Genome-Wide Analysis of the DC1 Domain Protein Gene Family in Tomatoes under Abiotic Stress
Abstract
:1. Introduction
2. Results
2.1. Identification of DC1 Domain Family Members in Tomatoes
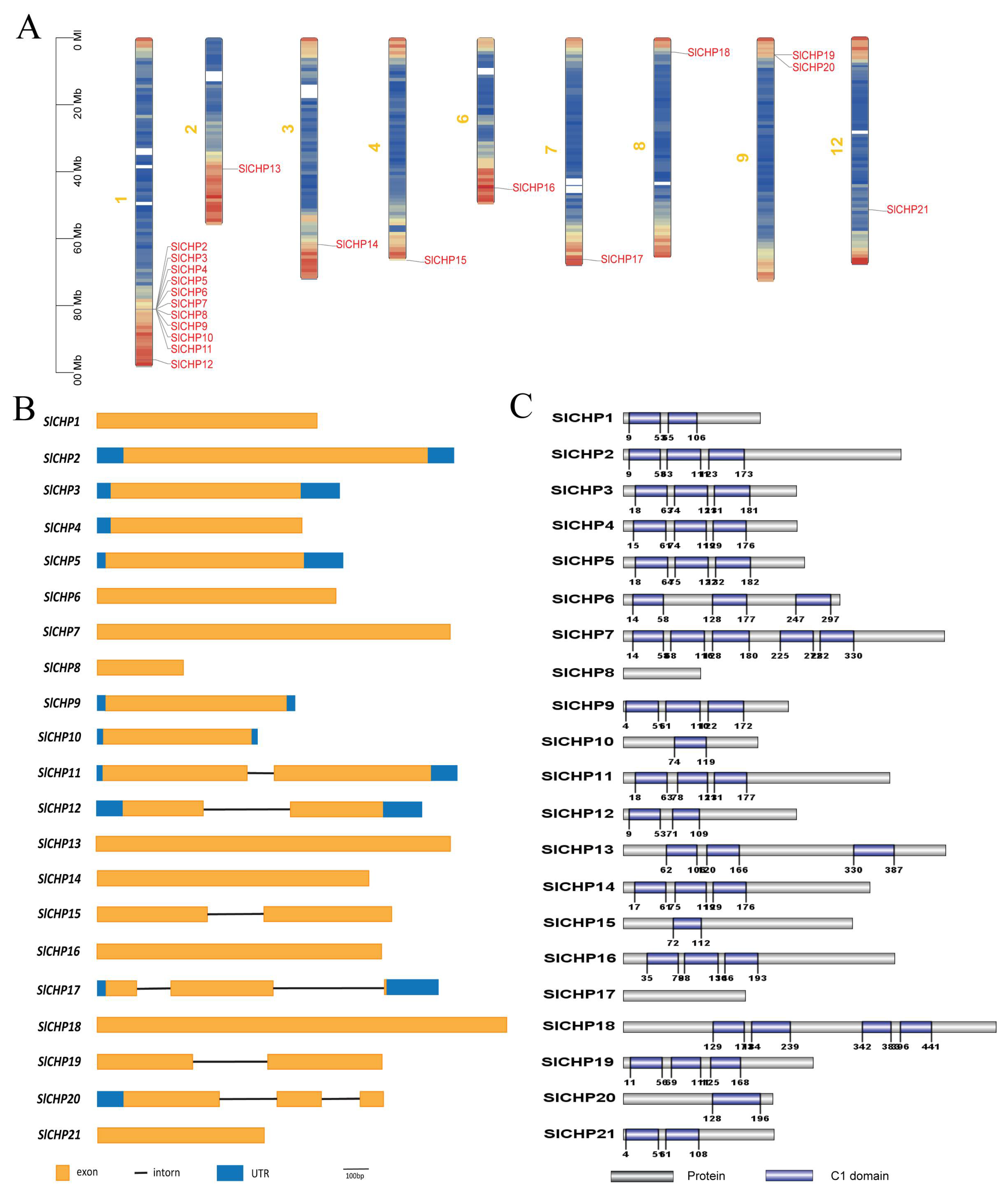
2.2. Chromosomal Distribution, Gene Structure, and Protein Conservation Domain Analysis of SlCHP Genes
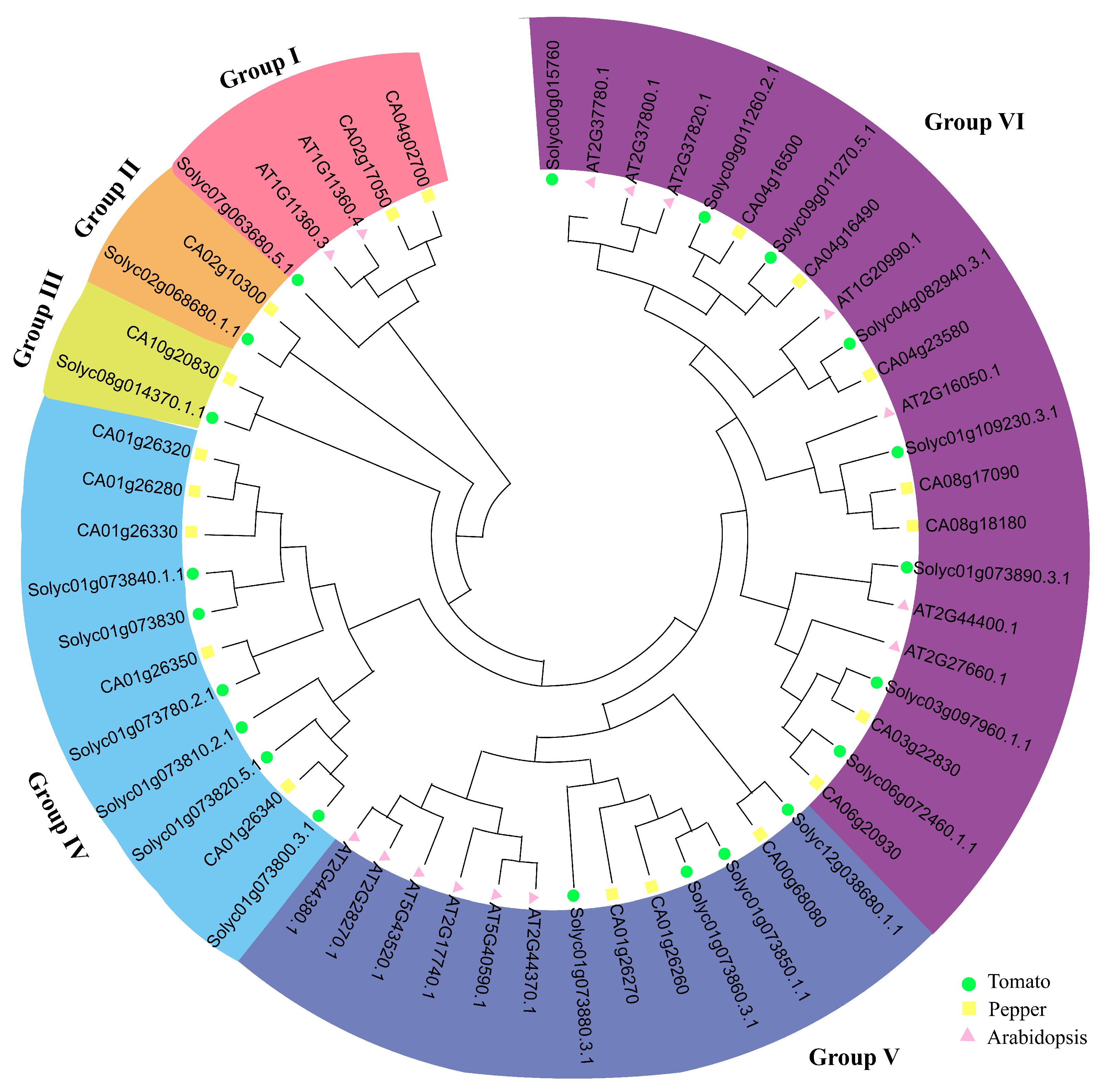
2.3. Phylogenetic Analysis of the SlCHP Genes in Tomatoes
2.4. Analysis of Cis-Elements in the Promoter of the SlCHP Gene Family
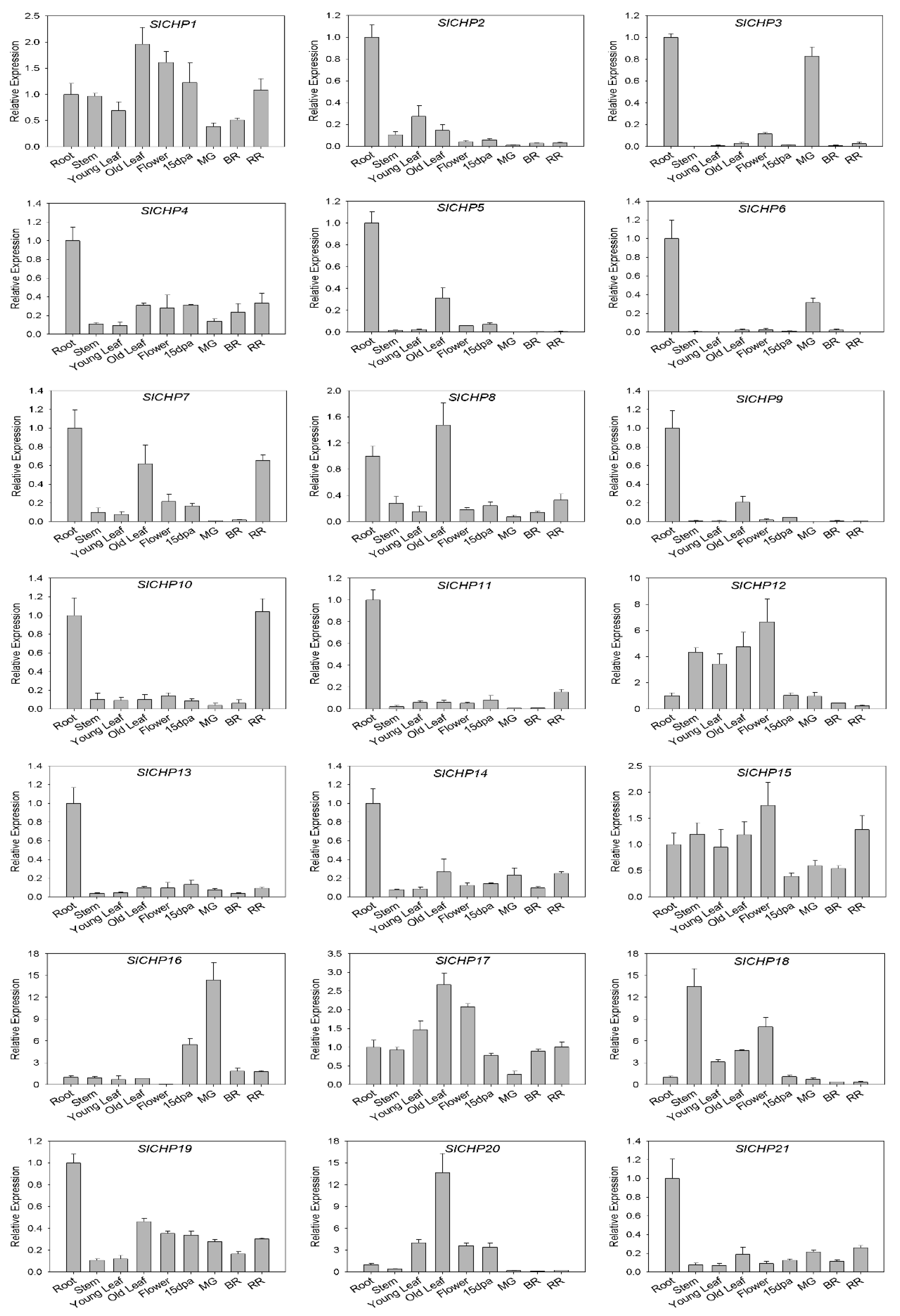
2.5. Analysis of SlCHP Family Genes Tissue Expression
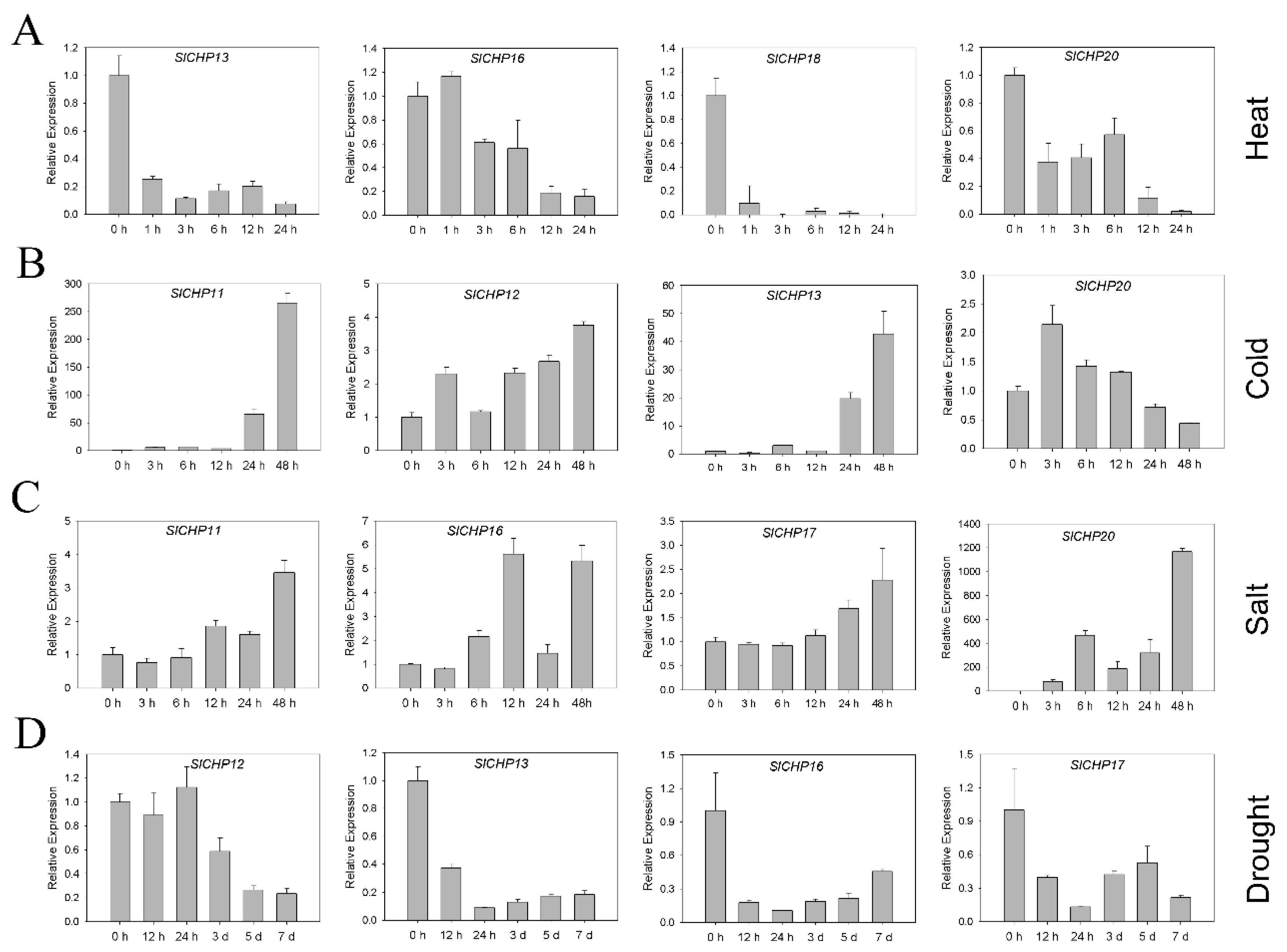
2.6. Expression Analysis of SlCHP Genes under Several Abiotic Stresses
2.7. Subcellular Localization of the SlCHP Proteins
2.8. Prediction Interaction Protein Prediction of SlCHP Proteins
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval and Identification of Tomato DC1 Domain Family Members
4.2. Physical and Chemical Properties Analysis of Tomato DC1 Domain Protein
4.3. Chromosomal Location, Gene Structure, and Conservative Domain Analysis
4.4. Phylogenetic Analysis
4.5. Promoter Analysis
4.6. Protein Interaction Network Prediction
4.7. Plant Material and Growth Condition
4.8. RNA Extraction and Real-Time Quantitative PCR Assay
4.9. Subcellular Localization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Willis, C.G.; Ruhfel, B.; Primack, R.B.; Miller-Rushing, A.J.; Davis, C.C. Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. Proc. Natl. Acad. Sci. USA 2008, 105, 17029–17033. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Peng, T.; Qu, F.; Wang, J.; Long, Y.; Hu, X. Bacillus methylotrophicus Could Improve the Tolerance and Recovery Ability of the Tomato to Low-Temperature Stress and Improve Fruit Quality. Agronomy 2023, 13, 1902. [Google Scholar] [CrossRef]
- Cao, Y.; Song, H.; Zhang, L. New Insight into Plant Saline-Alkali Tolerance Mechanisms and Application to Breeding. Int. J. Mol. Sci. 2022, 23, 16048. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Ni, L.; Xia, X.; Chen, S.; Zhang, Y.; Lang, M.; Li, M.; Liu, B.; Pan, Y.; Li, J.; et al. Genome-Wide Analysis of the Protein Phosphatase 2C Genes in Tomato. Genes 2022, 13, 604. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gao, K.; Ren, H.; Tang, W. Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, 757–779. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Bhaskar, R.V.; Mohanty, B.; Verma, V.; Wijaya, E.; Kumar, P.P. A hormone-responsive C1-domain-containing protein At5g17960 mediates stress response in Arabidopsis thaliana. PLoS ONE 2015, 10, e0115418. [Google Scholar] [CrossRef]
- Brose, N.; Betz, A.; Wegmeyer, H. Divergent and convergent signaling by the diacylglycerol second messenger pathway in mammals. Curr. Opin. Neurobiol. 2004, 14, 328–340. [Google Scholar] [CrossRef]
- Gomez-Fernandez, J.C.; Torrecillas, A.; Corbalan-Garcia, S. Diacylglycerols as activators of protein kinase C. Mol. Membr. Biol. 2004, 21, 339–349. [Google Scholar] [CrossRef]
- D’Ippolito, S.; Arias, L.A.; Casalongue, C.A.; Pagnussat, G.C.; Fiol, D.F. The DC1-domain protein VACUOLELESS GAMETOPHYTES is essential for development of female and male gametophytes in Arabidopsis. Plant J. 2017, 90, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Colon-Gonzalez, F.; Kazanietz, M.G. C1 domains exposed: From diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta 2006, 1761, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Quest, A.F.G.; Bloomenthal, J.; Bardes, E.S.G.; Bell, R.M. The regulatory domain of protein kinase C coordinates four atoms of zinc. J. Biol. Chem. 1992, 267, 10193–10197. [Google Scholar] [CrossRef] [PubMed]
- Buchner, K. Protein Kinase C in the Transduction of Signals Toward and within the Cell Nucleus. Eur. J. Biochem. 1995, 228, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Fujii, T.; Igarashi, K.; Kuno, T.; Tanaka, C.; Kikkawa, U.; Nishizuka, Y. Phorbol ester binding to protein kinase C requires a cysteine-rich__zinc-finger-like sequence. Proc. Natl. Acad. Sci. USA 1989, 86, 4868–4871. [Google Scholar] [CrossRef]
- Djafi, N.; Vergnolle, C.; Cantrel, C.; Wietrzynski, W.; Delage, E.; Cochet, F.; Puyaubert, J.; Soubigou-Taconnat, L.; Gey, D.; Collin, S.; et al. The Arabidopsis DREB2 genetic pathway is constitutively repressed by basal phosphoinositide-dependent phospholipase C coupled to diacylglycerol kinase. Front. Plant Sci. 2013, 4, 307. [Google Scholar] [CrossRef]
- Johnson, J.E.; Goulding, R.E.; Ding, Z.; Partovi, A.; Anthony, K.V.; Beaulieu, N.; Tazmini, G.; Cornell, R.B.; Kay, R.J. Differential membrane binding and diacylglycerol recognition by C1 domains of RasGRPs. Biochem. J. 2007, 406, 223–236. [Google Scholar] [CrossRef]
- Canagarajah, B.; Leskow, F.C.; Ho, J.Y.; Mischak, H.; Saidi, L.F.; Kazanietz, M.G.; Hurley, J.H. Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin. Cell 2004, 119, 407–418. [Google Scholar] [CrossRef]
- Suesslin, C.; Frohnmeyer, H. An Arabidopsis mutant defective in UV-B light-mediated responses. Plant J. 2003, 33, 591–601. [Google Scholar] [CrossRef]
- Brownfield, L. Pollen Helps Reveal a Role for DC1 Domain Proteins. Plant Cell Physiol. 2022, 63, 1761–1763. [Google Scholar] [CrossRef]
- Shinya, T.; Galis, I.; Narisawa, T.; Sasaki, M.; Fukuda, H.; Matsuoka, H.; Saito, M.; Matsuoka, K. Comprehensive analysis of glucan elicitor-regulated gene expression in tobacco BY-2 cells reveals a novel MYB transcription factor involved in the regulation of phenylpropanoid metabolism. Plant Cell Physiol. 2007, 48, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.S.; Choi, D.S.; Kim, N.H.; Kim, D.S.; Hwang, B.K. The pepper cysteine/histidine-rich DC1 domain protein CaDC1 binds both RNA and DNA and is required for plant cell death and defense response. New Phytol. 2014, 201, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lv, J.; Zhao, X.; Ai, X.; Zhu, X.; Wang, M.; Zhao, S.; Xia, G. TaCHP: A wheat zinc finger protein gene down-regulated by abscisic acid and salinity stress plays a positive role in stress tolerance. Plant Physiol. 2010, 154, 211–221. [Google Scholar] [CrossRef]
- Tanaka, N.; Itoh, J.; Nagato, Y. Role of rice PPS in late vegetative and reproductive growth. Plant Signal Behav. 2012, 7, 50–52. [Google Scholar] [CrossRef]
- Gao, S.; Yang, L.; Zeng, H.Q.; Zhou, Z.S.; Yang, Z.M.; Li, H.; Sun, D.; Xie, F.; Zhang, B. A cotton miRNA is involved in regulation of plant response to salt stress. Sci. Rep. 2016, 6, 19736. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 1994, 256, 119–124. [Google Scholar]
- Pich i Roselló, O.; Kondrashov, F.A. Long-Term Asymmetrical Acceleration of Protein Evolution after Gene Duplication. Genome Biol. Evol. 2014, 6, 1949–1955. [Google Scholar] [CrossRef]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef]
- Wang, R.; Ming, M.; Li, J.; Shi, D.; Qiao, X.; Li, L.; Zhang, S.; Wu, J. Genome-wide identification of theMADS-boxtranscription factor family in pear (Pyrus bretschneideri) reveals evolution and functional divergence. PeerJ 2017, 5, e3776. [Google Scholar] [CrossRef]
- Schaper, E.; Anisimova, M. The evolution and function of protein tandem repeats in plants. New Phytol. 2014, 206, 397–410. [Google Scholar] [CrossRef]
- Liu, M.; Gomes, B.L.; Mila, I.; Purgatto, E.; Peres, L.E.; Frasse, P.; Maza, E.; Zouine, M.; Roustan, J.P.; Bouzayen, M.; et al. Comprehensive Profiling of Ethylene Response Factor Expression Identifies Ripening-Associated ERF Genes and Their Link to Key Regulators of Fruit Ripening in Tomato. Plant Physiol. 2016, 170, 1732–1744. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- McCormack, M.L.; Dickie, I.A.; Eissenstat, D.M.; Fahey, T.J.; Fernandez, C.W.; Guo, D.; Helmisaari, H.S.; Hobbie, E.A.; Iversen, C.M.; Jackson, R.B.; et al. Redefining fine roots improves understanding of below-ground contributions to terrestrial biosphere processes. New Phytol. 2015, 207, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.; Testerink, C. Roots Withstanding their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef] [PubMed]
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Ma, W.; Liu, X.; Chen, K.; Yu, X.; Ji, D. Genome-Wide Re-Identification and Analysis of CrRLK1Ls in Tomato. Int. J. Mol. Sci. 2023, 24, 3142. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in sillco analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.-N.; Lee, T.-Y.; Hung, Y.-C.; Li, G.-Z.; Tseng, K.-C.; Liu, Y.-H.; Kuo, P.-L.; Zheng, H.-Q.; Chang, W.-C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.T.; Franz, M.; Kazi, F.; Donaldson, S.L.; Morris, Q.; Bader, G.D. Cytoscape Web: An interactive web-based network browser. Bioinformatics 2010, 26, 2347–2348. [Google Scholar] [CrossRef]
- Ai, G.; Zhang, D.; Huang, R.; Zhang, S.; Li, W.; Ahiakpa, J.K.; Zhang, J. Genome-Wide Identification and Molecular Characterization of the Growth-Regulating Factors-Interacting Factor Gene Family in Tomato. Genes 2020, 11, 1435. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Vandesompele, J.; Shipley, G.L.; Pfaffl, M.W.; Nolan, T.; Mueller, R.; Kubista, M.; Huggett, J.; Hellemans, J.; Garson, J.A.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Li, G.; Wang, J.; Zhang, C.; Ai, G.; Zhang, D.; Wei, J.; Cai, L.; Li, C.; Zhu, W.; Larkin, R.M.; et al. L2, a chloroplast metalloproteinase, regulates fruit ripening by participating in ethylene autocatalysis under the control of ethylene response factors. J. Exp. Bot. 2021, 72, 7035–7048. [Google Scholar] [CrossRef]



| No. | Gene Name | Gene ID | Chromosome Location | cDNA | AA | pI | MW (kDa) | Locali-Zation |
|---|---|---|---|---|---|---|---|---|
| 1 | SlCHP1 | Solyc00g015760.1.1 | Ch00: 11237737-11236877 (−) | 861 | 286 | 6.89 | 31.521 | Nucl |
| 2 | SlCHP2 | Solyc01g073780.2.1 | Ch01: 81154238-81152883 (−) | 1194 | 397 | 5.69 | 45.173 | Nucl |
| 3 | SlCHP3 | Solyc01g073800.3.1 | Ch01: 81158393-81157477 (−) | 747 | 248 | 6.58 | 27.623 | Nucl |
| 4 | SlCHP4 | Solyc01g073810.2.1 | Ch01: 81170807-81170011 (−) | 750 | 249 | 5.96 | 27.624 | Nucl |
| 5 | SlCHP5 | Solyc01g073820.5.1 | Ch01: 81173167-81172221 (−) | 780 | 259 | 6.65 | 29.144 | Nucl |
| 6 | SlCHP6 | Solyc01g073830.1 | Ch01: 81180355-81179423 (−) | 933 | 310 | 4.82 | 35.355 | Pero |
| 7 | SlCHP7 | Solyc01g073840.1.1 | Ch01: 81183692-81182313 (−) | 1380 | 459 | 5.69 | 52.643 | Nucl |
| 8 | SlCHP8 | Solyc01g073850.1.1 | Ch01: 81185584-81185249 (−) | 336 | 111 | 5.55 | 12.736 | Chlo |
| 9 | SlCHP9 | Solyc01g073860.3.1 | Ch01: 81194192-81193425 (−) | 711 | 236 | 5.51 | 27.207 | Extr |
| 10 | SlCHP10 | Solyc01g073880.3.1 | Ch01: 81211961-81211353 (−) | 582 | 193 | 5.32 | 21.907 | Cyto |
| 11 | SlCHP11 | Solyc01g073890.3.1 | Ch01: 81215509-81217001 (+) | 1146 | 381 | 7.07 | 42.222 | Nucl |
| 12 | SlCHP12 | Solyc01g109230.3.1 | Ch01: 96240517-96242373 (+) | 747 | 248 | 9.03 | 28.467 | Nucl or Cyto |
| 13 | SlCHP13 | Solyc02g068680.1.1 | Ch02: 38624575-38625960 (+) | 1386 | 461 | 7.31 | 53.235 | Nucl |
| 14 | SlCHP14 | Solyc03g097960.1.1 | Ch03: 60337788-60336727 (−) | 1062 | 353 | 7.88 | 38.769 | Chlo |
| 15 | SlCHP15 | Solyc04g082940.3.1 | Ch04: 66412733-66411463 (−) | 930 | 328 | 9.38 | 35.681 | Nucl |
| 16 | SlCHP16 | Solyc06g072460.1.1 | Ch06:44695610-44696722 (+) | 1167 | 370 | 8.29 | 40.426 | Nucl |
| 17 | SlCHP17 | Solyc07g063680.5.1 | Ch07: 66082531-66086025 (+) | 528 | 175 | 4.67 | 19.024 | Chlo |
| 18 | SlCHP18 | Solyc08g014370.1.1 | Ch08: 4293649-4295250 (+) | 1602 | 533 | 7.26 | 61.624 | Chlo |
| 19 | SlCHP19 | Solyc09g011260.2.1 | Ch09: 4592852-4594526 (+) | 819 | 272 | 8.27 | 31.079 | Cyto |
| 20 | SlCHP20 | Solyc09g011270.4.1 | Ch09: 4603003-4605126 (+) | 645 | 214 | 8.35 | 24.910 | Cyto |
| 21 | SlCHP21 | Solyc12g038680.1.1 | Ch12: 47165974-47165324 (−) | 651 | 216 | 8.80 | 24.740 | Nucl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Dang, J.; Pan, J.; Liu, J.; Peng, T.; Chen, G.; Wang, R.; Hu, S.; Li, X.; Hu, X. Genome-Wide Analysis of the DC1 Domain Protein Gene Family in Tomatoes under Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 16994. https://doi.org/10.3390/ijms242316994
Li G, Dang J, Pan J, Liu J, Peng T, Chen G, Wang R, Hu S, Li X, Hu X. Genome-Wide Analysis of the DC1 Domain Protein Gene Family in Tomatoes under Abiotic Stress. International Journal of Molecular Sciences. 2023; 24(23):16994. https://doi.org/10.3390/ijms242316994
Chicago/Turabian StyleLi, Guobin, Jiao Dang, Jiaqi Pan, Jingyi Liu, Tieli Peng, Guo Chen, Rongqun Wang, Songshen Hu, Xiaojing Li, and Xiaohui Hu. 2023. "Genome-Wide Analysis of the DC1 Domain Protein Gene Family in Tomatoes under Abiotic Stress" International Journal of Molecular Sciences 24, no. 23: 16994. https://doi.org/10.3390/ijms242316994
APA StyleLi, G., Dang, J., Pan, J., Liu, J., Peng, T., Chen, G., Wang, R., Hu, S., Li, X., & Hu, X. (2023). Genome-Wide Analysis of the DC1 Domain Protein Gene Family in Tomatoes under Abiotic Stress. International Journal of Molecular Sciences, 24(23), 16994. https://doi.org/10.3390/ijms242316994







