Enhancement of Radiation Sensitivity by Cathepsin L Suppression in Colon Carcinoma Cells
Abstract
:1. Introduction
2. Results
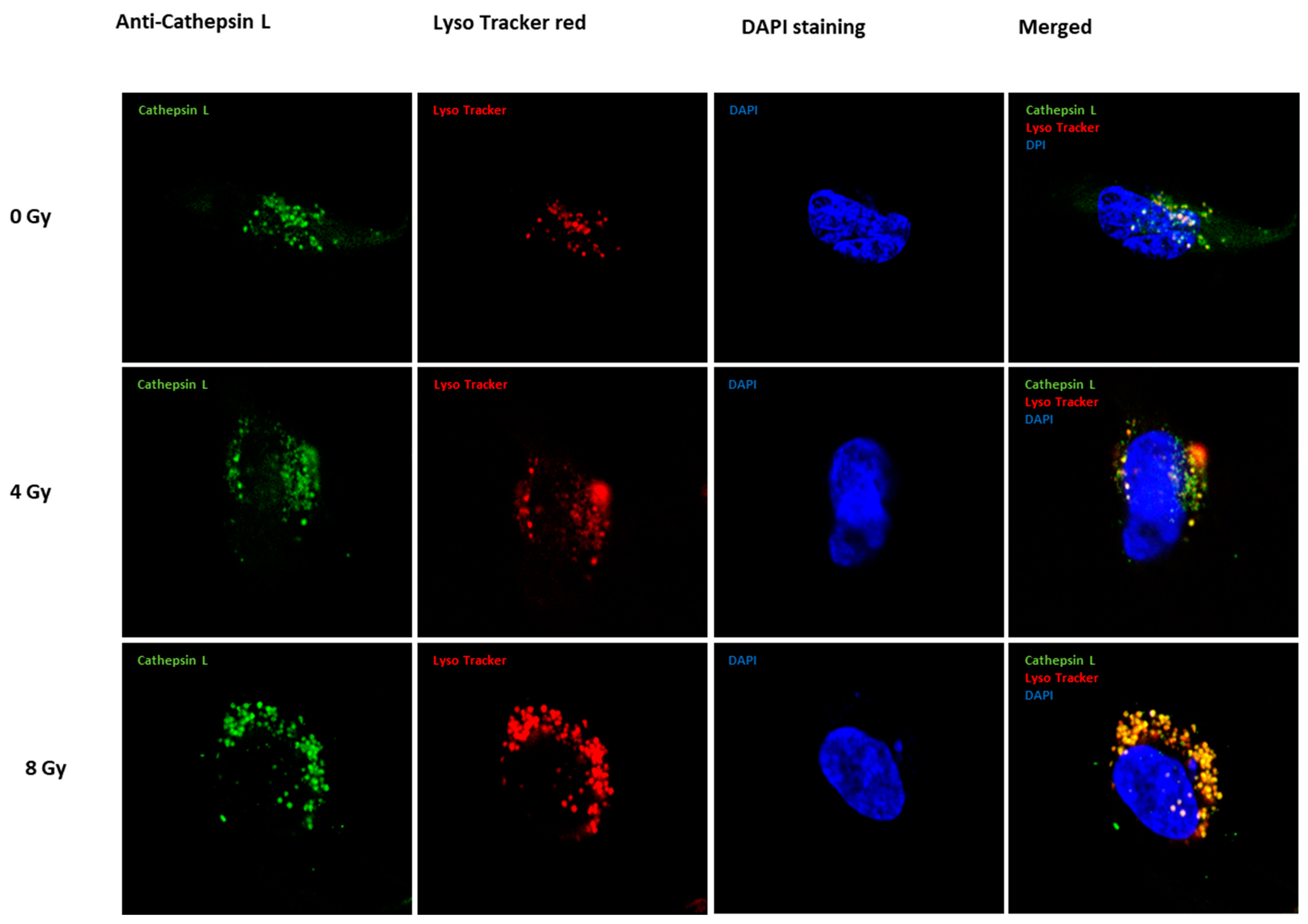
2.1. Expression of Cath L after Exposure to Ascending Doses of Ionizing Irradiation
2.2. In Vitro Labeling of Cysteine Cathepsins and Pull-Down of Cath L in Irradiated and Non-Irradiated Caco-2 Cells
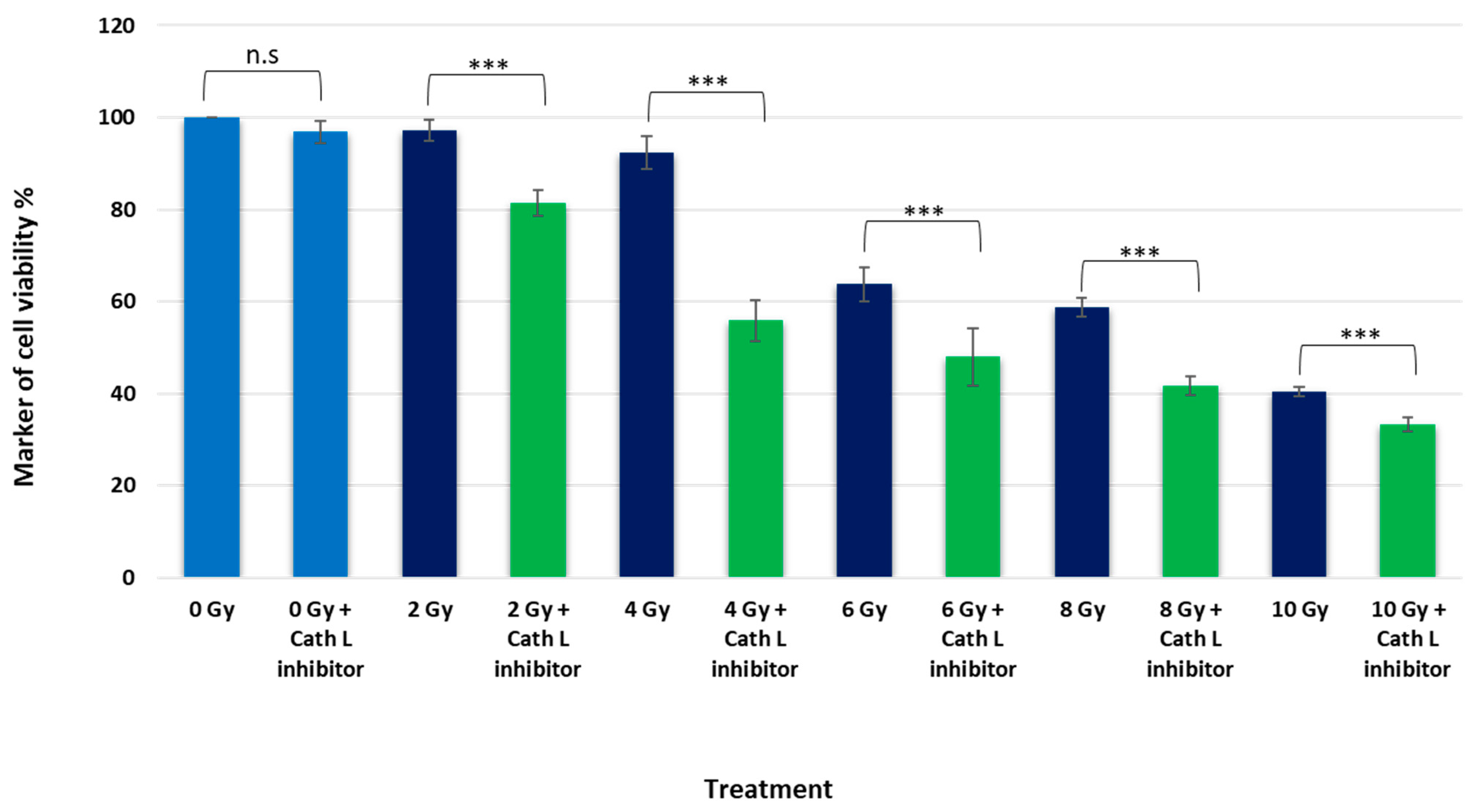
2.3. Unraveling the Impact of Cath L Inhibition on Ionizing Irradiation-Induced Cytotoxicity in Caco-2 Cells
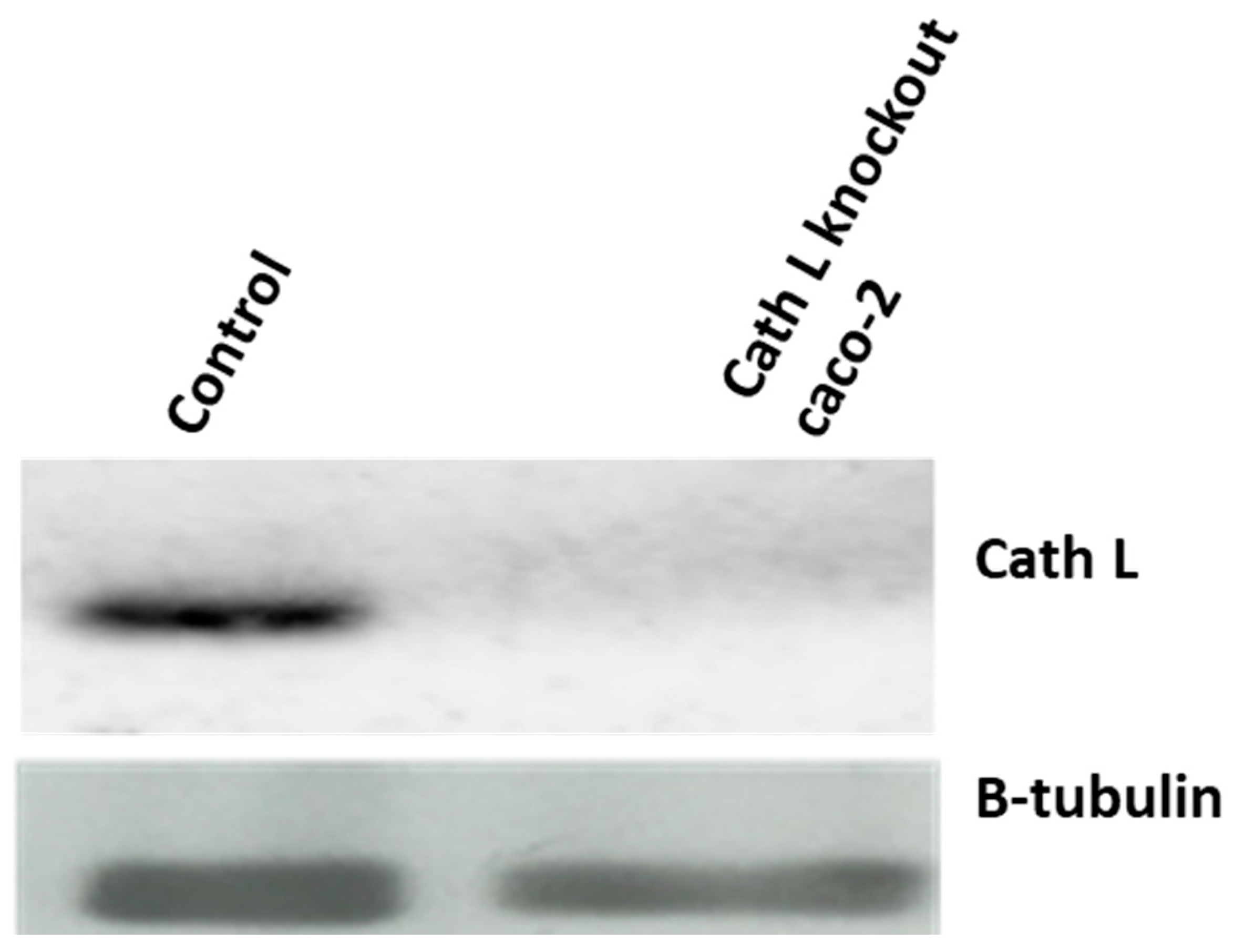
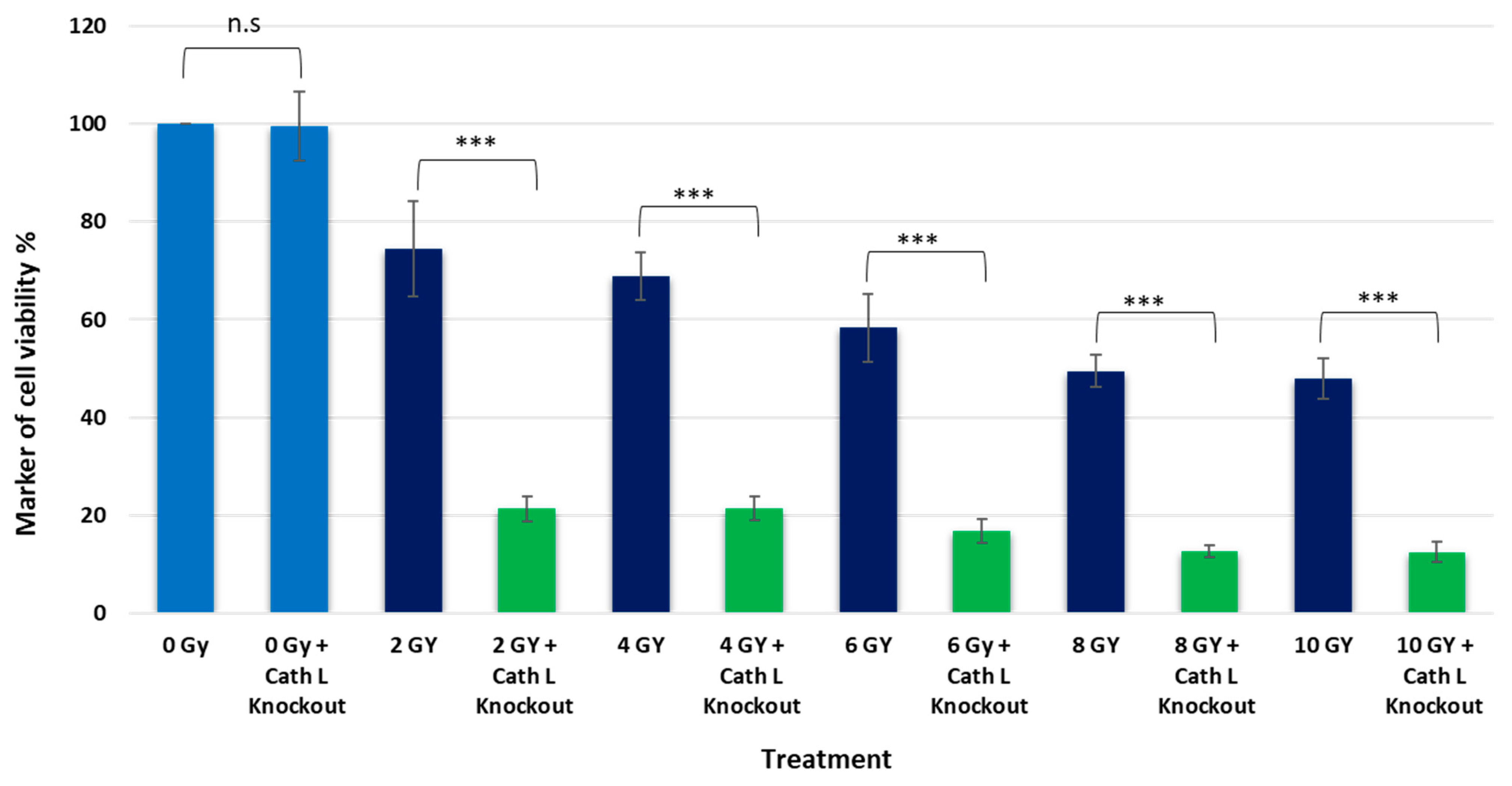
2.4. Cath L Knockout by CRISPR/Cas9
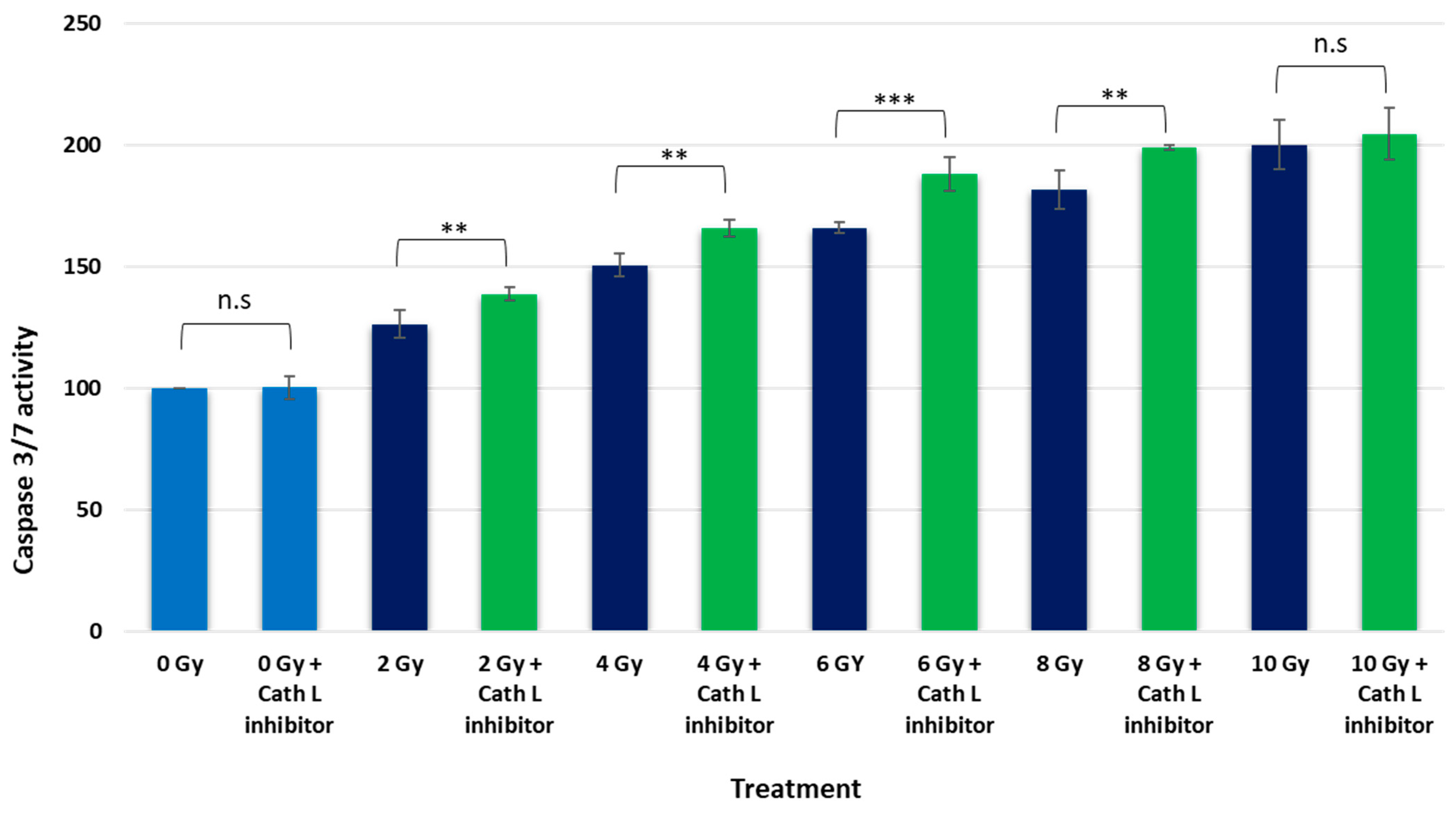
2.5. Caspase 3/7 Activity Assay upon Ionizing Irradiation in the Presence and Absence of Cath L Inhibitor
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Radiation Treatment
4.3. Western Blotting
4.4. Fluorescent Immunocytochemistry Detection and Microscopy
4.5. Active Site Labeling of Cysteine Cathepsins
4.6. Pull-Down of DCG04-Labeled Cysteine Cathepsins
4.7. Cell Viability & Cytotoxicity Test
4.8. CRISPR Cas9 and Cath L Gene Knockout
4.9. Apoptosis Assay
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Caths | Cathepsins |
| RT | Radiation therapy |
| Caco-2 | Colon Carcinoma Cells |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CRC | Colorectal cancer |
| SSBs | Single-strand breaks |
| DSBs | Double-strand breaks |
| IR | Ionizing radiation |
| Gy | Gray |
| SDS-PAGE | Sodium dodecyl-sulfate polyacrylamide gel electrophoresis |
| SE | Standard error |
| DAPI | 4′,6-diamidino-2-phenylindole |
| kDa | Kilodalton |
| WT | Wild type |
| DMEM | Dulbecco’s Modified Eagle Medium |
| ROS | Reactive Oxygen Species |
| DDR | DNA damage response |
| NHEJ | Non-homologous end-joining |
| HR | Homologous recombination |
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; D’Ippolito, E.; Grassi, R.; Sangiovanni, A.; Gagliardi, F.; De Marco, G.; Menditti, V.S.; D’Ambrosio, L.; Cioce, F.; Boldrini, L.; et al. Non-Oncological Radiotherapy: A Review of Modern Approaches. J. Pers. Med. 2022, 12, 1677. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Baskar, R.; Itahana, K. Radiation therapy and cancer control in developing countries: Can we save more lives? Int. J. Med. Sci. 2017, 14, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.M.; Lynam-Lennon, N.; O’Neill, H.; O’Sullivan, J. Targeting hallmarks of cancer to enhance radiosensitivity in gastrointestinal cancers. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 298–313. [Google Scholar] [CrossRef]
- Nakamura, K.; Sasaki, T.; Ohga, S.; Yoshitake, T.; Terashima, K.; Asai, K.; Matsumoto, K.; Shioyama, Y.; Honda, H. Recent advances in radiation oncology: Intensity-modulated radiotherapy, a clinical perspective. Int. J. Clin. Oncol. 2014, 19, 564–569. [Google Scholar] [CrossRef]
- Park, J.M.; Wu, H.G.; Kim, H.J.; Choi, C.H.; Kim, J.I. Comparison of treatment plans between IMRT with MR-linac and VMAT for lung SABR. Radiat. Oncol. 2019, 14, 105. [Google Scholar] [CrossRef]
- West, C.M.; Huddart, R.A. Biomarkers and Imaging for Precision Radiotherapy. Clin. Oncol. 2015, 27, 545–546. [Google Scholar] [CrossRef]
- Beaton, L.; Bandula, S.; Gaze, M.N.; Sharma, R.A. How rapid advances in imaging are defining the future of precision radiation oncology. Br. J. Cancer 2019, 120, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Tree, A.C.; Khoo, V.S.; Eeles, R.A.; Ahmed, M.; Dearnaley, D.P.; Hawkins, M.A.; Huddart, R.A.; Nutting, C.M.; Ostler, P.J.; van As, N.J. Stereotactic body radiotherapy for oligometastases. Lancet Oncol. 2013, 14, e28–e37. [Google Scholar] [CrossRef] [PubMed]
- Corbin, K.S.; Hellman, S.; Weichselbaum, R.R. Extracranial oligometastases: A subset of metastases curable with stereotactic radiotherapy. J. Clin. Oncol. 2013, 31, 1384–1390. [Google Scholar] [CrossRef] [PubMed]
- Schaue, D.; McBride, W.H. Counteracting tumor radioresistance by targeting DNA repair. Mol. Cancer Ther. 2005, 4, 1548–1550. [Google Scholar] [CrossRef] [PubMed]
- Fowler, J.F.; Adams, G.E.; Denekamp, J. Radiosensitizers of hypoxic cells in solid tumors. Cancer Treat. Rev. 1976, 3, 227–256. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Zhang, Y.; Liu, C.; Zhang, M.; Han, S. Application of Radiosensitizers in Cancer Radiotherapy. Int. J. Nanomed. 2021, 16, 1083–1102. [Google Scholar] [CrossRef] [PubMed]
- De Ruysscher, D.; Niedermann, G.; Burnet, N.G.; Siva, S.; Lee, A.W.M.; Hegi-Johnson, F. Radiotherapy toxicity. Nat. Rev. Dis. Primers 2019, 5, 13. [Google Scholar] [CrossRef]
- Wang, H.; Mu, X.; He, H.; Zhang, X.D. Cancer Radiosensitizers. Trends Pharmacol. Sci. 2018, 39, 24–48. [Google Scholar] [CrossRef]
- Mahamud, O.; So, J.; Chua, M.L.K.; Bristow, R.G. Targeting DNA repair for precision radiotherapy: Balancing the therapeutic ratio. Curr. Probl. Cancer 2017, 41, 265–272. [Google Scholar] [CrossRef]
- Morgan, M.A.; Lawrence, T.S. Molecular Pathways: Overcoming Radiation Resistance by Targeting DNA Damage Response Pathways. Clin. Cancer Res. 2015, 21, 2898–2904. [Google Scholar] [CrossRef]
- Nahas, S.A.; Gatti, R.A. DNA double strand break repair defects, primary immunodeficiency disorders, and ‘radiosensitivity’. Curr. Opin. Allergy Clin. Immunol. 2009, 9, 510–516. [Google Scholar] [CrossRef]
- Fred, C.L. The DNA damage response—From cell biology to human disease. J. Transl. Genet. Genom. 2022, 6, 204–222. [Google Scholar] [CrossRef]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Guirouilh-Barbat, J.; Lambert, S.; Bertrand, P.; Lopez, B.S. Is homologous recombination really an error-free process? Front. Genet. 2014, 5, 175. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T.; Rogakou, E.P.; Yamazaki, V.; Kirchgessner, C.U.; Gellert, M.; Bonner, W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000, 10, 886–895. [Google Scholar] [CrossRef]
- Lanz, M.C.; Dibitetto, D.; Smolka, M.B. DNA damage kinase signaling: Checkpoint and repair at 30 years. EMBO J. 2019, 38, e101801. [Google Scholar] [CrossRef] [PubMed]
- Maier, P.; Hartmann, L.; Wenz, F.; Herskind, C. Cellular Pathways in Response to Ionizing Radiation and Their Targetability for Tumor Radiosensitization. Int. J. Mol. Sci. 2016, 17, 102. [Google Scholar] [CrossRef]
- Powell, S.N.; Kachnic, L.A. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene 2003, 22, 5784–5791. [Google Scholar] [CrossRef]
- Wu, Q.; Allouch, A.; Martins, I.; Brenner, C.; Modjtahedi, N.; Deutsch, E.; Perfettini, J.L. Modulating Both Tumor Cell Death and Innate Immunity Is Essential for Improving Radiation Therapy Effectiveness. Front. Immunol. 2017, 8, 613. [Google Scholar] [CrossRef]
- Huang, R.-X.; Zhou, P.-K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Santivasi, W.L.; Xia, F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid. Redox Signal 2014, 21, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Ranawat, P.; Rawat, S. Radiation resistance in thermophiles: Mechanisms and applications. World J. Microbiol. Biotechnol. 2017, 33, 112. [Google Scholar] [CrossRef] [PubMed]
- Abbott, E.M.; Falzone, N.; Lenzo, N.; Vallis, K.A. Combining External Beam Radiation and Radionuclide Therapies: Rationale, Radiobiology, Results and Roadblocks. Clin. Oncol. 2021, 33, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Ghaderi, N.; Jung, J.; Brüningk, S.C.; Subramanian, A.; Nassour, L.; Peacock, J. A Century of Fractionated Radiotherapy: How Mathematical Oncology Can Break the Rules. Int. J. Mol. Sci. 2022, 23, 1316. [Google Scholar] [CrossRef] [PubMed]
- Biau, J.; Chautard, E.; Verrelle, P.; Dutreix, M. Altering DNA Repair to Improve Radiation Therapy: Specific and Multiple Pathway Targeting. Front. Oncol. 2019, 9, 1009. [Google Scholar] [CrossRef] [PubMed]
- Hubenak, J.R.; Zhang, Q.; Branch, C.D.; Kronowitz, S.J. Mechanisms of injury to normal tissue after radiotherapy: A review. Plast. Reconstr. Surg. 2014, 133, 49e–56e. [Google Scholar] [CrossRef]
- Pawlik, T.M.; Keyomarsi, K. Role of cell cycle in mediating sensitivity to radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 928–942. [Google Scholar] [CrossRef]
- Bouleftour, W.; Rowinski, E.; Louati, S.; Sotton, S.; Wozny, A.S.; Moreno-Acosta, P.; Mery, B.; Rodriguez-Lafrasse, C.; Magne, N. A Review of the Role of Hypoxia in Radioresistance in Cancer Therapy. Med. Sci. Monit. 2021, 27, e934116. [Google Scholar] [CrossRef]
- Komorowska, D.; Radzik, T.; Kalenik, S.; Rodacka, A. Natural Radiosensitizers in Radiotherapy: Cancer Treatment by Combining Ionizing Radiation with Resveratrol. Int. J. Mol. Sci. 2022, 23, 10627. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, C.; Chen, H.; Ren, M.; Liu, X. Cathepsins Trigger Cell Death and Regulate Radioresistance in Glioblastoma. Cells 2022, 11, 4108. [Google Scholar] [CrossRef]
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016, 129, 4329–4339. [Google Scholar] [CrossRef] [PubMed]
- Chwieralski, C.E.; Welte, T.; Bühling, F. Cathepsin-regulated apoptosis. Apoptosis 2006, 11, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Lyu, X.; Yuan, S.; Wang, S.; Li, W.; Chen, Z.; Yu, H.; Li, f.; Jiang, Q. Oxidative stress: A critical hint in ionizing radiation induced pyroptosis. Radiat. Med. Prot. 2020, 1, 179–185. [Google Scholar] [CrossRef]
- Nagakannan, P.; Eftekharpour, E. Differential redox sensitivity of cathepsin B and L holds the key to autophagy-apoptosis interplay after Thioredoxin reductase inhibition in nutritionally stressed SH-SY5Y cells. Free Radic. Biol. Med. 2017, 108, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Thirusangu, P.; Pathoulas, C.L.; Ray, U.; Xiao, Y.; Staub, J.; Jin, L.; Khurana, A.; Shridhar, V. Quinacrine-Induced Autophagy in Ovarian Cancer Triggers Cathepsin-L Mediated Lysosomal/Mitochondrial Membrane Permeabilization and Cell Death. Cancers 2021, 13, 2004. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.-F.; Wu, C.-L.; Huang, S.-C.; Wu, C.-M.; Hsiao, J.-R.; Yo, Y.-T.; Chen, Y.-H.; Shiau, A.-L.; Chou, C.-Y. Cathepsin L mediates resveratrol-induced autophagy and apoptotic cell death in cervical cancer cells. Autophagy 2009, 5, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, X.; Xu, S.; Li, X.; Ma, X. Cathepsin B contributes to radioresistance by enhancing homologous recombination in glioblastoma. Biomed. Pharmacother. 2018, 107, 390–396. [Google Scholar] [CrossRef]
- Abdelaziz, R.F.; Hussein, A.M.; Kotob, M.H.; Weiss, C.; Chelminski, K.; Studenik, C.R.; Aufy, M. The Significance of Cathepsin B in Mediating Radiation Resistance in Colon Carcinoma Cell Line (Caco-2). Int. J. Mol. Sci. 2023, 24, 16146. [Google Scholar] [CrossRef]
- Seo, H.R.; Bae, S.; Lee, Y.S. Radiation-induced cathepsin S is involved in radioresistance. Int. J. Cancer 2009, 124, 1794–1801. [Google Scholar] [CrossRef]
- Yang, N.; Wang, P.; Wang, W.J.; Song, Y.Z.; Liang, Z.Q. Inhibition of cathepsin L sensitizes human glioma cells to ionizing radiation in vitro through NF-kappa B signaling pathway. Acta Pharmacol. Sin. 2015, 36, 400–410. [Google Scholar] [CrossRef]
- Aufy, M.; Abdelaziz, R.F.; Hussein, A.M.; Topcagic, N.; Shamroukh, H.; Abdel-Maksoud, M.A.; Salem, T.Z.; Studenik, C.R. Impact of Enniatin B and Beauvericin on Lysosomal Cathepsin B Secretion and Apoptosis Induction. Int. J. Mol. Sci. 2023, 24, 2030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-q.; Wang, W.-j.; Li, J.; Yang, N.; Chen, G.; Wang, Z.; Liang, Z.-q. Cathepsin L suppression increases the radiosensitivity of human glioma U251 cells via G2/M cell cycle arrest and DNA damage. Acta Pharmacol. Sin. 2015, 36, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yang, P.; Li, C.; Wang, W.-a.; Zhang, T.; Li, J.; Li, H.; Dong, C.; Meng, W.; Zhou, H. Ionizing radiation triggers mitophagy to enhance DNA damage in cancer cells. Cell Death Discov. 2023, 9, 267. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, D.; Stigbrand, T. Radiation-induced cell death mechanisms. Tumour Biol. 2010, 31, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.S. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef]
- Fan, T.J.; Han, L.H.; Cong, R.S.; Liang, J. Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. 2005, 37, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Vinod, S.K.; Hau, E. Radiotherapy treatment for lung cancer: Current status and future directions. Respirology 2020, 25 (Suppl. S2), 61–71. [Google Scholar] [CrossRef]
- Natoli, M.; Leoni, B.D.; D’Agnano, I.; Zucco, F.; Felsani, A. Good Caco-2 cell culture practices. Toxicol. Vitr. 2012, 26, 1243–1246. [Google Scholar] [CrossRef]
- Lea, T. Caco-2 Cell Line. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 103–111. [Google Scholar]
- Hernández-Bojórquez, M.; Trejo-Solis, C.; Lárraga-Gutiérrez, J.M.; Martínez-Dávalos, A. Monte Carlo dosimetry of a cell culture irradiation model using a 6 MV X-ray beam. Radiat. Phys. Chem. 2021, 180, 109251. [Google Scholar] [CrossRef]
- Hao, J.; Magnelli, A.; Godley, A.; Yu, J.S. Use of a Linear Accelerator for Conducting In Vitro Radiobiology Experiments. J. Vis. Exp. 2019, 147, e59514. [Google Scholar] [CrossRef]
- Ahmad, F.; Leake, D.S. Lysosomal oxidation of LDL alters lysosomal pH, induces senescence, and increases secretion of pro-inflammatory cytokines in human macrophages[S]. J. Lipid Res. 2019, 60, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.M.; Aufy, M.; Shabbir, W.; Lemmens-Gruber, R. Identification of phosphorylation sites and binding pockets for modulation of Na(V) 1.5 channel by Fyn tyrosine kinase. FEBS J. 2018, 285, 2520–2530. [Google Scholar] [CrossRef] [PubMed]
- Willam, A.; Aufy, M.; Tzotzos, S.; Evanzin, H.; Chytracek, S.; Geppert, S.; Fischer, B.; Fischer, H.; Pietschmann, H.; Czikora, I.; et al. Restoration of Epithelial Sodium Channel Function by Synthetic Peptides in Pseudohypoaldosteronism Type 1B Mutants. Front. Pharmacol. 2017, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Poreba, M.; Groborz, K.; Vizovisek, M.; Maruggi, M.; Turk, D.; Turk, B.; Powis, G.; Drag, M.; Salvesen, G.S. Fluorescent probes towards selective cathepsin B detection and visualization in cancer cells and patient samples. Chem. Sci. 2019, 10, 8461–8477. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.; Gantchev, J.; Ghazawi, F.M.; Litvinov, I.V. Protocol for adhesion and immunostaining of lymphocytes and other non-adherent cells in culture. Biotechniques 2017, 63, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, D.; Medzihradszky, K.F.; Burlingame, A.; Bogyo, M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 2000, 7, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Shabbir, W.; Topcagic, N.; Aufy, M. Activation of autosomal recessive Pseudohypoaldosteronism1 ENaC with aldosterone. Eur. J. Pharmacol. 2021, 901, 174090. [Google Scholar] [CrossRef]
- Nagahama, M.; Kobayashi, K.; Takehara, M. Cathepsin Release from Lysosomes Promotes Endocytosis of Clostridium perfringens Iota-Toxin. Toxins 2021, 13, 721. [Google Scholar] [CrossRef]
- Ghasemi, M.; Liang, S.; Luu, Q.M.; Kempson, I. The MTT Assay: A Method for Error Minimization and Interpretation in Measuring Cytotoxicity and Estimating Cell Viability. Methods Mol. Biol. 2023, 2644, 15–33. [Google Scholar] [CrossRef]
- Labuda, R.; Bacher, M.; Gratzl, H.; Doppler, M.; Parich, A.; Aufy, M.; Lemmens-Gruber, R.; Schuhmacher, R.; Rychli, K.; Wagner, M.; et al. Luteapyrone, a Novel ƴ-Pyrone Isolated from the Filamentous Fungus Metapochonia lutea. Molecules 2021, 26, 6589. [Google Scholar] [CrossRef] [PubMed]
- Moreira, V.P.; da Silva Mela, M.F.; Anjos, L.R.d.; Saraiva, L.F.; Arenas Velásquez, A.M.; Kalaba, P.; Fabisiková, A.; Clementino, L.d.C.; Aufy, M.; Studenik, C.; et al. Novel Selective and Low-Toxic Inhibitor of LmCPB2.8ΔCTE (CPB) One Important Cysteine Protease for Leishmania Virulence. Biomolecules 2022, 12, 1903. [Google Scholar]
- Shabbir, W.; Topcagic, N.; Aufy, M.; Oz, M. CRISPR/Cas9 Mediated Knock Down of δ-ENaC Blunted the TNF-Induced Activation of ENaC in A549 Cells. Int. J. Mol. Sci. 2021, 22, 1858. [Google Scholar] [CrossRef] [PubMed]
- Gnanamony, M.; Gondi, C.S. Targeting the Expression of Cathepsin B Using CRISPR/Cas9 System in Mammalian Cancer Cells. Methods Mol. Biol. 2018, 1731, 123–131. [Google Scholar] [CrossRef]
- Wesierska-Gadek, J.; Gueorguieva, M.; Ranftler, C.; Zerza-Schnitzhofer, G. A new multiplex assay allowing simultaneous detection of the inhibition of cell proliferation and induction of cell death. J. Cell Biochem. 2005, 96, 1–7. [Google Scholar] [CrossRef]
- Kuete, V.; Sandjo, L.P.; Ouete, J.L.; Fouotsa, H.; Wiench, B.; Efferth, T. Cytotoxicity and modes of action of three naturally occurring xanthones (8-hydroxycudraxanthone G, morusignin I and cudraxanthone I) against sensitive and multidrug-resistant cancer cell lines. Phytomedicine 2014, 21, 315–322. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelaziz, R.F.; Hussein, A.M.; Kotob, M.H.; Weiss, C.; Chelminski, K.; Stojanovic, T.; Studenik, C.R.; Aufy, M. Enhancement of Radiation Sensitivity by Cathepsin L Suppression in Colon Carcinoma Cells. Int. J. Mol. Sci. 2023, 24, 17106. https://doi.org/10.3390/ijms242317106
Abdelaziz RF, Hussein AM, Kotob MH, Weiss C, Chelminski K, Stojanovic T, Studenik CR, Aufy M. Enhancement of Radiation Sensitivity by Cathepsin L Suppression in Colon Carcinoma Cells. International Journal of Molecular Sciences. 2023; 24(23):17106. https://doi.org/10.3390/ijms242317106
Chicago/Turabian StyleAbdelaziz, Ramadan F., Ahmed M. Hussein, Mohamed H. Kotob, Christina Weiss, Krzysztof Chelminski, Tamara Stojanovic, Christian R. Studenik, and Mohammed Aufy. 2023. "Enhancement of Radiation Sensitivity by Cathepsin L Suppression in Colon Carcinoma Cells" International Journal of Molecular Sciences 24, no. 23: 17106. https://doi.org/10.3390/ijms242317106
APA StyleAbdelaziz, R. F., Hussein, A. M., Kotob, M. H., Weiss, C., Chelminski, K., Stojanovic, T., Studenik, C. R., & Aufy, M. (2023). Enhancement of Radiation Sensitivity by Cathepsin L Suppression in Colon Carcinoma Cells. International Journal of Molecular Sciences, 24(23), 17106. https://doi.org/10.3390/ijms242317106




