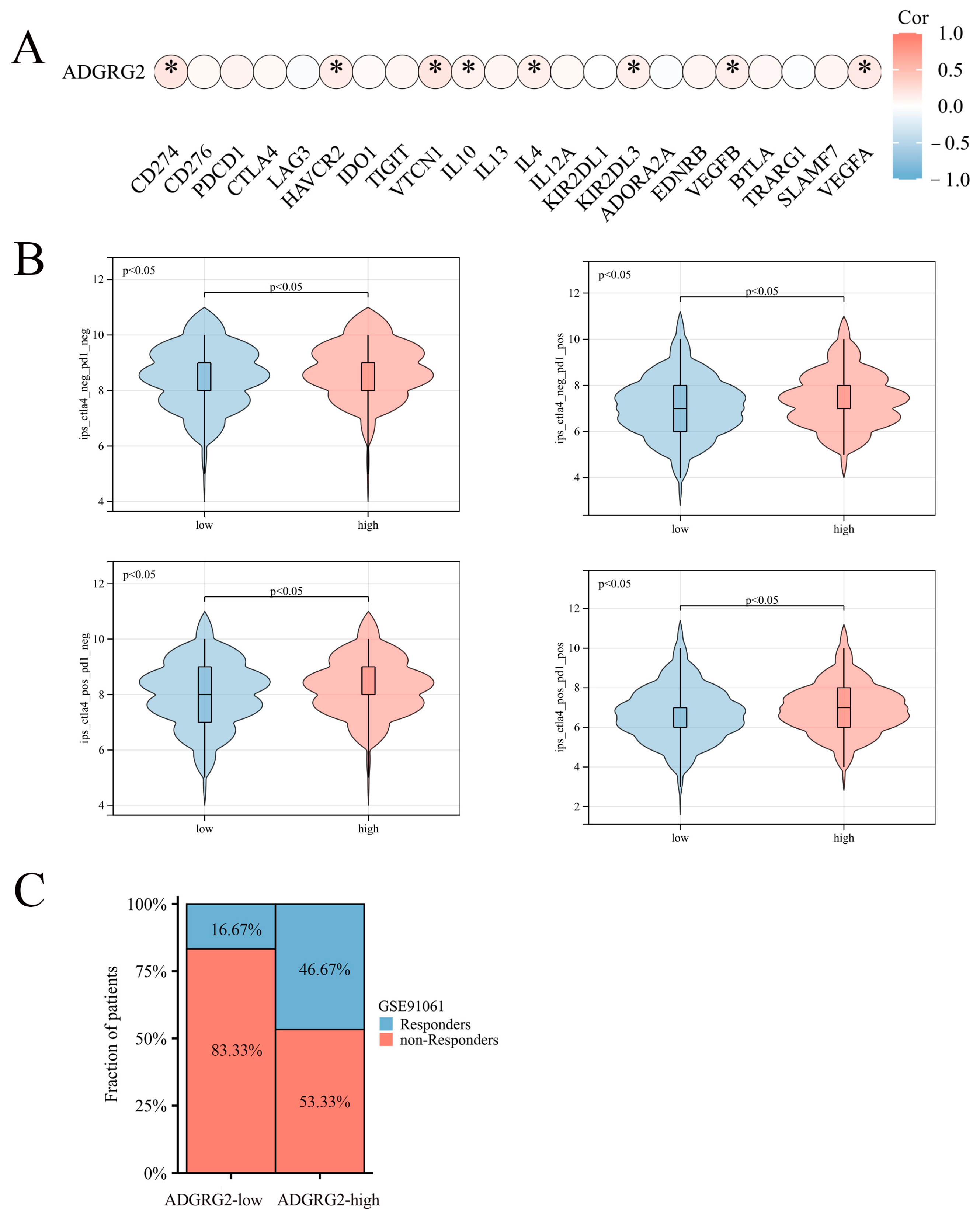
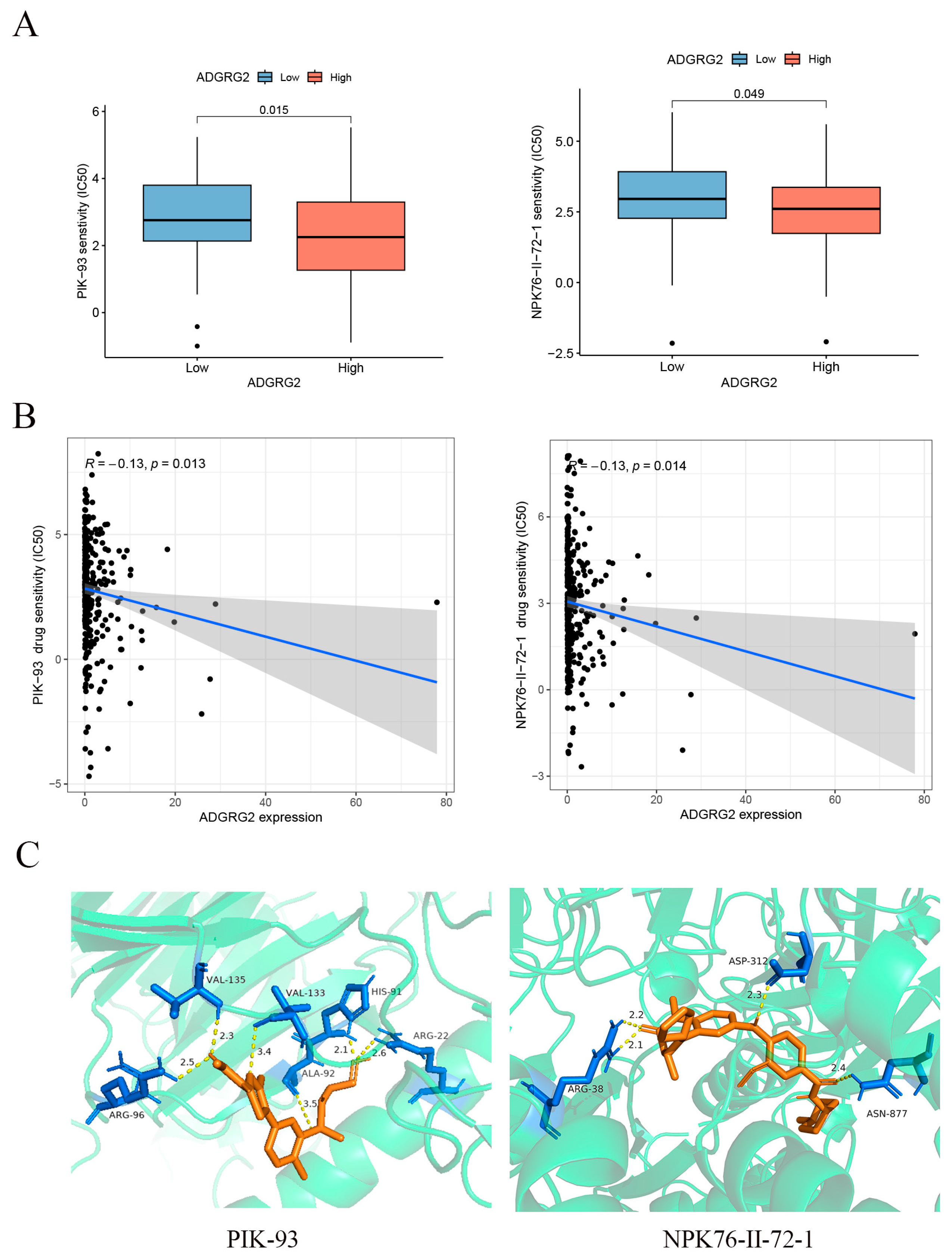
2.7. Correlation of ADGRG2 Expression with Immunotherapy and Drug Sensitivity
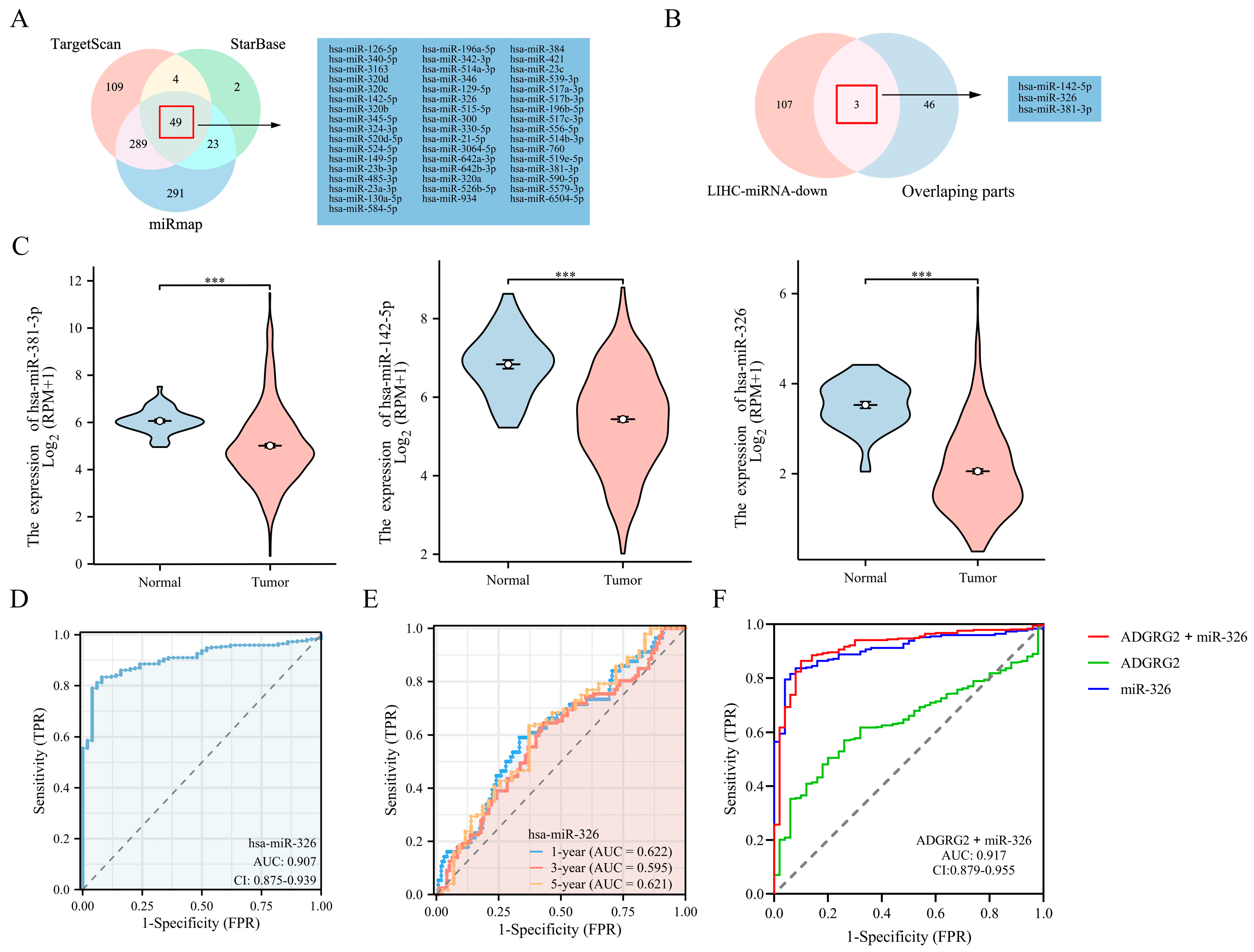
2.8. Prediction of miRNAs Targeting ADGRG2
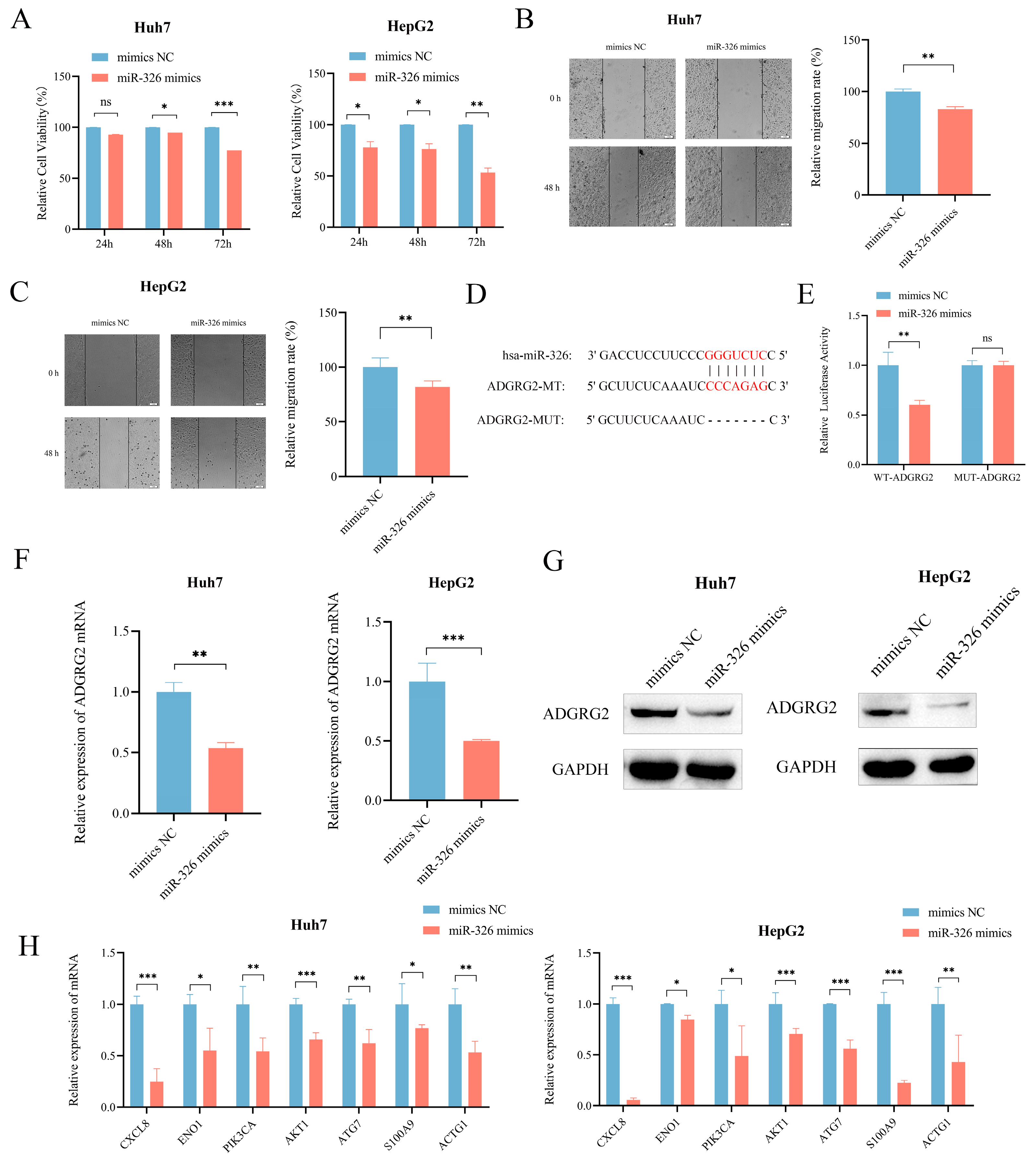
2.9. MiR-326 Suppressed the Proliferation and Migration of Liver Cancer Cells and Directly Targeted ADGRG2
3. Discussion
4. Materials and Methods
4.1. Data Download and Processing
4.2. Identification of ADGRG2-Related Genes
4.3. Enrichment Analysis of ADGRG2-Related DEGs in HCC
4.4. Identification of Potential miRNAs Targeting ADGRG2
4.5. Correlation Analysis of ADGRG2 and Immunity Characteristics
4.6. Prediction of Response to Immunotherapy
4.7. Drug Sensitivity Analysis and Molecular Docking
4.8. Cell Culture and Transfection
4.9. Reverse Transcription and Quantitative PCR (RT-qPCR)
4.10. CCK8 and Wound Healing Assays
4.11. Dual Luciferase Reporter Assay
4.12. Western Blot Analysis
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef]
- Farazi, P.A.; DePinho, R.A. Hepatocellular Carcinoma Pathogenesis: From Genes to Environment. Nat. Rev. Cancer 2006, 6, 674–687. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Gomaa, A.I.; Khan, S.A.; Toledano, M.B.; Waked, I.; Taylor-Robinson, S.D. Hepatocellular Carcinoma: Epidemiology, Risk Factors and Pathogenesis. World J. Gastroenterol. 2008, 14, 4300–4308. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Zou, H.; Liao, W. Prospect of Circular RNA in Hepatocellular Carcinoma: A Novel Potential Biomarker and Therapeutic Target. Front. Oncol. 2018, 8, 332. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-T.; Ma, W.K.; Scharner, J.; Liu, Y.-R.; Krainer, A.R. A Human-Specific Switch of Alternatively Spliced AFMID Isoforms Contributes to TP53 Mutations and Tumor Recurrence in Hepatocellular Carcinoma. Genome Res. 2018, 28, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Ghavimi, S.; Apfel, T.; Azimi, H.; Persaud, A.; Pyrsopoulos, N.T. Management and Treatment of Hepatocellular Carcinoma with Immunotherapy: A Review of Current and Future Options. J. Clin. Transl. Hepatol. 2020, 8, 168–176. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Zhang, J.; Zhang, Y.; Li, Y.; Liu, Z.; Li, Q.; Luo, M.; Liang, R.; Ye, J. Molecular Targeted and Immune Checkpoint Therapy for Advanced Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 447. [Google Scholar] [CrossRef]
- Feng, M.Y.; Chan, L.L.; Chan, S.L. Drug Treatment for Advanced Hepatocellular Carcinoma: First-Line and Beyond. Curr. Oncol. 2022, 29, 5489–5507. [Google Scholar] [CrossRef]
- Jiang, L.; Li, L.; Liu, Y.; Lu, L.; Zhan, M.; Yuan, S.; Liu, Y. Drug Resistance Mechanism of Kinase Inhibitors in the Treatment of Hepatocellular Carcinoma. Front. Pharmacol. 2023, 14, 1097277. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, B.P.; Robson, S.C.; Burnstock, G. Pathological Roles of Purinergic Signaling in the Liver. J. Hepatol. 2012, 57, 916–920. [Google Scholar] [CrossRef]
- Pang, Y.-L.; Zhang, H.-G.; Peng, J.-R.; Pang, X.-W.; Yu, S.; Xing, Q.; Yu, X.; Gong, L.; Yin, Y.-H.; Zhang, Y.; et al. The Immunosuppressive Tumor Microenvironment in Hepatocellular Carcinoma. Cancer Immunol. Immunother. 2009, 58, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Osterhoff, C.; Ivell, R.; Kirchhoff, C. Cloning of a Human Epididymis-Specific MRNA, HE6, Encoding a Novel Member of the Seven Transmembrane-Domain Receptor Superfamily. DNA Cell Biol. 1997, 16, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, Y.; Lin, H.; Sun, Y.; Wang, M.; Jia, Y.; Yu, X.; Jiang, H.; Xu, W.; Sun, J.-P.; et al. Function and Therapeutic Potential of G Protein-Coupled Receptors in Epididymis. Br. J. Pharmacol. 2020, 177, 5489–5508. [Google Scholar] [CrossRef] [PubMed]
- Richter, G.H.S.; Fasan, A.; Hauer, K.; Grunewald, T.G.P.; Berns, C.; Rössler, S.; Naumann, I.; Staege, M.S.; Fulda, S.; Esposito, I.; et al. G-Protein Coupled Receptor 64 Promotes Invasiveness and Metastasis in Ewing Sarcomas through PGF and MMP1. J. Pathol. 2013, 230, 70–81. [Google Scholar] [CrossRef]
- Balenga, N.; Azimzadeh, P.; Hogue, J.A.; Staats, P.N.; Shi, Y.; Koh, J.; Dressman, H.; Olson, J.A. Orphan Adhesion GPCR GPR64/ADGRG2 Is Overexpressed in Parathyroid Tumors and Attenuates Calcium-Sensing Receptor-Mediated Signaling. J. Bone Miner. Res. 2017, 32, 654–666. [Google Scholar] [CrossRef]
- Xie, T.; Tang, Y.; Luo, R.; Zhang, X.; Wu, S.; Gu, Y.; Liu, T.; Hu, F. GPR64 Promotes CAMP Pathway in Tumor Aggressiveness in Sparsely Granulated Growth Hormone Cell Adenomas. Endocrine 2020, 68, 629–639. [Google Scholar] [CrossRef]
- Ahn, J.I.; Yoo, J.-Y.; Kim, T.H.; Kim, Y.I.; Broaddus, R.R.; Ahn, J.Y.; Lim, J.M.; Jeong, J.-W. G-Protein Coupled Receptor 64 (GPR64) Acts as a Tumor Suppressor in Endometrial Cancer. BMC Cancer 2019, 19, 810. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, B.; Wu, S.; Liu, Y. Prognostic Immune-Related Genes of Patients With Ewing’s Sarcoma. Front. Genet. 2021, 12, 669549. [Google Scholar] [CrossRef]
- Nakamura, K.; Asanuma, K.; Okamoto, T.; Yoshida, K.; Matsuyama, Y.; Kita, K.; Hagi, T.; Nakamura, T.; Sudo, A. GPR64, Screened from Ewing Sarcoma Cells, Is a Potential Target for Antibody-Based Therapy for Various Sarcomas. Cancers 2022, 14, 814. [Google Scholar] [CrossRef]
- Jeon, T.-W.; Yang, H.; Lee, C.G.; Oh, S.T.; Seo, D.; Baik, I.H.; Lee, E.H.; Yun, I.; Park, K.R.; Lee, Y.-H. Electro-Hyperthermia up-Regulates Tumour Suppressor Septin 4 to Induce Apoptotic Cell Death in Hepatocellular Carcinoma. Int. J. Hyperth. 2016, 32, 648–656. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR Drug Discovery: New Agents, Targets and Indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Chaudhary, P.K.; Kim, S. An Insight into GPCR and G-Proteins as Cancer Drivers. Cells 2021, 10, 3288. [Google Scholar] [CrossRef]
- Zenlander, R.; Havervall, S.; Magnusson, M.; Engstrand, J.; Ågren, A.; Thålin, C.; Stål, P. Neutrophil Extracellular Traps in Patients with Liver Cirrhosis and Hepatocellular Carcinoma. Sci. Rep. 2021, 11, 18025. [Google Scholar] [CrossRef]
- Mossanen, J.C.; Tacke, F. Role of Lymphocytes in Liver Cancer. Oncoimmunology 2013, 2, e26468. [Google Scholar] [CrossRef] [PubMed]
- Lazennec, G.; Richmond, A. Chemokines and Chemokine Receptors: New Insights into Cancer-Related Inflammation. Trends Mol. Med. 2010, 16, 133–144. [Google Scholar] [CrossRef]
- Chu, H.X.; Arumugam, T.V.; Gelderblom, M.; Magnus, T.; Drummond, G.R.; Sobey, C.G. Role of CCR2 in Inflammatory Conditions of the Central Nervous System. J. Cereb. Blood Flow Metab. 2014, 34, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-Q.; Zhou, S.-L.; Li, J.; Zhou, Z.-J.; Wang, P.-C.; Xin, H.-Y.; Mao, L.; Luo, C.-B.; Yu, S.-Y.; Huang, X.-W.; et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology 2020, 72, 906–922. [Google Scholar] [CrossRef] [PubMed]
- Oura, K.; Morishita, A.; Masaki, T. Molecular and Functional Roles of MicroRNAs in the Progression of Hepatocellular Carcinoma-A Review. Int. J. Mol. Sci. 2020, 21, 8362. [Google Scholar] [CrossRef]
- Yilmaz, M.; Christofori, G. EMT, the Cytoskeleton, and Cancer Cell Invasion. Cancer Metastasis Rev. 2009, 28, 15–33. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xun, M.; Li, C.; Chen, Y. The O-GlcNAcylation and Its Promotion to Hepatocellular Carcinoma. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188806. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-K.; Li, C.; Zhang, R.-Y.; Wei, D.; Shang, Y.-K.; Yong, Y.-L.; Kong, L.-M.; Zheng, N.-S.; Liu, K.; Lu, M.; et al. EYA2 Suppresses the Progression of Hepatocellular Carcinoma via SOCS3-Mediated Blockade of JAK/STAT Signaling. Mol. Cancer 2021, 20, 79. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Liu, D.; Chen, L.; He, Y.; Tian, X.; Qi, L.; Chen, L.; Luo, Y.; Chen, Z.; Hu, X.; et al. PNO1 Regulates Autophagy and Apoptosis of Hepatocellular Carcinoma via the MAPK Signaling Pathway. Cell Death Dis. 2021, 12, 552. [Google Scholar] [CrossRef]
- Ishtiaq, S.M.; Arshad, M.I.; Khan, J.A. PPARγ Signaling in Hepatocarcinogenesis: Mechanistic Insights for Cellular Reprogramming and Therapeutic Implications. Pharmacol. Ther. 2022, 240, 108298. [Google Scholar] [CrossRef]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in Cancer: Neutral No More. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Demkow, U. Neutrophil Extracellular Traps (NETs) in Cancer Invasion, Evasion and Metastasis. Cancers 2021, 13, 4495. [Google Scholar] [CrossRef]
- Yang, L.-Y.; Luo, Q.; Lu, L.; Zhu, W.-W.; Sun, H.-T.; Wei, R.; Lin, Z.-F.; Wang, X.-Y.; Wang, C.-Q.; Lu, M.; et al. Increased Neutrophil Extracellular Traps Promote Metastasis Potential of Hepatocellular Carcinoma via Provoking Tumorous Inflammatory Response. J. Hematol. Oncol. 2020, 13, 3. [Google Scholar] [CrossRef]
- Huang, C.K.; Sun, Y.; Lv, L.; Ping, Y. ENO1 and Cancer. Mol. Ther. Oncolytics 2022, 24, 288–298. [Google Scholar] [CrossRef]
- Jiang, K.; Dong, C.; Yin, Z.; Li, R.; Mao, J.; Wang, C.; Zhang, J.; Gao, Z.; Liang, R.; Wang, Q.; et al. Exosome-Derived ENO1 Regulates Integrin A6β4 Expression and Promotes Hepatocellular Carcinoma Growth and Metastasis. Cell Death Dis. 2020, 11, 972. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Zhang, Y.; Xiang, Y.; Tang, Y.; Cui, F.; Cao, J.; Zhou, L.; You, Y.; Duan, L. Association between Serum S100A9 Levels and Liver Necroinflammation in Chronic Hepatitis B. J. Transl. Med. 2018, 16, 83. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Wu, R.; Kong, X.-H.; You, Y.; He, K.; Sun, X.-Y.; Huang, Y.; Chen, W.-X.; Duan, L. Elevated Neutrophil Extracellular Traps by HBV-Mediated S100A9-TLR4/RAGE-ROS Cascade Facilitate the Growth and Metastasis of Hepatocellular Carcinoma. Cancer Commun. 2023, 43, 225–245. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Yan, J.; Xia, X.; Zhou, S.; Chen, J.; Mishra, L.; Li, S. IL6-Mediated Inflammatory Loop Reprograms Normal to Epithelial-Mesenchymal Transition+ Metastatic Cancer Stem Cells in Preneoplastic Liver of Transforming Growth Factor Beta-Deficient Β2-Spectrin+/− Mice. Hepatology 2017, 65, 1222–1236. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, Inflammation, and Cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int. J. Mol. Sci. 2020, 21, 7619. [Google Scholar] [CrossRef]
- Sadhukhan, P.; Seiwert, T.Y. The Role of Macrophages in the Tumor Microenvironment and Tumor Metabolism. Semin. Immunopathol. 2023, 45, 187–201. [Google Scholar] [CrossRef]
- Zou, G.; Zhang, X.; Wang, L.; Li, X.; Xie, T.; Zhao, J.; Yan, J.; Wang, L.; Ye, H.; Jiao, S.; et al. Herb-Sourced Emodin Inhibits Angiogenesis of Breast Cancer by Targeting VEGFA Transcription. Theranostics 2020, 10, 6839–6853. [Google Scholar] [CrossRef]
- Robertson, C.L.; Mendoza, R.G.; Jariwala, N.; Dozmorov, M.; Mukhopadhyay, N.D.; Subler, M.A.; Windle, J.J.; Lai, Z.; Fisher, P.B.; Ghosh, S.; et al. Astrocyte Elevated Gene-1 Regulates Macrophage Activation in Hepatocellular Carcinogenesis. Cancer Res. 2018, 78, 6436–6446. [Google Scholar] [CrossRef]
- Matsui, T.; Nagai, H.; Sumino, Y.; Miki, K. Relationship of Peripheral Blood CD4-Positive T Cells to Carcinogenesis in Patients with HCV-Related Chronic Hepatitis and Liver Cirrhosis. Cancer Chemother. Pharmacol. 2008, 62, 401–406. [Google Scholar] [CrossRef]
- Ilan, Y. Immune Therapy for Hepatocellular Carcinoma. Hepatol. Int. 2014, 8 (Suppl. 2), 499–504. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Ikeda, M.; Zhu, A.X.; Sung, M.W.; Baron, A.D.; Kudo, M.; Okusaka, T.; Kobayashi, M.; Kumada, H.; Kaneko, S.; et al. Phase Ib Study of Lenvatinib Plus Pembrolizumab in Patients With Unresectable Hepatocellular Carcinoma. J. Clin. Oncol. 2020, 38, 2960–2970. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-Y.; Huang, K.-Y.; Kao, S.-H.; Lin, M.-S.; Lin, C.-C.; Yang, S.-C.; Chung, W.-C.; Chang, Y.-H.; Chein, R.-J.; Yang, P.-C. Small-Molecule PIK-93 Modulates the Tumor Microenvironment to Improve Immune Checkpoint Blockade Response. Sci. Adv. 2023, 9, eade9944. [Google Scholar] [CrossRef]
- Chen, W.; Chen, L.; Guo, L.; Liu, N.; Wu, T.; Cheng, Y.; Xu, P.; Li, Y.; Yang, X.; Xu, R.; et al. The Signature of Immune-Subtype Specific Driving Transcription Factors Suggest Potential Drugs for Refractory Glioblastoma. Am. J. Cancer Res. 2023, 13, 1278–1294. [Google Scholar]
- Turato, C.; Simonato, D.; Quarta, S.; Gatta, A.; Pontisso, P. MicroRNAs and SerpinB3 in Hepatocellular Carcinoma. Life Sci. 2014, 100, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.-Q.; Li, L.; Lu, C.; Liu, J.; Chen, Y.; Wu, H. Involvement of H19/MiR-326 Axis in Hepatocellular Carcinoma Development through Modulating TWIST1. J. Cell. Physiol. 2019, 234, 5153–5162. [Google Scholar] [CrossRef]
- Varghese, F.; Bukhari, A.B.; Malhotra, R.; De, A. IHC Profiler: An Open Source Plugin for the Quantitative Evaluation and Automated Scoring of Immunohistochemistry Images of Human Tissue Samples. PLoS ONE 2014, 9, e96801. [Google Scholar] [CrossRef]
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martín-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949.e16. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Shen, X.-T.; Xie, S.-Z.; Xu, J.; Yang, L.-Y.; Qin, L.-X. Pan-Cancer Analysis Reveals a Distinct Neutrophil Extracellular Trap-Associated Regulatory Pattern. Front. Immunol. 2022, 13, 798022. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, W.; Li, Q.; Mao, J.; Chen, X. A Novel Neutrophil Extracellular Trap Signature to Predict Prognosis and Immunotherapy Response in Head and Neck Squamous Cell Carcinoma. Front. Immunol. 2022, 13, 1019967. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand Docking and Binding Site Analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]

| Characteristics | Low Expression of ADGRG2 | High Expression of ADGRG2 | p Value |
|---|---|---|---|
| Pathologic stage, n (%) | 0.03174037 | ||
| Stage I | 96 (36.9%) | 77 (29.6%) | |
| Stage II | 36 (13.8%) | 51 (19.6%) | |
| AFP (ng/mL), n (%) | 0.00295382 | ||
| ≤400 | 97 (34.6%) | 118 (42.1%) | |
| >400 | 43 (15.4%) | 22 (7.9%) | |
| OS event, n (%) | 0.02987853 | ||
| Alive | 132 (35.3%) | 112 (29.9%) | |
| Dead | 55 (14.7%) | 75 (20.1%) |
| Marker | Marker Type | Correlation | p Value | Marker | Marker Type | Correlation | p Value |
|---|---|---|---|---|---|---|---|
| CD15 | TAN | 0.374 | 9.95 × 10−14 | CD54 | TAN | 0.243 | 2.08 × 10−6 |
| CXCR1 | inflammation | 0.126 | 1.51 × 10−2 | CD217 | inflammation | 0.165 | 1.44 × 10−3 |
| CXCR2 | TAN, inflammation | 0.219 | 2.02 × 10−5 | ITGB2 | inflammation | 0.147 | 4.68 × 10−3 |
| CD11b | TAN, inflammation | 0.265 | 2.33 × 10−7 | PTPRC | TAN | 0.154 | 2.99 × 10−3 |
| CD11c | inflammation | 0.143 | 5.90 × 10−3 | CD43 | inflammation | 0.113 | 2.95 × 10−2 |
| FCGR3A | TAN, inflammation | 0.189 | 2.55 × 10−4 | TLR2 | inflammation | 0.253 | 7.67 × 10−7 |
| HLA-DRA | TAN | 0.228 | 9.39 × 10−6 | TLR4 | inflammation | 0.26 | 3.64 × 10−7 |
| HLA-DRB1 | TAN | 0.143 | 5.91 × 10−3 | TLR5 | inflammation | 0.348 | 4.96 × 10−12 |
| HLA-DRB3 | TAN | 0.181 | 4.43 × 10−4 | TLR7 | inflammation | 0.243 | 2.15 × 10−6 |
| CD49d | inflammation | 0.169 | 1.05 × 10−3 | TLR8 | inflammation | 0.14 | 7.07 × 10−3 |
| ARG1 | TAN | 0.108 | 3.79 × 10−2 | TLR9 | inflammation | 0.165 | 1.42 × 10−3 |
| CD86 | TAN | 0.147 | 4.65 × 10−3 | CD63 | inflammation | 0.144 | 5.58 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Wang, P.; Peng, Q.; Kang, Z.; Deng, Y.; Li, J.; Chen, Y.; Li, J.; Ge, F. Adhesion G Protein-Coupled Receptor G2 Promotes Hepatocellular Carcinoma Progression and Serves as a Neutrophil-Related Prognostic Biomarker. Int. J. Mol. Sci. 2023, 24, 16986. https://doi.org/10.3390/ijms242316986
Wu Q, Wang P, Peng Q, Kang Z, Deng Y, Li J, Chen Y, Li J, Ge F. Adhesion G Protein-Coupled Receptor G2 Promotes Hepatocellular Carcinoma Progression and Serves as a Neutrophil-Related Prognostic Biomarker. International Journal of Molecular Sciences. 2023; 24(23):16986. https://doi.org/10.3390/ijms242316986
Chicago/Turabian StyleWu, Qian, Pei Wang, Qihang Peng, Zhongcui Kang, Yiting Deng, Jiayi Li, Ying Chen, Jin Li, and Feng Ge. 2023. "Adhesion G Protein-Coupled Receptor G2 Promotes Hepatocellular Carcinoma Progression and Serves as a Neutrophil-Related Prognostic Biomarker" International Journal of Molecular Sciences 24, no. 23: 16986. https://doi.org/10.3390/ijms242316986





