Neuronal Scaffold Protein ARMS Interacts with Synaptotagmin-4 C2AB through the Ankyrin Repeat Domain with an Unexpected Mode
Abstract
:1. Introduction
2. Results
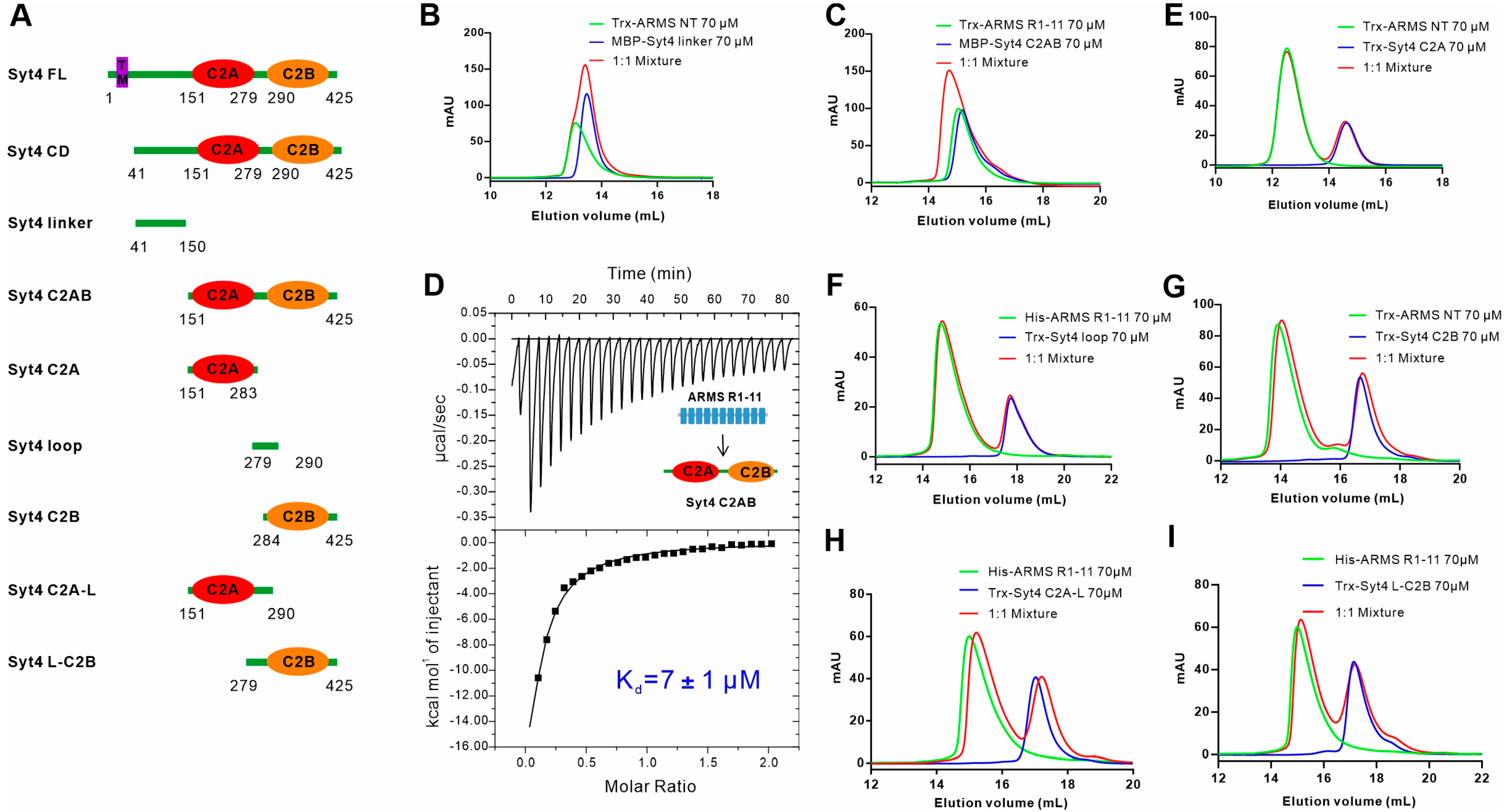
2.1. ARMS Interacts with the Cytoplasmic Region of Syt4 via the Ankyrin Repeat Domain
2.2. C2AB Domain of Syt4 Is the ARMS Binding Region
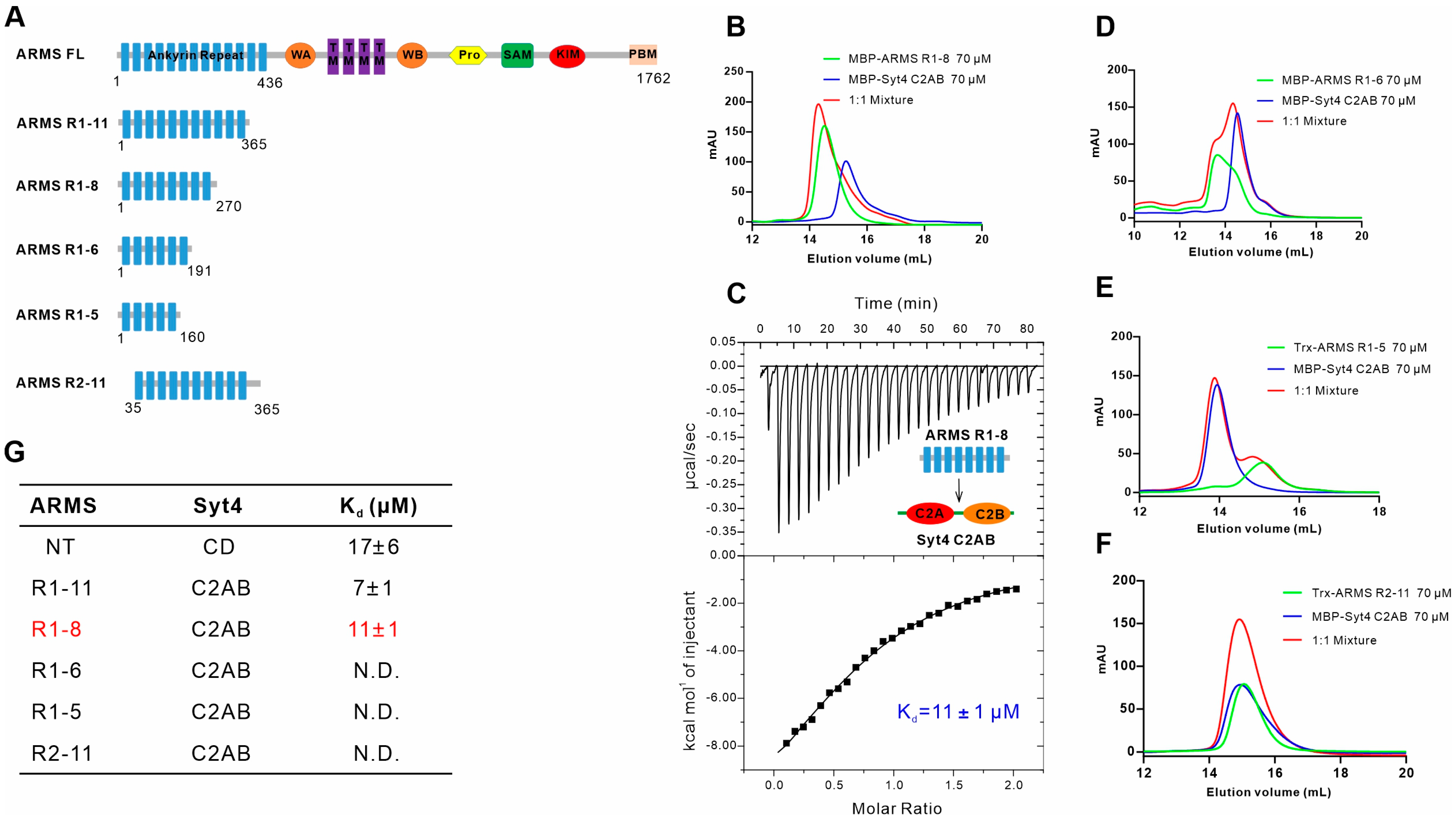
2.3. ARMS R1-8 Is Required for Binding with Syt4
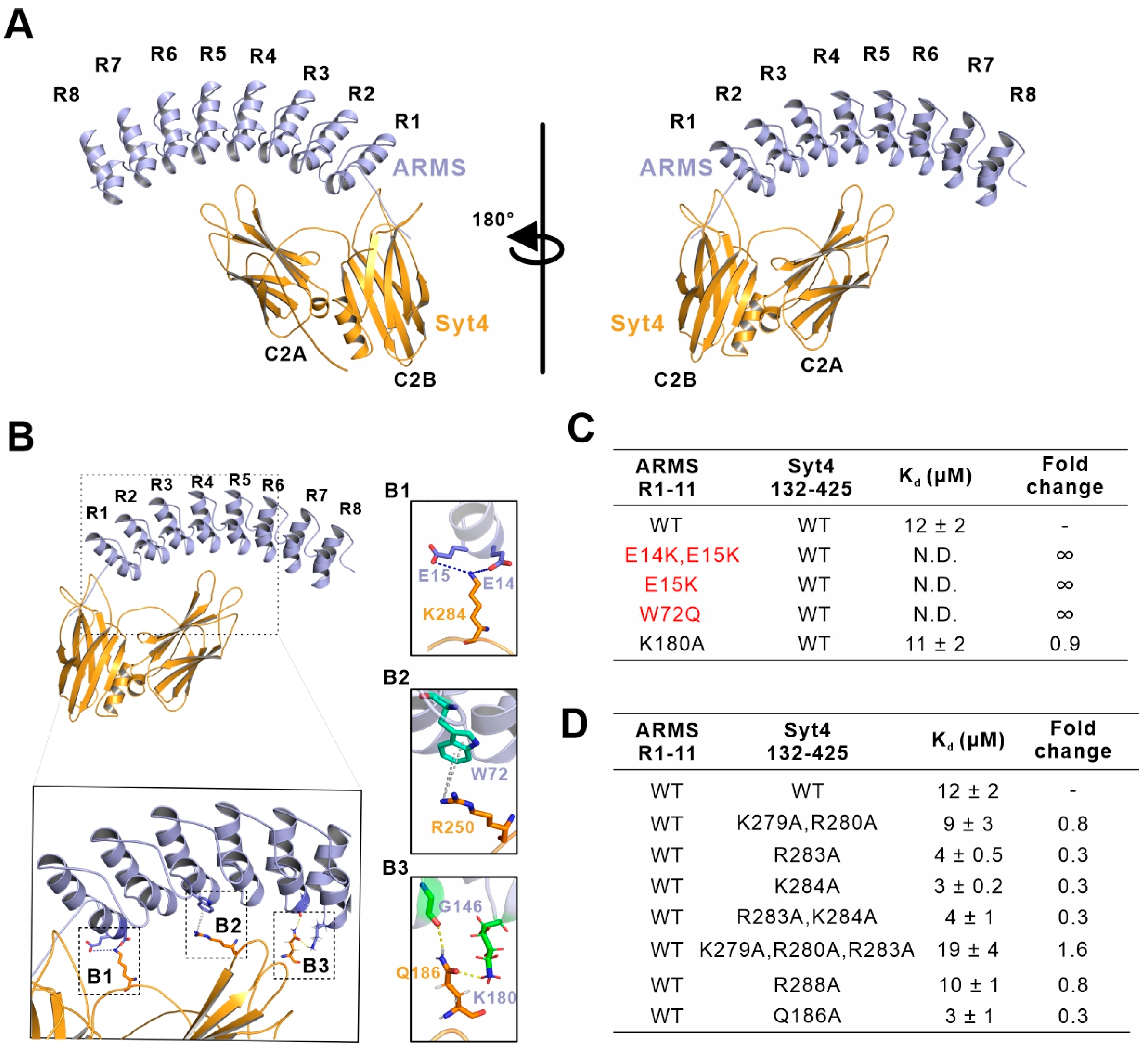
2.4. E15 and W72 of ARMS Are Key Residues Mediating the Assembly of the ARMS/Syt4 Complex
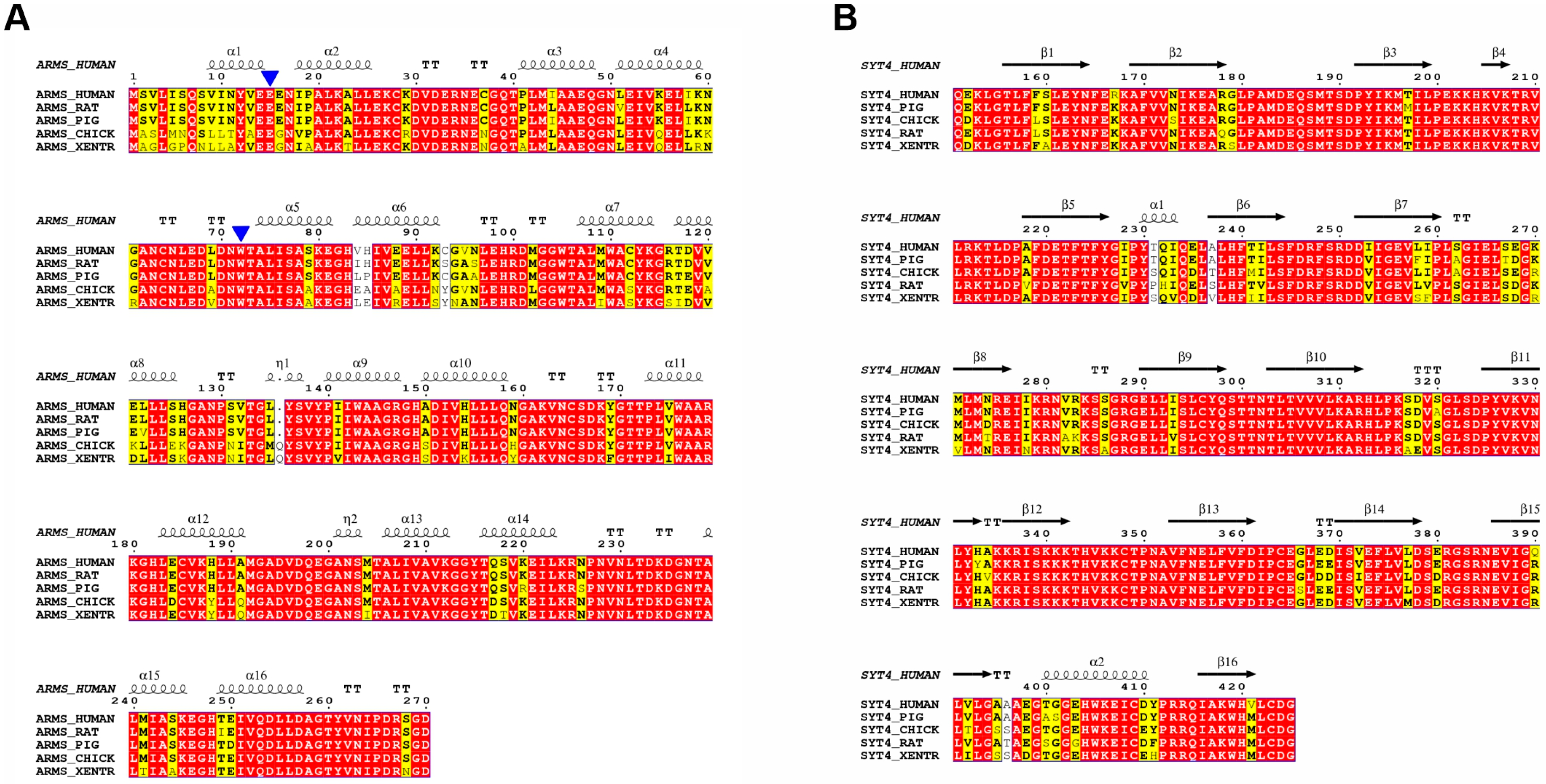
2.5. Multiple Sequence Alignment Reveals Highly Conserved Residues
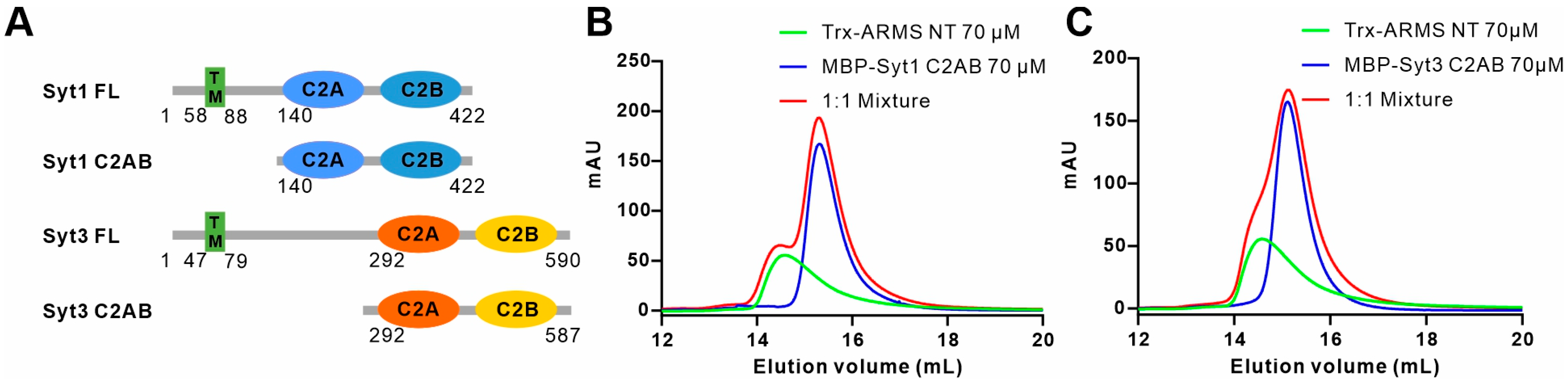
2.6. ARMS Does Not Bind to Syt1 or Syt3
3. Discussion
4. Materials and Methods
4.1. Constructs, Protein Expression, and Purification
4.2. Analytical Gel Filtration Chromatography Assay
4.3. Isothermal Titration Calorimetry Assay
4.4. Structure Prediction
4.5. Sequence Alignment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bracale, A.; Cesca, F.; Neubrand, V.E.; Newsome, T.P.; Way, M.; Schiavo, G. Kidins220/ARMS Is Transported by a Kinesin-1-Based Mechanism Likely to Be Involved in Neuronal Differentiation. Mol. Biol. Cell 2007, 18, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Higuero, A.M.; Sánchez-Ruiloba, L.; Doglio, L.E.; Portillo, F.; Abad-Rodríguez, J.; Dotti, C.G.; Iglesias, T. Kidins220/ARMS Modulates the Activity of Microtubule-Regulating Proteins and Controls Neuronal Polarity and Development. J. Biol. Chem. 2010, 285, 1343–1357. [Google Scholar] [CrossRef] [PubMed]
- Neubrand, V.E.; Thomas, C.; Schmidt, S.; Debant, A.; Schiavo, G. Kidins220/ARMS Regulates Rac1-Dependent Neurite Outgrowth by Direct Interaction with the RhoGEF Trio. J. Cell Sci. 2010, 123, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Scholz-Starke, J.; Cesca, F. Stepping Out of the Shade: Control of Neuronal Activity by the Scaffold Protein Kidins220/ARMS. Front. Cell. Neurosci. 2016, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Cai, J.; Jiang, W.G.; Ye, L. Kidins220 and Tumour Development: Insights into a Complexity of Cross-Talk among Signalling Pathways (Review). Int. J. Mol. Med. 2017, 40, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Fife, C.M.; McCarroll, J.A.; Kavallaris, M. Movers and Shakers: Cell Cytoskeleton in Cancer Metastasis. Br. J. Pharmacol. 2014, 171, 5507–5523. [Google Scholar] [CrossRef] [PubMed]
- López-Menéndez, C.; Gamir-Morralla, A.; Jurado-Arjona, J.; Higuero, A.M.; Campanero, M.R.; Ferrer, I.; Hernández, F.; Ávila, J.; Díaz-Guerra, M.; Iglesias, T. Kidins220 Accumulates with Tau in Human Alzheimer’s Disease and Related Models: Modulation of Its Calpain-Processing by GSK3β/PP1 Imbalance. Hum. Mol. Genet. 2013, 22, 466–482. [Google Scholar] [CrossRef]
- López-Benito, S.; Sánchez-Sánchez, J.; Brito, V.; Calvo, L.; Lisa, S.; Torres-Valle, M.; Palko, M.E.; Vicente-García, C.; Fernández-Fernández, S.; Bolaños, J.P.; et al. Regulation of BDNF Release by ARMS/Kidins220 through Modulation of Synaptotagmin-IV Levels. J. Neurosci. 2018, 38, 5415–5428. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, J.; Vicente-García, C.; Cañada-García, D.; Martín-Zanca, D.; Arévalo, J.C. ARMS/Kidins220 Regulates Nociception by Controlling Brain-Derived Neurotrophic Factor Secretion. Pain 2023, 164, 563–576. [Google Scholar] [CrossRef]
- Neubrand, V.E.; Cesca, F.; Benfenati, F.; Schiavo, G. Kidins220/ARMS as a Functional Mediator of Multiple Receptor Signalling Pathways. J. Cell Sci. 2012, 125, 1845–1854. [Google Scholar] [CrossRef]
- Cesca, F.; Yabe, A.; Spencer-Dene, B.; Scholz-Starke, J.; Medrihan, L.; Maden, C.H.; Gerhardt, H.; Orriss, I.R.; Baldelli, P.; Al-Qatari, M.; et al. Kidins220/ARMS Mediates the Integration of the Neurotrophin and VEGF Pathways in the Vascular and Nervous Systems. Cell Death Differ. 2012, 19, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Mero, I.-L.; Mørk, H.H.; Sheng, Y.; Blomhoff, A.; Opheim, G.L.; Erichsen, A.; Vigeland, M.D.; Selmer, K.K. Homozygous KIDINS220 Loss-of-Function Variants in Fetuses with Cerebral Ventriculomegaly and Limb Contractures. Hum. Mol. Genet. 2017, 26, 3792–3796. [Google Scholar] [CrossRef] [PubMed]
- El-Dessouky, S.H.; Issa, M.Y.; Aboulghar, M.M.; Gaafar, H.M.; Elarab, A.E.; Ateya, M.I.; Omar, H.H.; Beetz, C.; Zaki, M.S. Prenatal Delineation of a Distinct Lethal Fetal Syndrome Caused by a Homozygous Truncating KIDINS220 Variant. Am. J. Med. Genet. A 2020, 182, 2867–2876. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.H.; Arévalo, J.C.; Sarti, F.; Tessarollo, L.; Gan, W.-B.; Chao, M.V. ARMS/Kidins220 Regulates Dendritic Branching and Spine Stability in Vivo. Dev. Neurobiol. 2009, 69, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Colucci-D’Amato, L.; Speranza, L.; Volpicelli, F. Neurotrophic Factor BDNF, Physiological Functions and Therapeutic Potential in Depression, Neurodegeneration and Brain Cancer. IJMS 2020, 21, 7777. [Google Scholar] [CrossRef]
- Park, H.; Poo, M. Neurotrophin Regulation of Neural Circuit Development and Function. Nat. Rev. Neurosci. 2013, 14, 7–23. [Google Scholar] [CrossRef]
- Bartkowska, K.; Paquin, A.; Gauthier, A.S.; Kaplan, D.R.; Miller, F.D. Trk Signaling Regulates Neural Precursor Cell Proliferation and Differentiation during Cortical Development. Development 2007, 134, 4369–4380. [Google Scholar] [CrossRef]
- Snapyan, M.; Lemasson, M.; Brill, M.S.; Blais, M.; Massouh, M.; Ninkovic, J.; Gravel, C.; Berthod, F.; Götz, M.; Barker, P.A.; et al. Vasculature Guides Migrating Neuronal Precursors in the Adult Mammalian Forebrain via Brain-Derived Neurotrophic Factor Signaling. J. Neurosci. 2009, 29, 4172–4188. [Google Scholar] [CrossRef]
- Scharfman, H.; Goodman, J.; Macleod, A.; Phani, S.; Antonelli, C.; Croll, S. Increased Neurogenesis and the Ectopic Granule Cells after Intrahippocampal BDNF Infusion in Adult Rats. Exp. Neurol. 2005, 192, 348–356. [Google Scholar] [CrossRef]
- Lee, J.; Duan, W.; Mattson, M.P. Evidence That Brain-Derived Neurotrophic Factor Is Required for Basal Neurogenesis and Mediates, in Part, the Enhancement of Neurogenesis by Dietary Restriction in the Hippocampus of Adult Mice. J. Neurochem. 2002, 82, 1367–1375. [Google Scholar] [CrossRef]
- Rossi, C.; Angelucci, A.; Costantin, L.; Braschi, C.; Mazzantini, M.; Babbini, F.; Fabbri, M.E.; Tessarollo, L.; Maffei, L.; Berardi, N.; et al. Brain-Derived Neurotrophic Factor (BDNF) Is Required for the Enhancement of Hippocampal Neurogenesis Following Environmental Enrichment. Eur. J. Neurosci. 2006, 24, 1850–1856. [Google Scholar] [CrossRef] [PubMed]
- Hock, C.; Heese, K.; Hulette, C.; Rosenberg, C.; Otten, U. Region-Specific Neurotrophin Imbalances in Alzheimer Disease: Decreased Levels of Brain-Derived Neurotrophic Factor and Increased Levels of Nerve Growth Factor in Hippocampus and Cortical Areas. Arch. Neurol. 2000, 57, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Goutan, E.; Marín, C.; Rey, M.J.; Ribalta, T. Brain-Derived Neurotrophic Factor in Huntington Disease. Brain Res. 2000, 866, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Zuccato, C.; Marullo, M.; Conforti, P.; MacDonald, M.E.; Tartari, M.; Cattaneo, E. Systematic Assessment of BDNF and Its Receptor Levels in Human Cortices Affected by Huntington’s Disease. Brain Pathol. 2008, 18, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Narisawa-Saito, M.; Wakabayashi, K.; Tsuji, S.; Takahashi, H.; Nawa, H. Regional Specificity of Alterations in NGF, BDNF and NT-3 Levels in Alzheimer’s Disease. NeuroReport 1996, 7, 2925. [Google Scholar] [CrossRef]
- Dieni, S.; Matsumoto, T.; Dekkers, M.; Rauskolb, S.; Ionescu, M.S.; Deogracias, R.; Gundelfinger, E.D.; Kojima, M.; Nestel, S.; Frotscher, M.; et al. BDNF and Its Pro-Peptide Are Stored in Presynaptic Dense Core Vesicles in Brain Neurons. J. Cell Biol. 2012, 196, 775–788. [Google Scholar] [CrossRef]
- Berg, E.A.; Johnson, R.J.; Leeman, S.E.; Boyd, N.; Kimerer, L.; Fine, R.E. Isolation and Characterization of Substance P-Containing Dense Core Vesicles from Rabbit Optic Nerve and Termini. J. Neurosci. Res. 2000, 62, 830–839. [Google Scholar] [CrossRef]
- Canossa, M.; Gärtner, A.; Campana, G.; Inagaki, N.; Thoenen, H. Regulated Secretion of Neurotrophins by Metabotropic Glutamate Group I (mGluRI) and Trk Receptor Activation Is Mediated via Phospholipase C Signalling Pathways. EMBO J 2001, 20, 1640–1650. [Google Scholar] [CrossRef]
- Goodman, L.J.; Valverde, J.; Lim, F.; Geschwind, M.D.; Federoff, H.J.; Geller, A.I.; Hefti, F. Regulated Release and Polarized Localization of Brain-Derived Neurotrophic Factor in Hippocampal Neurons. Mol. Cell Neurosci. 1996, 7, 222–238. [Google Scholar] [CrossRef]
- Griesbeck, O.; Canossa, M.; Campana, G.; Gärtner, A.; Hoener, M.C.; Nawa, H.; Kolbeck, R.; Thoenen, H. Are There Differences between the Secretion Characteristics of NGF and BDNF? Implications for the Modulatory Role of Neurotrophins in Activity-Dependent Neuronal Plasticity. Microsc Res Tech 1999, 45, 262–275. [Google Scholar] [CrossRef]
- Matsuda, N.; Lu, H.; Fukata, Y.; Noritake, J.; Gao, H.; Mukherjee, S.; Nemoto, T.; Fukata, M.; Poo, M. Differential Activity-Dependent Secretion of Brain-Derived Neurotrophic Factor from Axon and Dendrite. J. Neurosci. 2009, 29, 14185–14198. [Google Scholar] [CrossRef] [PubMed]
- Arévalo, J.C.; Deogracias, R. Mechanisms Controlling the Expression and Secretion of BDNF. Biomolecules 2023, 13, 789. [Google Scholar] [CrossRef] [PubMed]
- Canossa, M.; Griesbeck, O.; Berninger, B.; Campana, G.; Kolbeck, R.; Thoenen, H. Neurotrophin Release by Neurotrophins: Implications for Activity-Dependent Neuronal Plasticity. Proc. Natl. Acad. Sci. USA 1997, 94, 13279–13286. [Google Scholar] [CrossRef]
- Krüttgen, A.; Möller, J.C.; Heymach, J.V.; Shooter, E.M. Neurotrophins Induce Release of Neurotrophins by the Regulated Secretory Pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 9614–9619. [Google Scholar] [CrossRef] [PubMed]
- Leßmann, V. Neurotrophin-Dependent Modulation of Glutamatergic Synaptic Transmission in the Mammalian CNS. Gen. Pharmacol. Vasc. Syst. 1998, 31, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Kuczewski, N.; Porcher, C.; Ferrand, N.; Fiorentino, H.; Pellegrino, C.; Kolarow, R.; Lessmann, V.; Medina, I.; Gaiarsa, J.-L. Backpropagating Action Potentials Trigger Dendritic Release of BDNF during Spontaneous Network Activity. J. Neurosci. 2008, 28, 7013–7023. [Google Scholar] [CrossRef]
- Edelmann, E.; Leßmann, V.; Brigadski, T. Pre- and Postsynaptic Twists in BDNF Secretion and Action in Synaptic Plasticity. Neuropharmacol. 2014, 76, 610–627. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Heumann, R.; Lessmann, V. Synaptic Secretion of BDNF after High-Frequency Stimulation of Glutamatergic Synapses. EMBO J 2001, 20, 5887–5897. [Google Scholar] [CrossRef]
- Brigadski, T.; Hartmann, M.; Lessmann, V. Differential Vesicular Targeting and Time Course of Synaptic Secretion of the Mammalian Neurotrophins. J. Neurosci. 2005, 25, 7601–7614. [Google Scholar] [CrossRef]
- Kojima, M.; Takei, N.; Numakawa, T.; Ishikawa, Y.; Suzuki, S.; Matsumoto, T.; Katoh-Semba, R.; Nawa, H.; Hatanaka, H. Biological Characterization and Optical Imaging of Brain-Derived Neurotrophic Factor-Green Fluorescent Protein Suggest an Activity-Dependent Local Release of Brain-Derived Neurotrophic Factor in Neurites of Cultured Hippocampal Neurons. J. Neurosci. Res. 2001, 64, 1–10. [Google Scholar] [CrossRef]
- Kolarow, R.; Brigadski, T.; Lessmann, V. Postsynaptic Secretion of BDNF and NT-3 from Hippocampal Neurons Depends on Calcium–Calmodulin Kinase II Signaling and Proceeds via Delayed Fusion Pore Opening. J. Neurosci. 2007, 27, 10350–10364. [Google Scholar] [CrossRef] [PubMed]
- Lochner, J.E.; Spangler, E.; Chavarha, M.; Jacobs, C.; McAllister, K.; Schuttner, L.C.; Scalettar, B.A. Efficient Copackaging and Cotransport Yields Postsynaptic Colocalization of Neuromodulators Associated with Synaptic Plasticity. Dev. Neurobiol. 2008, 68, 1243–1256. [Google Scholar] [CrossRef] [PubMed]
- Rind, H.B.; Butowt, R.; Bartheld, C.S. von Synaptic Targeting of Retrogradely Transported Trophic Factors in Motoneurons: Comparison of Glial Cell Line-Derived Neurotrophic Factor, Brain-Derived Neurotrophic Factor, and Cardiotrophin-1 with Tetanus Toxin. J. Neurosci. 2005, 25, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Dean, C.; Liu, H.; Mark Dunning, F.; Chang, P.Y.; Jackson, M.B.; Chapman, E.R. Synaptotagmin-IV Modulates Synaptic Function and Long-Term Potentiation by Regulating BDNF Release. Nat Neurosci 2009, 12, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Haubensak, W.; Narz, F.; Heumann, R.; Leβmann, V. BDNF-GFP Containing Secretory Granules Are Localized in the Vicinity of Synaptic Junctions of Cultured Cortical Neurons. J. Cell Sci. 1998, 111, 1483–1493. [Google Scholar] [CrossRef] [PubMed]
- Kohara, K.; Kitamura, A.; Morishima, M.; Tsumoto, T. Activity-Dependent Transfer of Brain-Derived Neurotrophic Factor to Postsynaptic Neurons. Science 2001, 291, 2419–2423. [Google Scholar] [CrossRef]
- Sadakata, T.; Shinoda, Y.; Oka, M.; Sekine, Y.; Sato, Y.; Saruta, C.; Miwa, H.; Tanaka, M.; Itohara, S.; Furuichi, T. Reduced Axonal Localization of a Caps2 Splice Variant Impairs Axonal Release of BDNF and Causes Autistic-like Behavior in Mice. Proc. Natl. Acad. Sci. USA 2012, 109, 21104–21109. [Google Scholar] [CrossRef]
- Scalettar, B.A.; Jacobs, C.; Fulwiler, A.; Prahl, L.; Simon, A.; Hilken, L.; Lochner, J.E. Hindered Submicron Mobility and Long-Term Storage of Presynaptic Dense-Core Granules Revealed by Single-Particle Tracking. Dev. Neurobiol. 2012, 72, 1181–1195. [Google Scholar] [CrossRef]
- Shinoda, Y.; Sadakata, T.; Nakao, K.; Katoh-Semba, R.; Kinameri, E.; Furuya, A.; Yanagawa, Y.; Hirase, H.; Furuichi, T. Calcium-Dependent Activator Protein for Secretion 2 (CAPS2) Promotes BDNF Secretion and Is Critical for the Development of GABAergic Interneuron Network. Proc. Natl. Acad. Sci. USA 2011, 108, 373–378. [Google Scholar] [CrossRef]
- Wolfes, A.C.; Dean, C. The Diversity of Synaptotagmin Isoforms. Curr. Opin. Neurobiol. 2020, 63, 198–209. [Google Scholar] [CrossRef]
- Ahras, M.; Otto, G.P.; Tooze, S.A. Synaptotagmin IV Is Necessary for the Maturation of Secretory Granules in PC12 Cells. J. Cell Biol. 2006, 173, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Shin, O.-H.; Machius, M.; Tomchick, D.R.; Südhof, T.C.; Rizo, J. Structural Basis for the Evolutionary Inactivation of Ca2+ Binding to Synaptotagmin 4. Nat. Struct. Mol. Biol. 2004, 11, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.M.; Ferguson, G.D.; Herschman, H.R.; Elferink, L.A. Functional and Biochemical Analysis of the C2 Domains of Synaptotagmin IV. MBoC 1999, 10, 2285–2295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chapman, E.R. Rat and Drosophila Synaptotagmin 4 Have Opposite Effects during SNARE-Catalyzed Membrane Fusion. J. Biol. Chem. 2010, 285, 30759–30766. [Google Scholar] [CrossRef]
- Bharat, V.; Siebrecht, M.; Burk, K.; Ahmed, S.; Reissner, C.; Kohansal-Nodehi, M.; Steubler, V.; Zweckstetter, M.; Ting, J.T.; Dean, C. Capture of Dense Core Vesicles at Synapses by JNK-Dependent Phosphorylation of Synaptotagmin-4. Cell Rep. 2017, 21, 2118–2133. [Google Scholar] [CrossRef] [PubMed]
- McVicker, D.P.; Awe, A.M.; Richters, K.E.; Wilson, R.L.; Cowdrey, D.A.; Hu, X.; Chapman, E.R.; Dent, E.W. Transport of a Kinesin-Cargo Pair along Microtubules into Dendritic Spines Undergoing Synaptic Plasticity. Nat. Commun. 2016, 7, 12741. [Google Scholar] [CrossRef]
- Weingarten, D.J.; Shrestha, A.; Juda-Nelson, K.; Kissiwaa, S.A.; Spruston, E.; Jackman, S.L. Fast Resupply of Synaptic Vesicles Requires Synaptotagmin-3. Nature 2022, 611, 320–325. [Google Scholar] [CrossRef]
- Van Westen, R.; Poppinga, J.; Díez Arazola, R.; Toonen, R.F.; Verhage, M. Neuromodulator Release in Neurons Requires Two Functionally Redundant Calcium Sensors. Proc. Natl. Acad. Sci. USA 2021, 118, e2012137118. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhou, P.; Wang, A.L.; Wu, D.; Zhao, M.; Südhof, T.C.; Brunger, A.T. The Primed SNARE–Complexin–Synaptotagmin Complex for Neuronal Exocytosis. Nature 2017, 548, 420–425. [Google Scholar] [CrossRef]
- Awasthi, A.; Ramachandran, B.; Ahmed, S.; Benito, E.; Shinoda, Y.; Nitzan, N.; Heukamp, A.; Rannio, S.; Martens, H.; Barth, J.; et al. Synaptotagmin-3 Drives AMPA Receptor Endocytosis, Depression of Synapse Strength, and Forgetting. Science 2019, 363, eaav1483. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Chen, J.; Li, Y.; Ye, J.; Wang, C. Neuronal Scaffold Protein ARMS Interacts with Synaptotagmin-4 C2AB through the Ankyrin Repeat Domain with an Unexpected Mode. Int. J. Mol. Sci. 2023, 24, 16993. https://doi.org/10.3390/ijms242316993
Zhang F, Chen J, Li Y, Ye J, Wang C. Neuronal Scaffold Protein ARMS Interacts with Synaptotagmin-4 C2AB through the Ankyrin Repeat Domain with an Unexpected Mode. International Journal of Molecular Sciences. 2023; 24(23):16993. https://doi.org/10.3390/ijms242316993
Chicago/Turabian StyleZhang, Fa, Jiasheng Chen, Yahong Li, Jin Ye, and Chao Wang. 2023. "Neuronal Scaffold Protein ARMS Interacts with Synaptotagmin-4 C2AB through the Ankyrin Repeat Domain with an Unexpected Mode" International Journal of Molecular Sciences 24, no. 23: 16993. https://doi.org/10.3390/ijms242316993




