miR-195-5p as Regulator of γ-Catenin and Desmosome Junctions in Colorectal Cancer
Abstract
:1. Introduction
2. Results
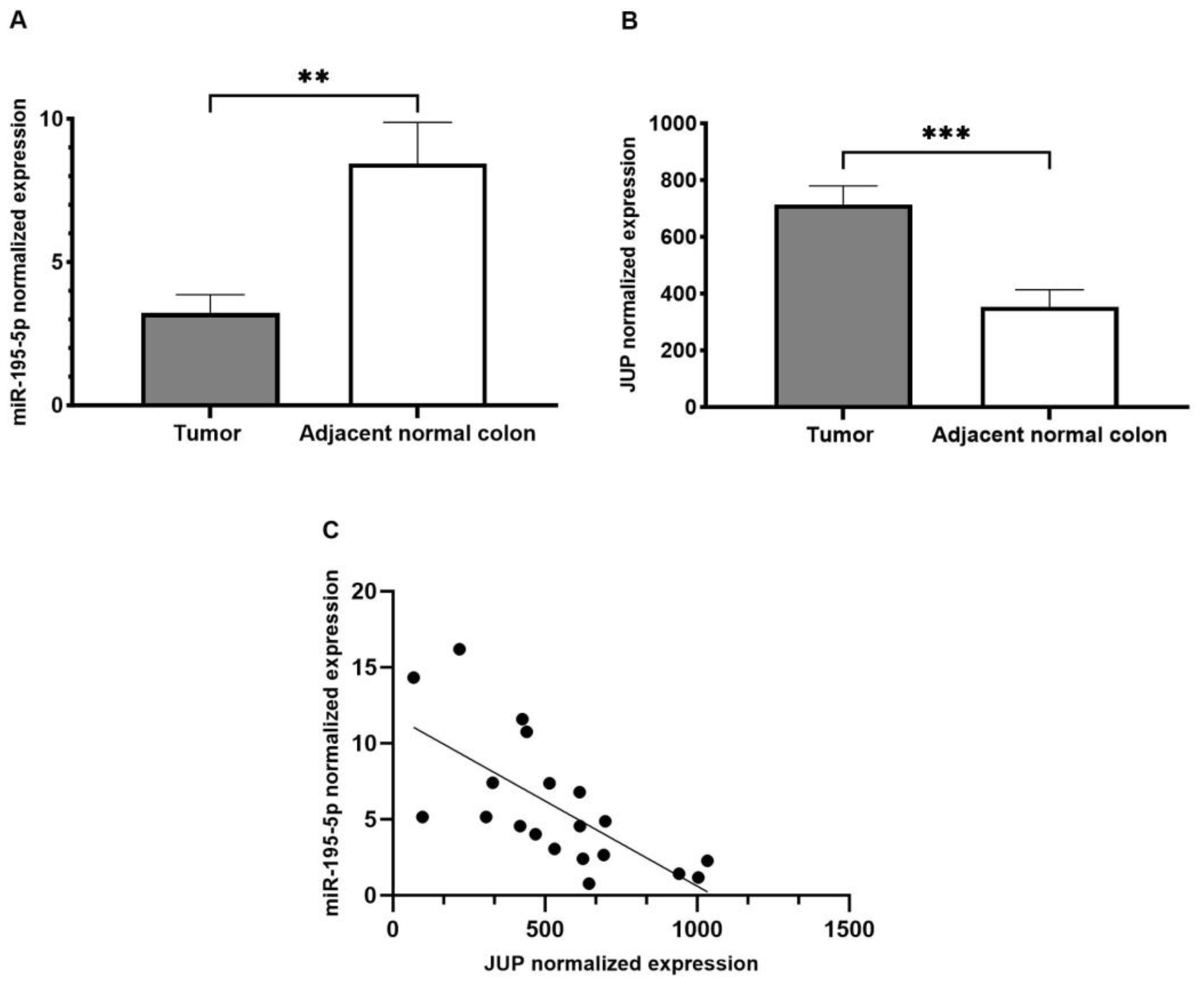
2.1. miR-195-5p Expression in CRC Patients
2.2. miR-195-5p as a Potential Modulator of JUP
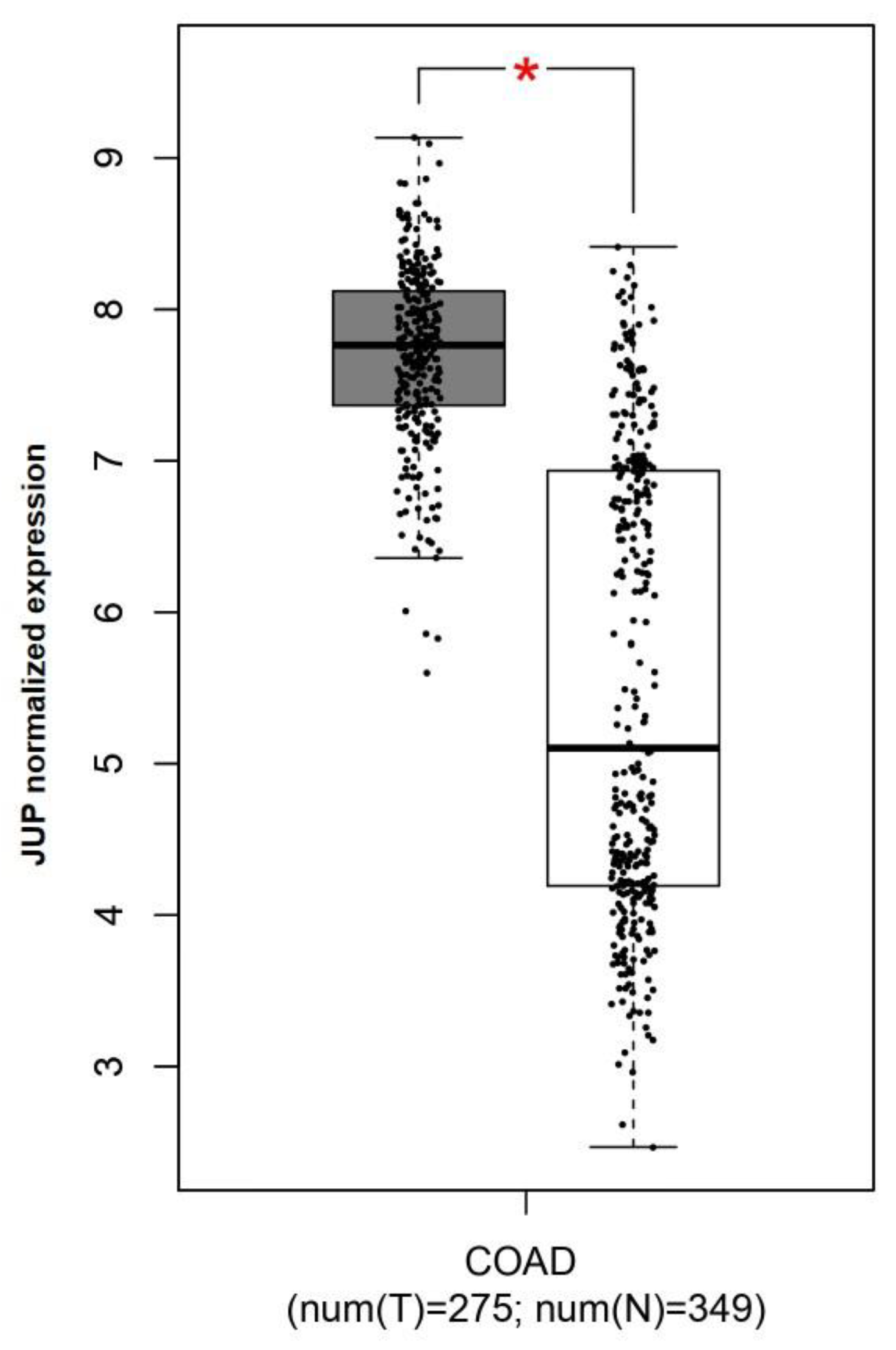
2.3. JUP Expression in CRC Patients
2.4. miR-195-5p Is an Effective Regulator of JUP mRNA
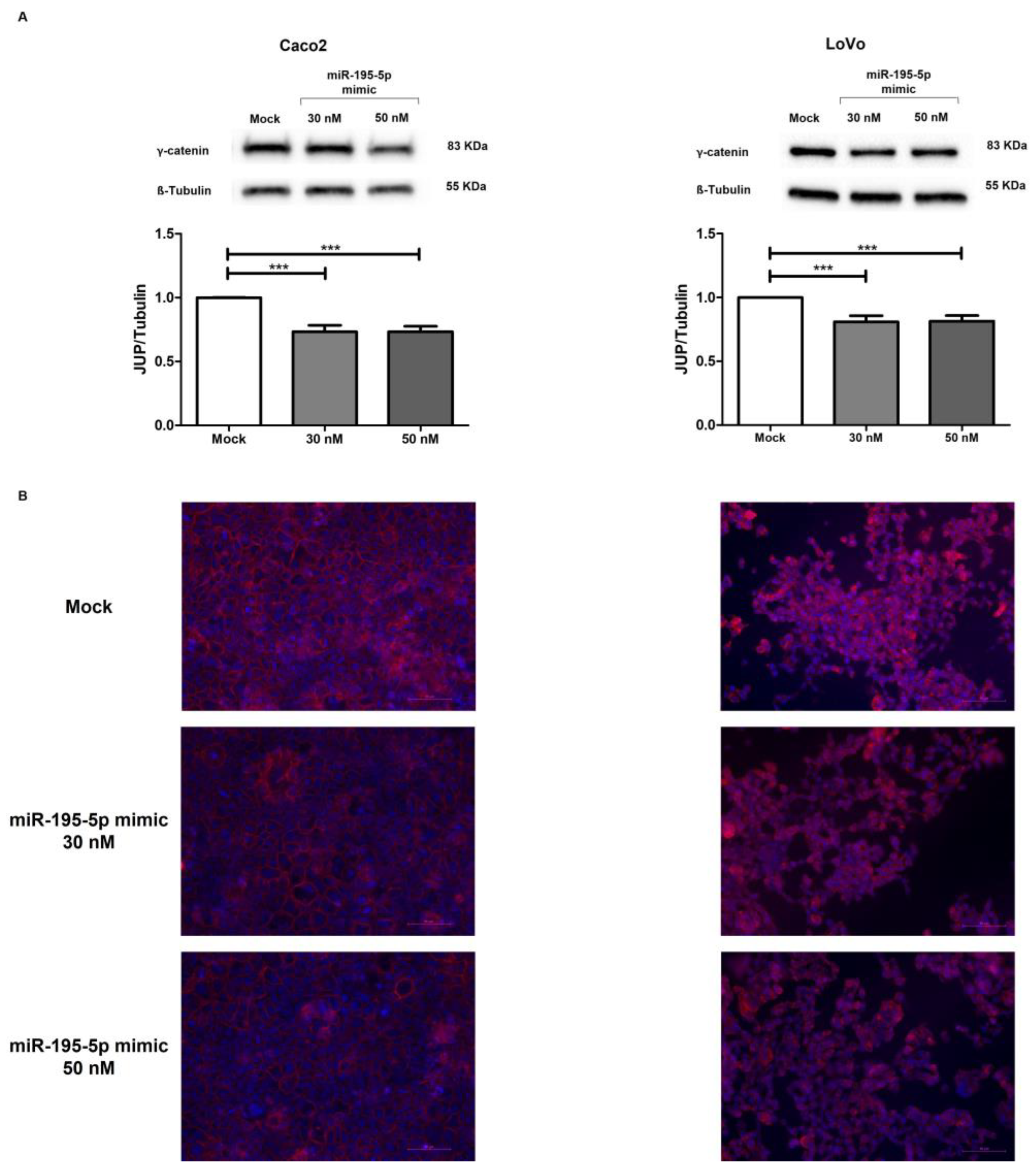
2.5. miR-195-5p Directly Regulates γ-Catenin Protein Expression
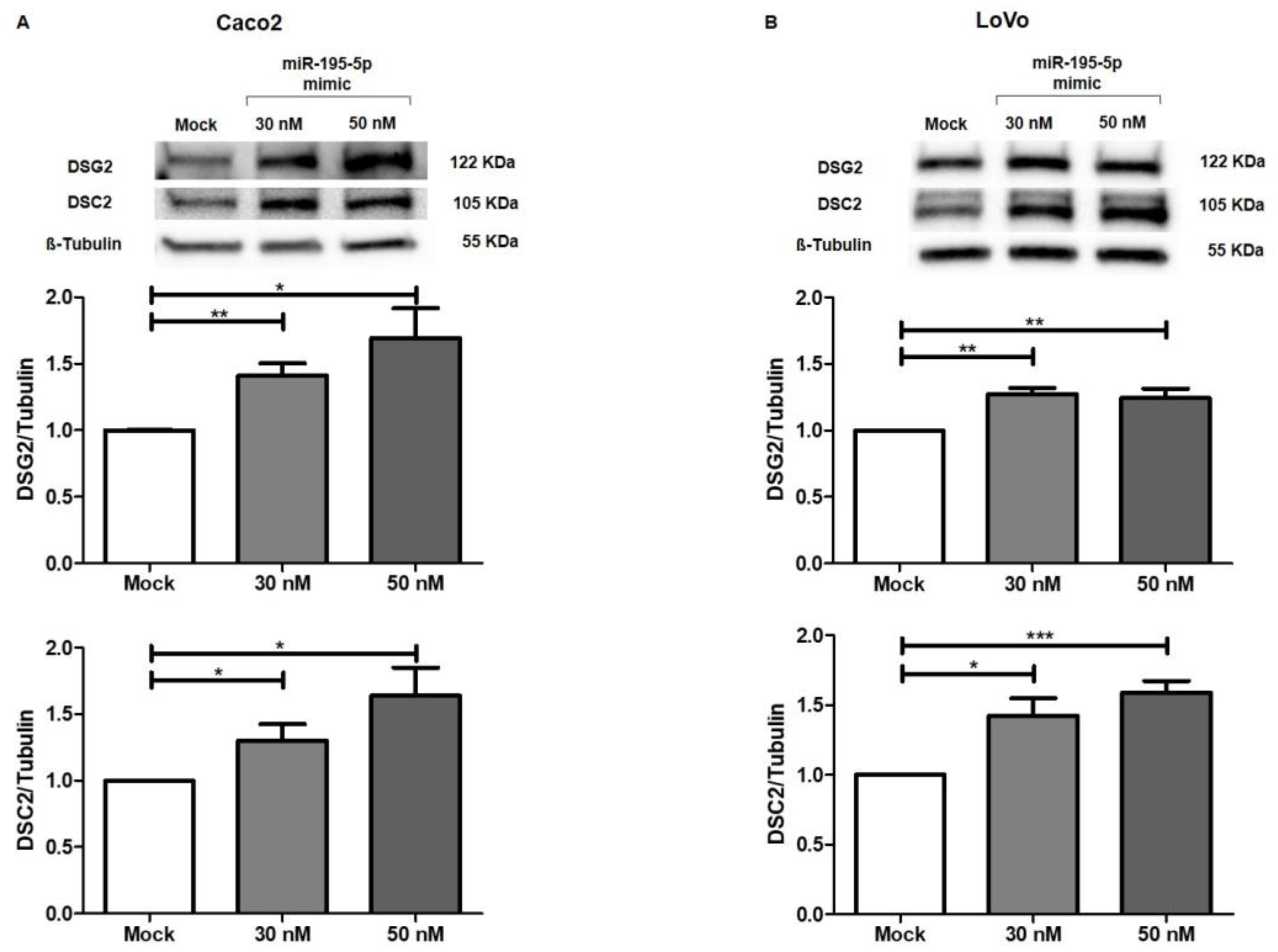
2.6. miR-195-5p Indirectly Modulates the Desmosome Cadherins DSG2 and DSC2
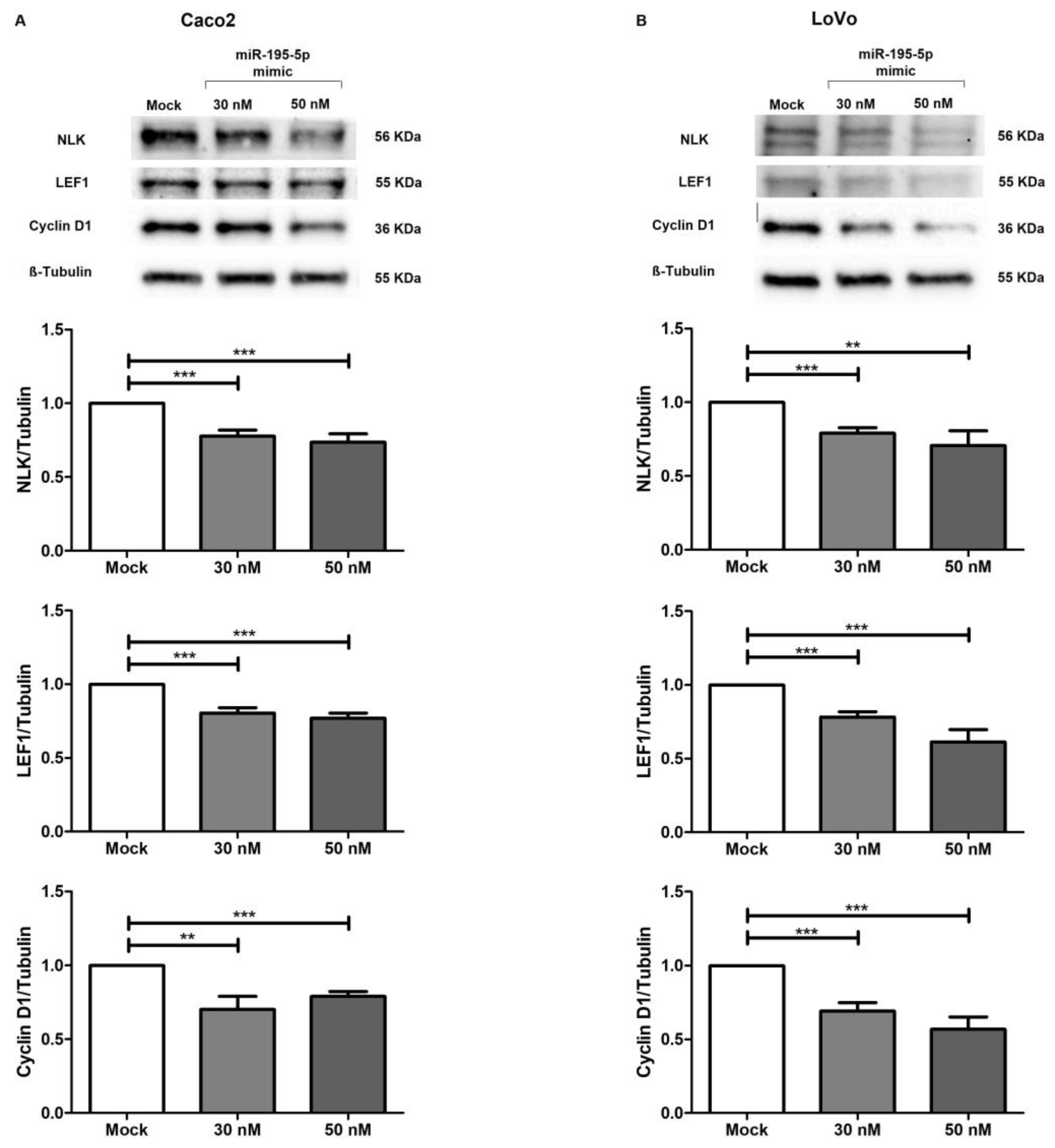
2.7. miR-195-5p Regulates Wnt Pathway Activation
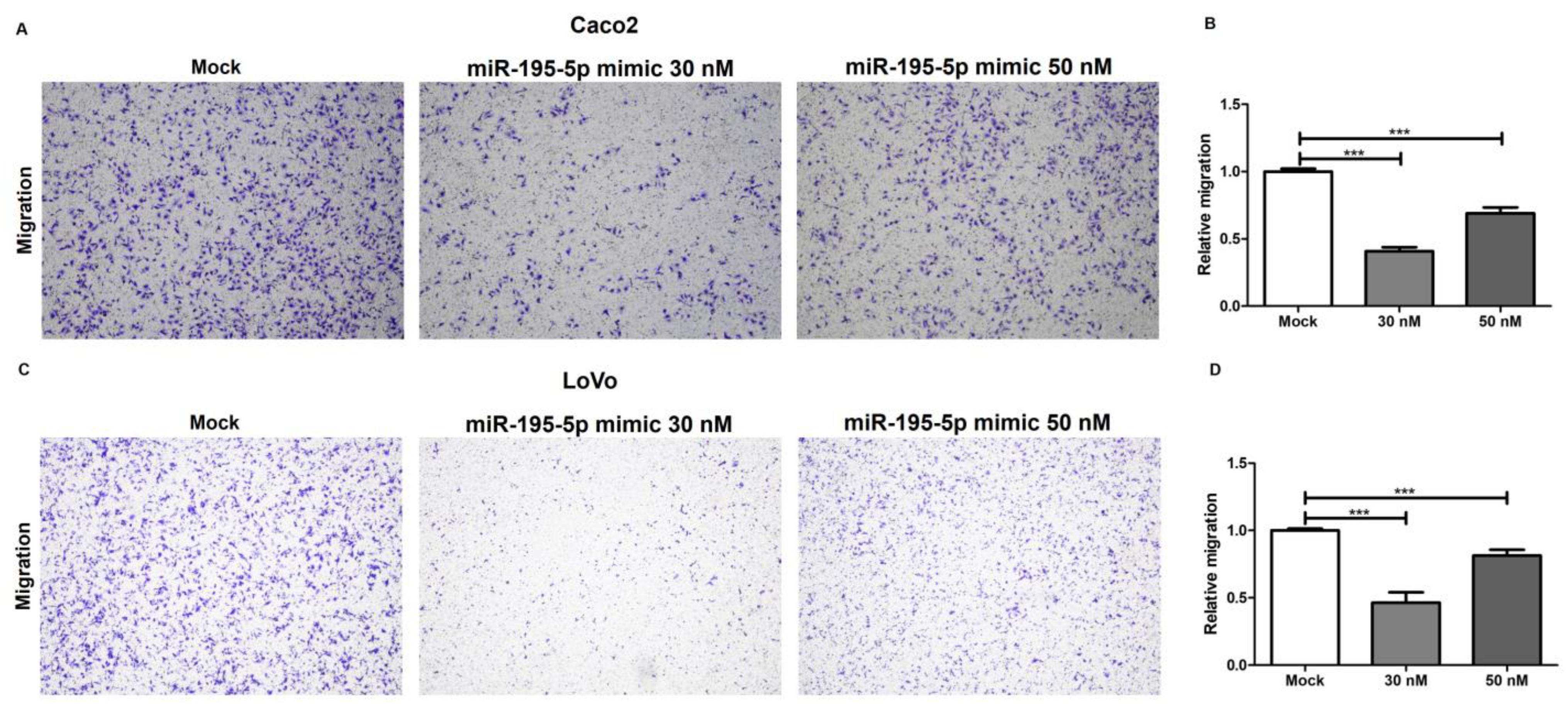
2.8. miR-195-5p Inhibits Cell Migration in CRC
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. In Vitro Transfection
4.3. RNA Extraction and Real-Time PCR
4.4. Western Blot
4.5. Immunofluorescence
4.6. Migration Assay
4.7. Bioinformatic and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Long, A.G.; Lundsmith, E.T.; Hamilton, K.E. Inflammation and colorectal cancer. Curr. Color. Cancer Rep. 2017, 13, 341–351. [Google Scholar] [CrossRef]
- Janakiram, N.B.; Rao, C.V. The role of inflammation in colon cancer. Adv. Exp. Med. Biol. 2014, 816, 25–52. [Google Scholar]
- Ullman, T.A.; Itzkowitz, S.H. Intestinal inflammation and cancer. Gastroenterology 2011, 140, 1807–1816.e1. [Google Scholar] [CrossRef] [PubMed]
- Olén, O.; Erichsen, R.; Sachs, M.C.; Pedersen, L.; Halfvarson, J.; Askling, J.; Ekbom, A.; Sørensen, H.T.; Ludvigsson, J.F. Colorectal cancer in ulcerative colitis: A Scandinavian population-based cohort study. Lancet 2020, 395, 123–131. [Google Scholar] [CrossRef] [PubMed]
- McGuckin, M.A.; Eri, R.; Simms, L.A.; Florin, T.H.; Radford-Smith, G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm. Bowel Dis. 2009, 15, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Yeruva, S.; Turner, J.R. Contributions of intestinal epithelial barriers to health and disease. Exp. Cell Res. 2017, 358, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Aktary, Z.; Alaee, M.; Pasdar, M. Beyond cell-cell adhesion: Plakoglobin and the regulation of tumorigenesis and metastasis. Oncotarget 2017, 8, 32270–32291. [Google Scholar] [CrossRef]
- Ben-Ze’ev, A.; Geiger, B. Differential molecular interactions of β-catenin and plakoglobin in adhesion, signaling and cancer. Curr. Opin. Cell Biol. 1998, 10, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Kodama, S.; Ikeda, S.; Asahara, T.; Kishida, M.; Kikuchi, A. Axin directly interacts with plakoglobin and regulates its stability. J. Biol. Chem. 1999, 274, 27682–27688. [Google Scholar] [CrossRef]
- Rubinfeld, B.; Souza, B.; Albert, I.; Munemitsu, S.; Polakis, P. The APC Protein and E-cadherin Form Similar but Independent Complexes with α-Catenin, β-Catenin, and Plakoglobin. J. Biol. Chem. 1995, 270, 5549–5555. [Google Scholar] [CrossRef]
- Hülsken, J.; Birchmeier, W.; Behrens, J. E-cadherin and APC compete for the interaction with beta-catenin and the cytoskeleton. J. Cell Biol. 1994, 127, 2061–2069. [Google Scholar] [CrossRef]
- Ozawa, M.; Terada, H.; Pedraza, C. The fourth armadillo repeat of plakoglobin (γ-catenin) is required for its high affinity binding to the cytoplasmic domains of E-cadherin and desmosomal cadherin Dsg2, and the tumor suppressor APC protein. J. Biochem. 1995, 118, 1077–1082. [Google Scholar] [CrossRef]
- Shibata, T.; Gotoh, M.; Ochiai, A.; Hirohashi, S. Association of Plakoglobin with APC, a Tumor-Suppressor Gene Product, and Its Regulation by Tyrosine Phosphorylation. Biochem. Biophys. Res. Commun. 1994, 203, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Lee, L.-Y.; Chao, Y.-K.; Chang, J.T.; Lu, Y.-C.; Li, H.-F.; Chiu, C.-C.; Li, Y.-C.; Li, Y.-L.; Chiou, J.-F. DSG3 facilitates cancer cell growth and invasion through the DSG3-plakoglobin-TCF/LEF-Myc/cyclin D1/MMP signaling pathway. PLoS ONE 2013, 8, e64088. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Scalavino, V.; Liso, M.; Cavalcanti, E.; Gigante, I.; Lippolis, A.; Mastronardi, M.; Chieppa, M.; Serino, G. miR-369-3p modulates inducible nitric oxide synthase and is involved in regulation of chronic inflammatory response. Sci. Rep. 2020, 10, 15942. [Google Scholar] [CrossRef]
- Scalavino, V.; Liso, M.; Serino, G. Role of microRNAs in the regulation of dendritic cell generation and function. Int. J. Mol. Sci. 2020, 21, 1319. [Google Scholar] [CrossRef]
- Scalavino, V.; Piccinno, E.; Valentini, A.M.; Mastronardi, M.; Armentano, R.; Giannelli, G.; Serino, G. A Novel Mechanism of Immunoproteasome Regulation via miR-369-3p in Intestinal Inflammatory Response. Int. J. Mol. Sci. 2022, 23, 13771. [Google Scholar] [CrossRef]
- Scalavino, V.; Piccinno, E.; Valentini, A.M.; Schena, N.; Armentano, R.; Giannelli, G.; Serino, G. miR-369-3p Modulates Intestinal Inflammatory Response via BRCC3/NLRP3 Inflammasome Axis. Cells 2023, 12, 2184. [Google Scholar] [CrossRef]
- Song, J.; Lin, Z.; Liu, Q.; Huang, S.; Han, L.; Fang, Y.; Zhong, P.; Dou, R.; Xiang, Z.; Zheng, J. MiR-192-5p/RB1/NF-κBp65 signaling axis promotes IL-10 secretion during gastric cancer EMT to induce Treg cell differentiation in the tumour microenvironment. Clin. Transl. Med. 2022, 12, e992. [Google Scholar] [CrossRef]
- Shi, L.; Zhu, W.; Huang, Y.; Zhuo, L.; Wang, S.; Chen, S.; Zhang, B.; Ke, B. Cancer-associated fibroblast-derived exosomal microRNA-20a suppresses the PTEN/PI3K-AKT pathway to promote the progression and chemoresistance of non-small cell lung cancer. Clin. Transl. Med. 2022, 12, e989. [Google Scholar] [CrossRef]
- Qian, F.; Wang, J.; Wang, Y.; Gao, Q.; Yan, W.; Lin, Y.; Shen, L.; Xie, Y.; Jiang, X.; Shen, B. MiR-378a-3p as a putative biomarker for hepatocellular carcinoma diagnosis and prognosis: Computational screening with experimental validation. Clin. Transl. Med. 2021, 11, e307. [Google Scholar] [CrossRef]
- Mohammadi, A.; Mansoori, B.; Baradaran, B. The role of microRNAs in colorectal cancer. Biomed. Pharmacother. 2016, 84, 705–713. [Google Scholar] [CrossRef]
- Luo, X.; Burwinkel, B.; Tao, S.; Brenner, H. MicroRNA Signatures: Novel Biomarker for Colorectal Cancer? MicroRNA and Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1272–1286. [Google Scholar] [CrossRef]
- Schetter, A.J.; Okayama, H.; Harris, C.C. The role of microRNAs in colorectal cancer. Cancer J. 2012, 18, 244. [Google Scholar] [CrossRef]
- Xu, J.; Meng, Q.; Li, X.; Yang, H.; Xu, J.; Gao, N.; Sun, H.; Wu, S.; Familiari, G.; Relucenti, M. Long noncoding RNA MIR17HG promotes colorectal cancer progression via miR-17-5p. Cancer Res. 2019, 79, 4882–4895. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Ye, L.; Han, Z.; Liu, Y.; Yang, Y.; Peng, Z.; Fan, J. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: Validation by bioinformatics and experimental analyses. J. Exp. Clin. Cancer Res. 2017, 36, 131. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-B.; Shi, Q.; Li, G.; Zheng, J.-H.; Lin, J.; Qiu, W. MicroRNA-488 inhibits progression of colorectal cancer via inhibition of the mitogen-activated protein kinase pathway by targeting claudin-2. Am. J. Physiol. Cell Physiol. 2019, 316, C33–C47. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Shao, W.; Jiang, Y.; Xu, J.; Li, F.; Qin, J. MicroRNA-103 regulates tumorigenesis in colorectal cancer by targeting ZO-1. Mol. Med. Rep. 2018, 17, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Algaber, A.; Madhi, R.; Hawez, A.; Rönnow, C.-F.; Rahman, M. Targeting FHL2-E-cadherin axis by miR-340-5p attenuates colon cancer cell migration and invasion. Oncol. Lett. 2021, 22, 637. [Google Scholar] [CrossRef] [PubMed]
- Paek, A.R.; Lee, C.H.; You, H.J. A role of zinc-finger protein 143 for cancer cell migration and invasion through ZEB1 and E-cadherin in colon cancer cells. Mol. Carcinog. 2014, 53, E161–E168. [Google Scholar] [CrossRef] [PubMed]
- Gulei, D.; Magdo, L.; Jurj, A.; Raduly, L.; Cojocneanu-Petric, R.; Moldovan, A.; Moldovan, C.; Florea, A.; Pasca, S.; Pop, L.-A. The silent healer: miR-205-5p up-regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up-regulating E-cadherin expression. Cell Death Dis. 2018, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-J.; Xiao, H.-X.; Tian, H.-P.; Liu, Z.-L.; Xia, S.-S.; Zhou, T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int. J. Mol. Med. 2013, 31, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Heydari, K.; Saidijam, M.; Sharifi, M.R.; Dermani, F.K.; Soleimani Asl, S.; Shabab, N.; Najafi, R. The effect of miR-200c inhibition on chemosensitivity (5-FluoroUracil) in colorectal cancer. Pathol. Oncol. Res. 2018, 24, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Scalavino, V.; Piccinno, E.; Lacalamita, A.; Tafaro, A.; Armentano, R.; Giannelli, G.; Serino, G. miR-195-5p Regulates Tight Junctions Expression via Claudin-2 Downregulation in Ulcerative Colitis. Biomedicines 2022, 10, 919. [Google Scholar] [CrossRef] [PubMed]
- Scalavino, V.; Piccinno, E.; Bianco, G.; Schena, N.; Armentano, R.; Giannelli, G.; Serino, G. The increase of miR-195-5p reduces intestinal permeability in ulcerative colitis, modulating tight junctions’ expression. Int. J. Mol. Sci. 2022, 23, 5840. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Herrick, J.S.; Pellatt, D.F.; Stevens, J.R.; Mullany, L.E.; Wolff, E.; Hoffman, M.D.; Samowitz, W.S.; Wolff, R.K. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: Variations in miRNA expression and disease progression. Carcinogenesis 2016, 37, 245–261. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Chen, Z.; Ren, R.; Wan, D.; Wang, Y.; Xue, X.; Jiang, M.; Shen, J.; Han, Y.; Liu, F.; Shi, J. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene 2019, 38, 6017–6034. [Google Scholar] [CrossRef]
- Fang, J.; Xiao, L.; Zhang, Q.; Peng, Y.; Wang, Z.; Liu, Y. Junction plakoglobin, a potential prognostic marker of oral squamous cell carcinoma, promotes proliferation, migration and invasion. J. Oral Pathol. Med. 2020, 49, 30–38. [Google Scholar] [CrossRef]
- Leick, K.M.; Rodriguez, A.B.; Melssen, M.M.; Benamar, M.; Lindsay, R.S.; Eki, R.; Du, K.-P.; Parlak, M.; Abbas, T.; Engelhard, V.H. The barrier molecules junction plakoglobin, filaggrin, and dystonin play roles in melanoma growth and angiogenesis. Ann. Surg. 2019, 270, 712. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Li, Q.; Mrdenovic, S.; Chu, G.C.-Y.; Wu, B.J.; Bu, H.; Duan, P.; Kim, J.; You, S.; Lewis, M.S. KRT13 promotes stemness and drives metastasis in breast cancer through a plakoglobin/c-Myc signaling pathway. Breast Cancer Res. 2022, 24, 7. [Google Scholar] [CrossRef] [PubMed]
- Nweke, E.; Ntwasa, M.; Brand, M.; Devar, J.; Smith, M.; Candy, G. Increased expression of plakoglobin is associated with upregulated MAPK and PI3K/AKT signalling pathways in early resectable pancreatic ductal adenocarcinoma. Oncol. Lett. 2020, 19, 4133–4141. [Google Scholar] [CrossRef]
- Aktary, Z.; Pasdar, M. Plakoglobin: Role in tumorigenesis and metastasis. Int. J. Cell Biol. 2012, 2012, 189521. [Google Scholar] [CrossRef] [PubMed]
- Kolligs, F.T.; Kolligs, B.; Hajra, K.M.; Hu, G.; Tani, M.; Cho, K.R.; Fearon, E.R. γ-Catenin is regulated by the APC tumor suppressor and its oncogenic activity is distinct from that of β-catenin. Gen. Dev. 2000, 14, 1319–1331. [Google Scholar] [CrossRef]
- Ye, D.; Guo, S.; Al–Sadi, R.; Ma, T.Y. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology 2011, 141, 1323–1333. [Google Scholar] [CrossRef]
- Zhi, X.; Tao, J.; Li, Z.; Jiang, B.; Feng, J.; Yang, L.; Xu, H.; Xu, Z. MiR-874 promotes intestinal barrier dysfunction through targeting AQP3 following intestinal ischemic injury. FEBS Lett. 2014, 588, 757–763. [Google Scholar] [CrossRef]
- Ren, L.-L.; Yan, T.-T.; Shen, C.-Q.; Tang, J.-Y.; Kong, X.; Wang, Y.-C.; Chen, J.; Liu, Q.; He, J.; Zhong, M. The distinct role of strand-specific miR-514b-3p and miR-514b-5p in colorectal cancer metastasis. Cell Death Dis. 2018, 9, 687. [Google Scholar] [CrossRef]
- Tang, W.; Zhu, Y.; Gao, J.; Fu, J.; Liu, C.; Liu, Y.; Song, C.; Zhu, S.; Leng, Y.; Wang, G. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br. J. Cancer 2014, 110, 450–458. [Google Scholar] [CrossRef]
- Maeda, O.; Usami, N.; Kondo, M.; Takahashi, M.; Goto, H.; Shimokata, K.; Kusugami, K.; Sekido, Y. Plakoglobin (γ-catenin) has TCF/LEF family-dependent transcriptional activity in β-catenin-deficient cell line. Oncogene 2004, 23, 964–972. [Google Scholar] [CrossRef]
- Zhurinsky, J.; Shtutman, M.; Ben-Ze’ev, A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by β-catenin and plakoglobin. Mol. Cell. Biol. 2000, 20, 4238–4252. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.O.; Barish, G.D.; Klymkowsky, M.W.; Varmus, H.E. A comparative evaluation of β-catenin and plakoglobin signaling activity. Oncogene 2000, 19, 5720–5728. [Google Scholar] [CrossRef] [PubMed]
- Quillet, A.; Saad, C.; Ferry, G.; Anouar, Y.; Vergne, N.; Lecroq, T.; Dubessy, C. Improving bioinformatics prediction of microRNA targets by ranks aggregation. Front. Gen. 2020, 10, 1330. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piccinno, E.; Scalavino, V.; Armentano, R.; Giannelli, G.; Serino, G. miR-195-5p as Regulator of γ-Catenin and Desmosome Junctions in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 17084. https://doi.org/10.3390/ijms242317084
Piccinno E, Scalavino V, Armentano R, Giannelli G, Serino G. miR-195-5p as Regulator of γ-Catenin and Desmosome Junctions in Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(23):17084. https://doi.org/10.3390/ijms242317084
Chicago/Turabian StylePiccinno, Emanuele, Viviana Scalavino, Raffaele Armentano, Gianluigi Giannelli, and Grazia Serino. 2023. "miR-195-5p as Regulator of γ-Catenin and Desmosome Junctions in Colorectal Cancer" International Journal of Molecular Sciences 24, no. 23: 17084. https://doi.org/10.3390/ijms242317084





