Anti-Biofilm Activity of Cocultimycin A against Candida albicans
Abstract
1. Introduction
2. Results
2.1. Cocultimycin A Inhibited Biofilm Formation and Disrupted Mature Biofilms of C. albicans
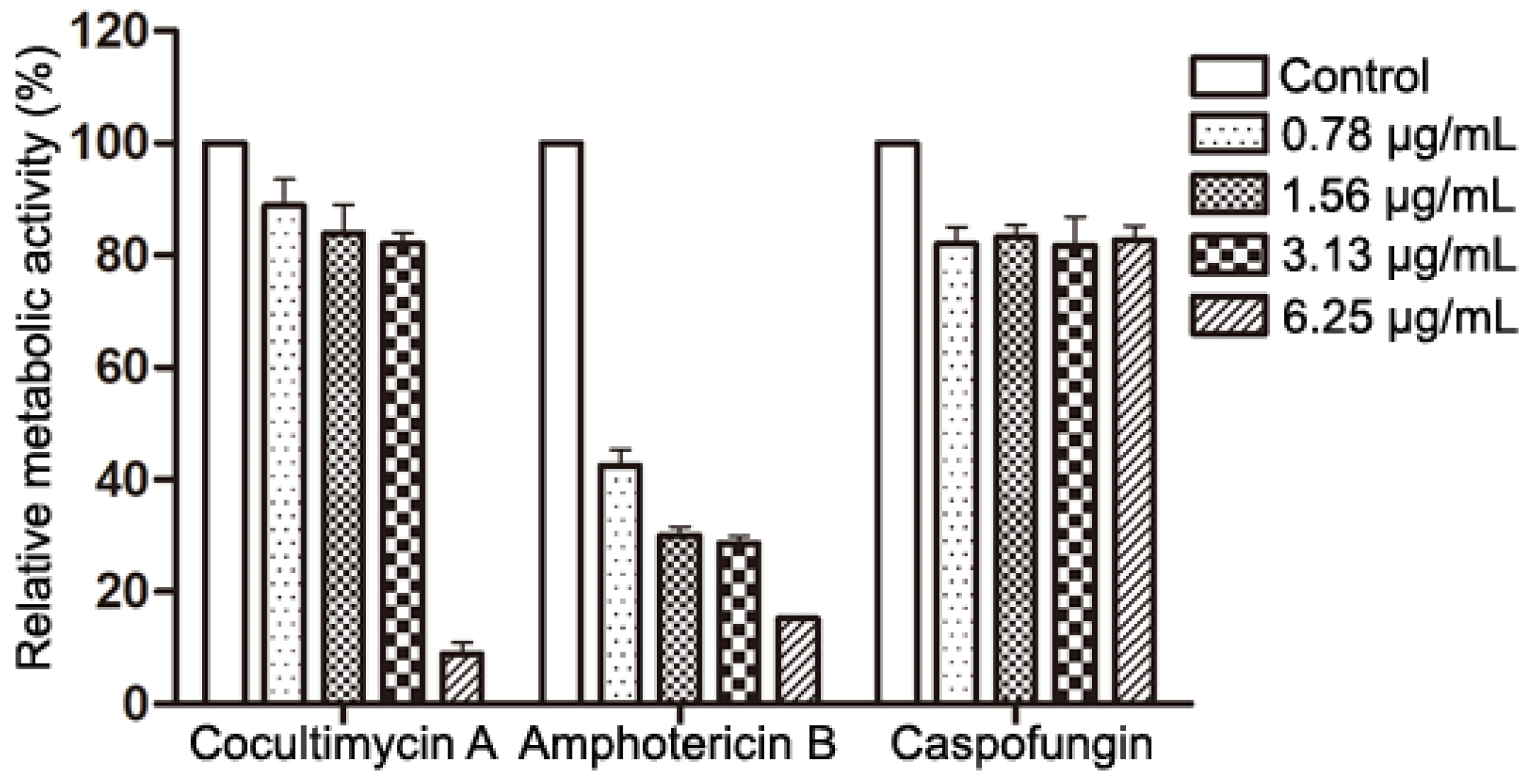
2.2. Effects of Cocultimycin A on Metabolic Activity of Mature Biofilm
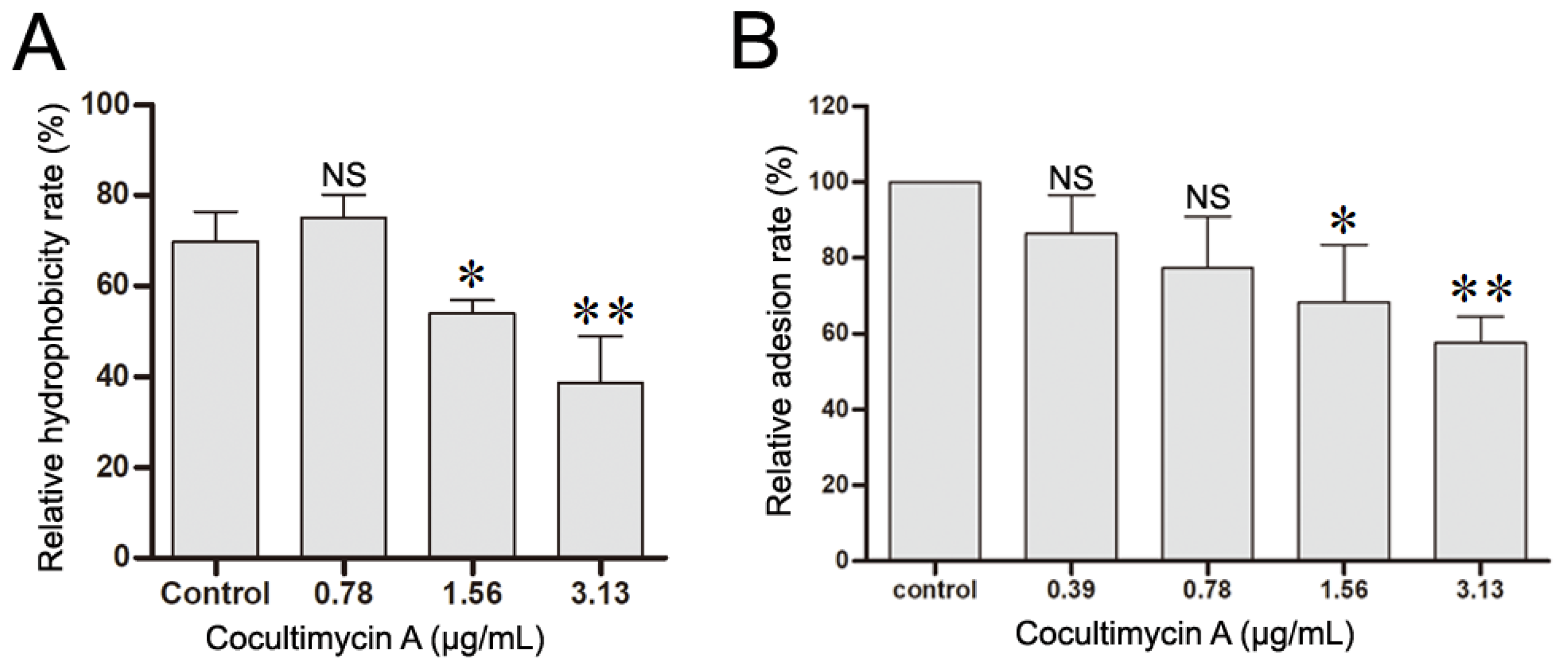
2.3. Cocultimycin A Decreased the Hydrophobicity of C. albicans and HOEC Cell Adhesion
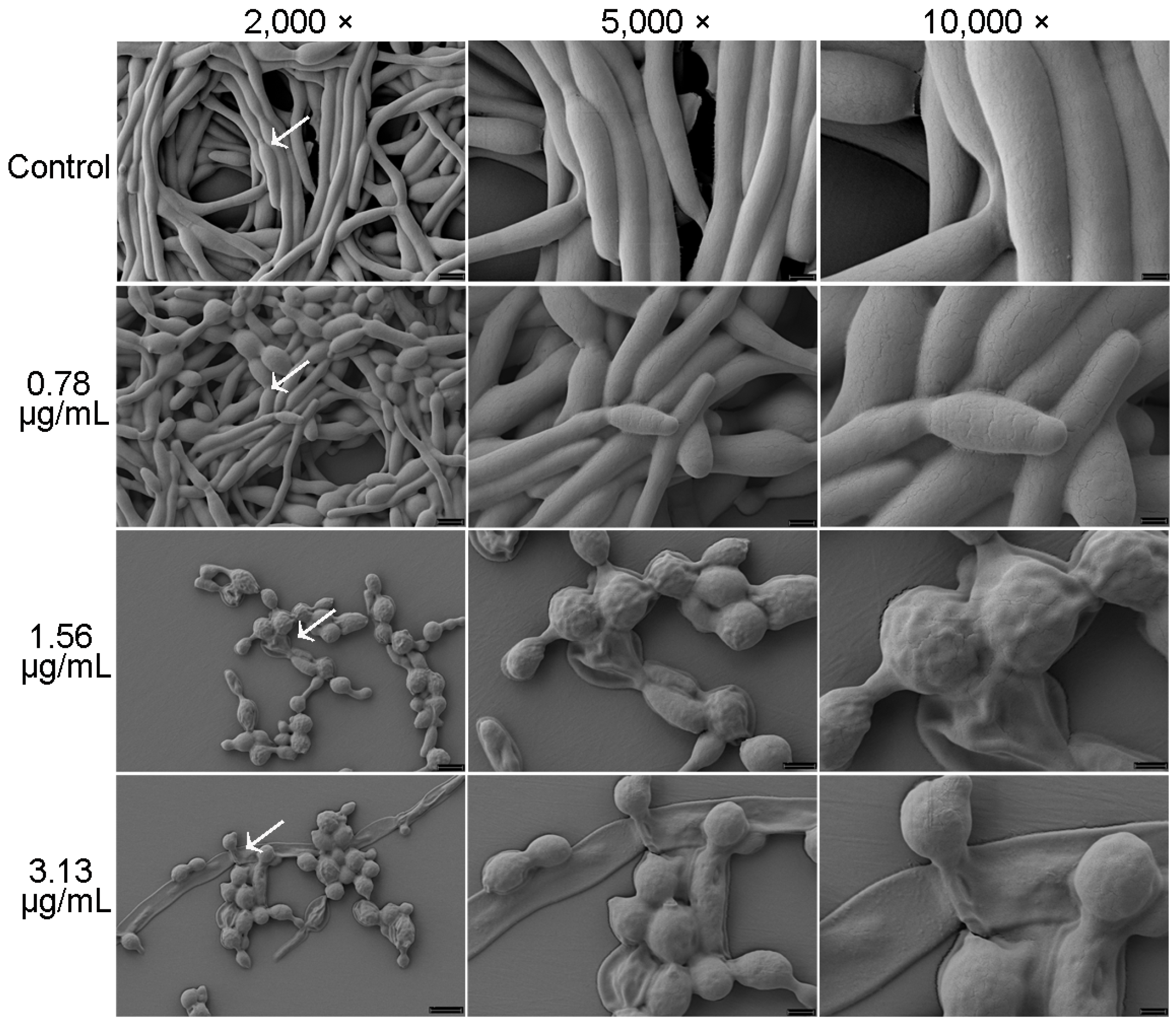
2.4. Cocultimycin A Weakens the Filamentous Development of C. albicans
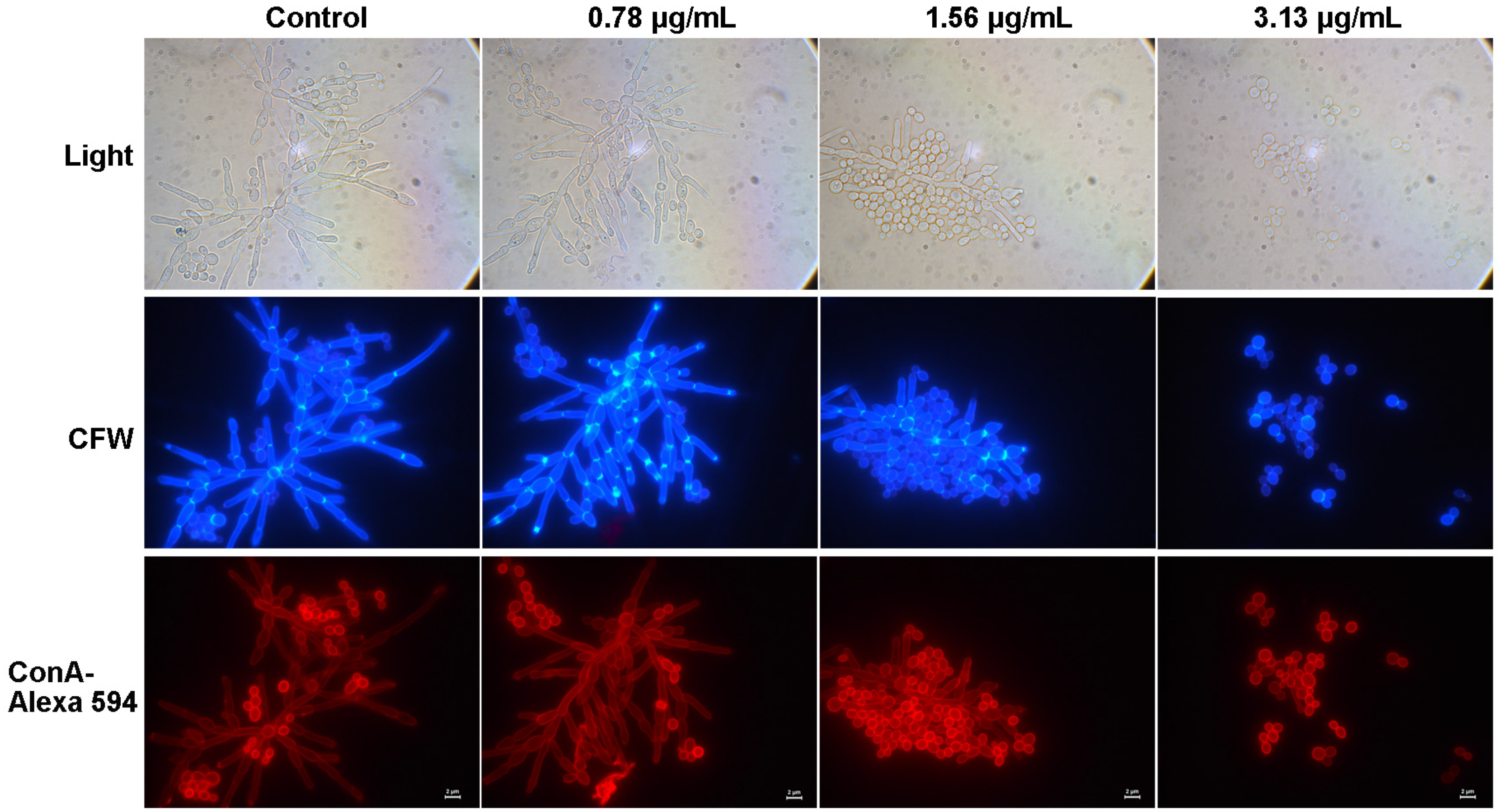
2.5. Cocultimycin A Influenced the Cell Wall Components of C. albicans
2.6. Cocultimycin A Affected Gene Expression Related to C. albicans Biofilm Formation
2.7. Inhibitory Activity of Cocultimycin A against Planktonic C. albicans
3. Materials and Methods
3.1. Fungal Strains and Growth Conditions
3.2. Effects on Biofilm
3.3. XTT Reduction Assay
3.4. Crystal Violet Staining
3.5. Cell Surface Hydrophobicity Assay
3.6. Human Oral Epithelial Cells (HOEC) Adhesion Assay
3.7. Detection of Yeast-to-Hypha Phase Transition
3.8. Colony Morphology
3.9. Scanning Electron Microscopy (SEM)
3.10. Cell Wall Staining Assay
3.11. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Assay
CT (DMSO β-actin), ΔΔCT = Δ CT (test sample) − ΔCT (calibration sample)
3.12. Detection of the Inhibitory Activity of Cocultimycin A against Planktonic C. albicans Cells
3.13. Statistical Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- WHO. Who Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization (WHO): Geneva, Switzerland, 2022.
- Wächtler, B.; Wilson, D.; Haedicke, K.; Dalle, F.; Hube, B. From attachment to damage: Defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS ONE 2011, 6, e17046. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Finnegan, M.; Özkan, S.; Kim, Y.; Lillehoj, P.; Ho, C.; Lux, R.; Mito, R.; Loewy, Z.; Shi, W. In vitro study of biofilm formation and effectiveness of antimicrobial treatment on various dental material surfaces. Mol. Oral Microbiol. 2010, 25, 384–390. [Google Scholar] [CrossRef]
- Ramage, G.; Martãnez, J.P.; LoPez-Ribot, J.L. Candida biofilms on implanted biomaterials: A clinically significant problem. FEMS Yeast Res. 2006, 6, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Kojic, E.M.; Darouiche, R.O. Candida infections of medical devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Costa-Orlandi, C.B.; Sardi, J.C.O.; Pitangui, N.S.; De Oliveira, H.C.; Scorzoni, L.; Galeane, M.C.; Medina-Alarcón, K.P.; Melo, W.C.M.A.; Marcelino, M.Y.; Braz, J.D.; et al. Fungal biofilms and polymicrobial diseases. J. Fungi 2017, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Gulati, M.; Nobile, C.J. Candida albicans biofilms: Development, regulation, and molecular mechanisms. Microbes Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef]
- Finkel, J.S.; Mitchell, A.P. Genetic control of Candida albicans biofilm development. Nat. Rev. Genet. 2011, 9, 109–118. [Google Scholar] [CrossRef]
- Araújo, D.; Henriques, M.; Silva, S. Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 2017, 25, 62–75. [Google Scholar] [CrossRef]
- Chandra, J.; Mukherjee, P.K. Candida biofilms: Development, architecture, and resistance. Microbiol. Spectr. 2015, 3, 115–134. [Google Scholar] [CrossRef]
- Naglik, J.R.; Moyes, D.L.; Wächtler, B.; Hube, B. Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 2011, 13, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Rodrigues, C.F.; Araújo, D.; Rodrigues, M.E.; Henriques, M. Candida species biofilms’ antifungal resistance. J. Fungi 2017, 3, 8. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Patterson, T.F. Multidrug-resistant Candida: Epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 2017, 216, S445–S451. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Nobile, C.J. Antifungal drug-resistance mechanisms in Candida biofilms. Curr. Opin. Microbiol. 2023, 71, 102237. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Liu, Y.; Liu, Y.; Lv, R.; Sun, W.; Ding, W.; Cai, Y.; Li, W.; Liu, X.; Qu, W. Candida albicans biofilms: Antifungal resistance, immune evasion, and emerging therapeutic strategies. Int. J. Antimicrob. Agents 2022, 60, 106673. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Kuhn, D.M.; Mukherjee, P.K.; Hoyer, L.L.; McCormick, T.; Ghannoum, M.A. Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. J. Bacteriol. 2001, 183, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- McCall, A.D.; Pathirana, R.U.; Prabhakar, A.; Cullen, P.J.; Edgerton, M. Candida albicans biofilm development is governed by cooperative attachment and adhesion maintenance proteins. NPJ Biofilms Microbiomes 2019, 5, 21. [Google Scholar] [CrossRef]
- Houšť, J.; Spížek, J.; Havlíček, V. Antifungal drugs. Metabolites 2020, 10, 106. [Google Scholar] [CrossRef]
- Heard, S.C.; Wu, G.; Winter, J.M. Antifungal natural products. Curr. Opin. Biotechnol. 2021, 69, 232–241. [Google Scholar] [CrossRef]
- Rauseo, A.M.; Coler-Reilly, A.; Larson, L.; Spec, A. Hope on the horizon: Novel fungal treatments in development. Open Forum Infect. Dis. 2020, 7, ofaa016. [Google Scholar] [CrossRef]
- Knowles, S.L.; Raja, H.A.; Roberts, C.D.; Oberlies, N.H. Fungal-fungal co-culture: A primer for generating chemical diversity. Nat. Prod. Rep. 2022, 39, 1557–1573. [Google Scholar] [CrossRef] [PubMed]
- Marmann, A.; Aly, A.H.; Lin, W.; Wang, B.; Proksch, P. Co-cultivation—A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef] [PubMed]
- Li, J. Study on Mining Secondary Metabolic Potential of Marine-Derived Fungi. Master’s Thesis, Peking Union Medical College, Beijing, China, 2019; p. 175. [Google Scholar]
- Gan, M.; Li, J.; Chen, M.; Hao, X.; Li, S.; Li, F. Antifungal Compounds from Marine Aspergillus sp. and Preparation Method Thereof. CN201911156556.9, filed 2019, and Issued. Patent. Available online: https://pss-system.cponline.cnipa.gov.cn/retrieveList?prevPageTit=changgui (accessed on 9 October 2023).
- Li, H.; Gilchrist, C.L.M.; Lacey, H.J.; Crombie, A.; Vuong, D.; Pitt, J.I.; Lacey, E.; Chooi, Y.-H.; Piggott, A.M. Discovery and heterologous biosynthesis of the burnettramic acids: Rare pks-nrps-derived bolaamphiphilic pyrrolizidinediones from an Australian fungus, Aspergillus burnettii. Org. Lett. 2019, 21, 1287–1291. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, M.; Hao, X.; Li, S.; Li, F.; Yu, L.; Xiao, C.; Gan, M. Structural revision and absolute configuration of burnettramic acid A. Org. Lett. 2020, 22, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-D.; Zhao, L.-X.; Mylonakis, E.; Hu, G.-H.; Zou, Y.; Huang, T.-K.; Yan, L.; Wang, Y.; Jiang, Y.-Y. In vitro andin vivo activities of pterostilbene against Candida albicans biofilms. Antimicrob. Agents Chemother. 2014, 58, 2344–2355. [Google Scholar] [CrossRef] [PubMed]
- Samaranayake, Y.H.; Wu, P.C.; Samaranayake, L.P.; So, M. Relationship between the cell surface hydrophobicity and adherence of Candida krusei and Candida albicans to epithelial and denture acrylic surfaces. Apmis 1995, 103, 707–713. [Google Scholar] [CrossRef]
- Luo, G.; Samaranayake, L.P. Candida glabrata, an emerging fungal pathogen, exhibits superior relative cell surface hydrophobicity and adhesion to denture acrylic surfaces compared with Candida albicans. Apmis 2002, 110, 601–610. [Google Scholar] [CrossRef]
- Sundstrom, P. Adhesion in Candida spp. Cell. Microbiol. 2002, 4, 461–469. [Google Scholar] [CrossRef]
- Pompilio, A.; Piccolomini, R.; Picciani, C.; D’Antonio, D.; Savini, V.; Di Bonaventura, G. Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: The role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 2010, 287, 41–47. [Google Scholar] [CrossRef]
- Zhong, H.; Hu, D.D.; Hu, G.H.; Su, J.; Bi, S.; Zhang, Z.E.; Wang, Z.; Zhang, R.L.; Xu, Z.; Jiang, Y.Y.; et al. Activity of sanguinarine against Candida albicans biofilms. Antimicrob. Agents Chemother. 2017, 61, 10–128. [Google Scholar] [CrossRef]
- Nobile, C.J.; Mitchell, A.P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor bcr1p. Curr. Biol. 2005, 15, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Mahto, K.K.; Singh, A.; Khandelwal, N.K.; Bhardwaj, N.; Jha, J.; Prasad, R. An assessment of growth media enrichment on lipid metabolome and the concurrent phenotypic properties of Candida albicans. PLoS ONE 2014, 9, e113664. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Henriques, M.; Martins, A.; Oliveira, R.; Williams, D.; Azeredo, J. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med. Mycol. 2009, 47, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Cantón, E.; Pemán, J. Antifungal susceptibility testing of filamentous fungi. Curr. Fungal Infect. Rep. 2012, 6, 41–50. [Google Scholar] [CrossRef]
- Pierce, C.G.; Uppuluri, P.; Tristan, A.R.; Wormley, F.L., Jr.; Mowat, E.; Ramage, G.; Lopez-Ribot, J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008, 3, 1494–1500. [Google Scholar] [CrossRef]
- Lal, P.; Agarwal, V.; Pruthi, P.; Pereira, B.M.J.; Kural, M.R.; Pruthi, V. Biofilm formation by Candida albicans isolated from intrauterine devices. Indian J. Microbiol. 2008, 48, 438–444. [Google Scholar] [CrossRef][Green Version]
- Yang, L.; Liu, X.; Lv, L.; Ma, Z.; Feng, X.; Ma, T. Dracorhodin perchlorate inhibits biofilm formation and virulence factors of Candida albicans. J. Med. Mycol. 2018, 28, 36–44. [Google Scholar] [CrossRef]
- Yadav, B.; Bhatnagar, S.; Ahmad, M.F.; Jain, P.; Pratyusha, V.A.; Kumar, P.; Komath, S.S. First step of glycosylphosphatidylinositol (gpi) biosynthesis cross-talks with ergosterol biosynthesis and ras signaling in Candida albicans. J. Biol. Chem. 2014, 289, 3365–3382. [Google Scholar] [CrossRef]
- Liu, H.; Köhler, J.; Fink, G.R. Suppression of hyphal formation in Candida albicans by mutation of a ste12 homolog. Science 1994, 266, 1723–1726. [Google Scholar] [CrossRef]
- Gong, J.; Hu, D.; He, J.; Zou, L.; Chen, Z.; Li, M. Effect of longzhang gargle on dual-species biofilm of Candida albicans and Streptococcus mutans. BioMed Res. Int. 2021, 2021, 6654793. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−δδct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fothergill, A.W. Antifungal Susceptibility Testing: Clinical Laboratory and Standards Institute (Clsi) Methods; Humana Press: Totowa, NJ, USA, 2011; pp. 65–74. [Google Scholar]
- Klepser, M.E.; Wolfe, E.J.; Jones, R.N.; Nightingale, C.H.; Pfaller, M.A. Antifungal pharmacodynamic characteristics of fluconazole and amphotericin b tested against Candida albicans. Antimicrob. Agents Chemother. 1997, 41, 1392–1395. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Chen, Y.; Chen, I.; Chen, F.; Chien, C. Impact of biofilm production by Candida species and antifungal therapy on mortality of patients with candidemia. Mycoses 2020, 63, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.S.; Motta, F.A.; Picharski, G.L.; Vasconcelos, T.M.; Riccieri, M.C.; Dalla-Costa, L.M. Invasive candidiasis: Risk factor for mortality in a pediatric tertiary care hospital in south of brazil. Medicine 2019, 98, e15933. [Google Scholar] [CrossRef] [PubMed]
- de Barros, P.P.; Rossoni, R.D.; de Souza, C.M.; Scorzoni, L.; Fenley, J.D.C.; Junqueira, J.C. Candida biofilms: An update on developmental mechanisms and therapeutic challenges. Mycopathologia 2020, 185, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Wall, G.; Montelongo-Jauregui, D.; Bonifacio, B.V.; Lopez-Ribot, J.L.; Uppuluri, P. Candida albicans biofilm growth and dispersal: Contributions to pathogenesis. Curr. Opin. Microbiol. 2019, 52, 1–6. [Google Scholar] [CrossRef]
- Brunetti, G.; Navazio, A.S.; Giuliani, A.; Giordano, A.; Proli, E.M.; Antonelli, G.; Raponi, G. Candida blood stream infections observed between 2011 and 2016 in a large italian university hospital: A time-based retrospective analysis on epidemiology, biofilm production, antifungal agents consumption and drug-susceptibility. PLoS ONE 2019, 14, e0224678. [Google Scholar] [CrossRef]
- Li, C.X.; Gleason, J.E.; Zhang, S.X.; Bruno, V.M.; Cormack, B.P.; Culotta, V.C. Candida albicans adapts to host copper during infection by swapping metal cofactors for superoxide dismutase. Proc. Natl. Acad. Sci. USA 2015, 112, E5336–E5342. [Google Scholar] [CrossRef]
- Bink, A.; Vandenbosch, D.; Coenye, T.; Nelis, H.; Cammue, B.P.A.; Thevissen, K. Superoxide dismutases are involved in Candida albicans biofilm persistence against miconazole. Antimicrob. Agents Chemother. 2011, 55, 4033–4037. [Google Scholar] [CrossRef]
- Pasrija, R.; Krishnamurthy, S.; Prasad, T.; Ernst, J.F.; Prasad, R. Squalene epoxidase encoded by erg1 affects morphogenesis and drug susceptibilities of Candida albicans. J. Antimicrob. Chemother. 2005, 55, 905–913. [Google Scholar] [CrossRef]
- Dižová, S.; Černáková, L.; Bujdáková, H. The impact of farnesol in combination with fluconazole on Candida albicans biofilm: Regulation of erg20, erg9, and erg11 genes. Folia Microbiol. 2018, 63, 363–371. [Google Scholar] [CrossRef] [PubMed]


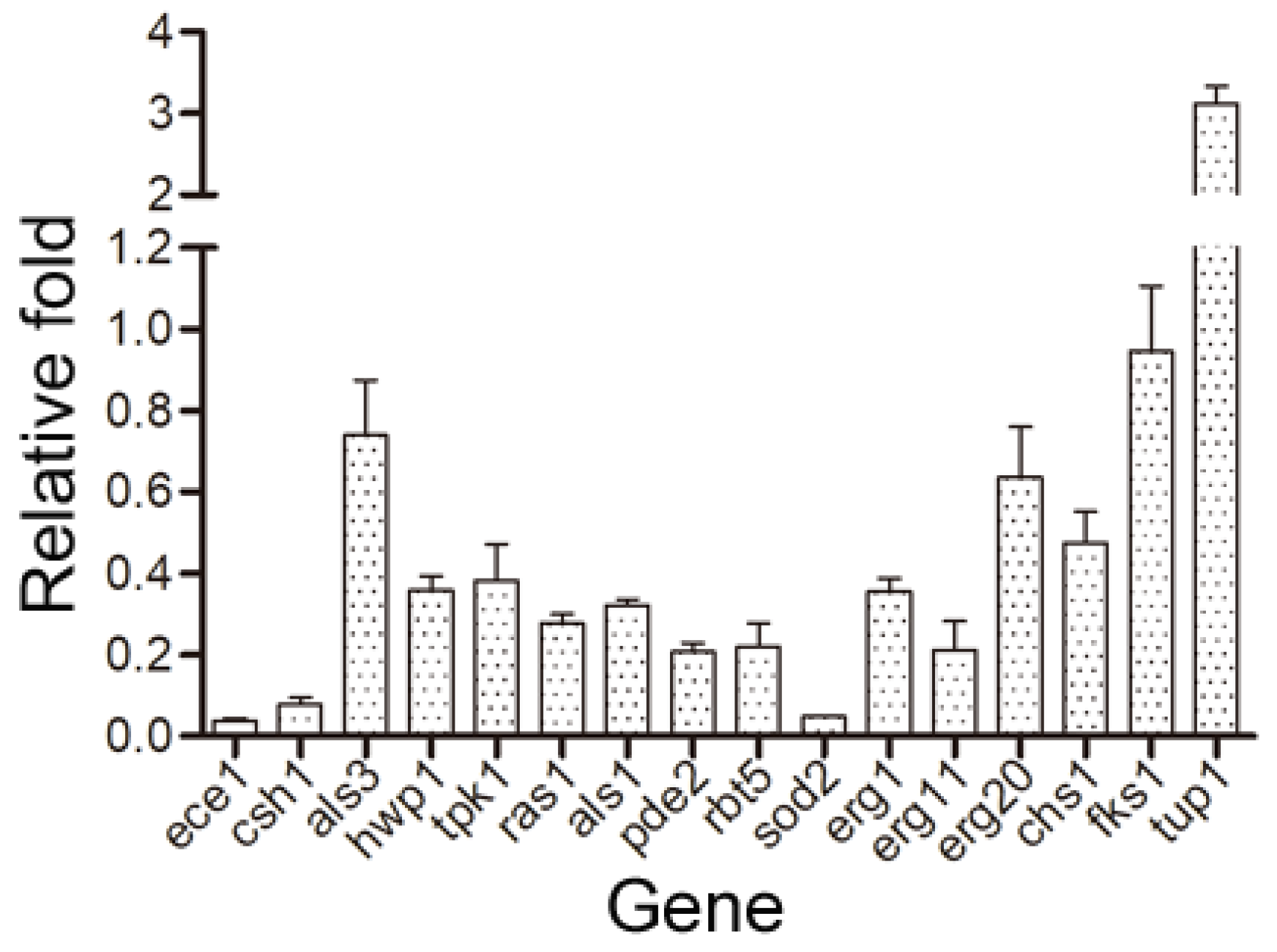
| Gene | Mean | SEM | Gene Description |
|---|---|---|---|
| ece1 | 0.038 | 0.006 | Candidalysin, cytolytic peptide toxin essential for mucosal infection; hypha-specific protein; regulated by Rfg1, Nrg1, Tup1, Cph1, Efg1, Hog1, farnesol, phagocytosis; fluconazole-induced; rat catheter and Spider biofilm induced |
| ras1 | 0.277 | 0.023 | RAS signal transduction GTPase; regulates cAMP and MAP kinase pathways; role in hyphal induction, virulence, apoptosis, heat-shock sensitivity; nonessential; plasma membrane-localized; complements viability of S. cerevisiae ras1 ras2 mutant |
| sod2 | 0.048 | 0.002 | Mitochondrial Mn-containing superoxide dismutase; protection against oxidative stress; homotetramer active; N-terminal 34 amino acids removed on mitochondrial import; H2O2-induced -, alkaline-downregulated, farnesol-induced |
| erg1 | 0.355 | 0.032 | Squalene epoxidase, epoxidation of squalene to 2,3(S)-oxidosqualene; ergosterol biosynthesis; allylamine antifungal drug target; NADH-reducing cofactor but S. cerevisiae Erg1 uses NADPH; flow model biofilm induced; Spider biofilm repressed |
| erg11 | 0.212 | 0.070 | Lanosterol 14-alpha-demethylase; cytochrome P450 family; role in ergosterol biosynthesis; target of azole antifungals; may contribute to drug resistance; azole or flow model biofilm induced; drug treated biofilm induced; hypoxia-regulated |
| csh1 | 0.078 | 0.016 | Aldo-keto reductase; role in fibronectin adhesion, cell surface hydrophobicity; regulated by temperature, growth phase, benomyl, macrophage interaction; azole resistance associated; Spider biofilm induced; rat catheter biofilm repressed |
| als1 | 0.323 | 0.000 | Cell-surface adhesin; adhesion, virulence, immunoprotective roles; band at hyphal base; Rfg1, Ssk1, Spider biofilm induced; flow model biofilm repressed; CAI-4 strain background effects; promoter bound Bcr1, Tec1, Efg1, Ndt80, and Brg1 |
| erg20 | 0.636 | 0.123 | Putative farnesyl pyrophosphate synthetase involved in isoprenoid and sterol biosynthesis, based on similarity to S. cerevisiae Erg20p; likely to be essential for growth, based on an insertional mutagenesis strategy |
| tpk1 | 0.383 | 0.089 | cAMP-dependent protein kinase catalytic subunit; Tpk2 isoform; control of morphogenesis and stress response; WT nuclear localization requires Bcy1; produced during stationary, not exponential growth; rat catheter and Spider biofilm induced |
| chs1 | 0.475 | 0.077 | Chitin synthase; essential; for primary septum synthesis in yeast and hyphae; one of several chitin synthases; enzymatically activated by proteolytic processing; complements defects of S. cerevisiae chs1 or chs2; Spider biofilm repressed |
| rbt5 | 0.220 | 0.058 | GPI-linked cell wall protein; hemoglobin utilization; Rfg1, Rim101, Tbf1, Fe regulated; Sfu1, Hog1, Tup1, serum, alkaline pH, antifungal drugs, geldamycin repressed; Hap43 induced; required for RPMI biofilms; Spider biofilm induced |
| fks1 | 0.945 | 0.016 | Catalytic subunit of 1,3-beta-D-glucan synthase; involved in cell wall synthesis and maintenance; localizes to sites of cell wall remodeling; FKS1 has a paralog, GSC2, which arose from the whole-genome duplication |
| tup1 | 3.117 | 0.022 | Transcriptional corepressor; represses filamentous growth; regulates switching; role in germ tube induction, farnesol response; in repression pathways with Nrg1, Rfg1; farnesol upregulated in biofilm; rat catheter, Spider biofilm repressed |
| pde2 | 0.207 | 0.022 | High-affinity cyclic nucleotide phosphodiesterase; moderates signaling by cAMP; required for virulence, switching, cell wall, hyphal, not pseudohyphal growth; expressed shortly after hyphal induction; rat catheter and Spider biofilm induced |
| als3 | 0.739 | 0.136 | Cell wall adhesin; epithelial adhesion, endothelial invasion; alleles vary in adhesiveness; immunoprotective in mice; binds SspB adhesin of S. gordonii in mixed biofilm; induced in/required for Spider biofilm; flow model biofilm repressed |
| hwp1 | 0.359 | 0.034 | Hyphal cell wall protein; host transglutaminase substrate; opaque-, a-specific, alpha-factor induced; at MTLa side of conjugation tube; virulence complicated by URA3 effects; Bcr1-repressed in RPMI a/a biofilms; Spider biofilm induced |
| Drug | MIC (μg/mL) | MFC (μg/mL) |
|---|---|---|
| Cocultimycin A | 1.56 | 6.25 |
| Amphotericin B | 0.39 | 0.78 |
| Caspofungin | 0.10 | 0.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Wang, A.; Zheng, Y.; Li, D.; Wei, Y.; Gan, M.; Li, Y.; Si, S. Anti-Biofilm Activity of Cocultimycin A against Candida albicans. Int. J. Mol. Sci. 2023, 24, 17026. https://doi.org/10.3390/ijms242317026
Zhu X, Wang A, Zheng Y, Li D, Wei Y, Gan M, Li Y, Si S. Anti-Biofilm Activity of Cocultimycin A against Candida albicans. International Journal of Molecular Sciences. 2023; 24(23):17026. https://doi.org/10.3390/ijms242317026
Chicago/Turabian StyleZhu, Xiaohong, Anqi Wang, Yifan Zheng, Dan Li, Yuanjuan Wei, Maoluo Gan, Yan Li, and Shuyi Si. 2023. "Anti-Biofilm Activity of Cocultimycin A against Candida albicans" International Journal of Molecular Sciences 24, no. 23: 17026. https://doi.org/10.3390/ijms242317026
APA StyleZhu, X., Wang, A., Zheng, Y., Li, D., Wei, Y., Gan, M., Li, Y., & Si, S. (2023). Anti-Biofilm Activity of Cocultimycin A against Candida albicans. International Journal of Molecular Sciences, 24(23), 17026. https://doi.org/10.3390/ijms242317026





