A New RNA-Dependent Cas12g Nuclease
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification and In Silico Characterization of Cas12g Nuclease
2.2. Comparison of the Found Cas12g Variant with the Known Homologue
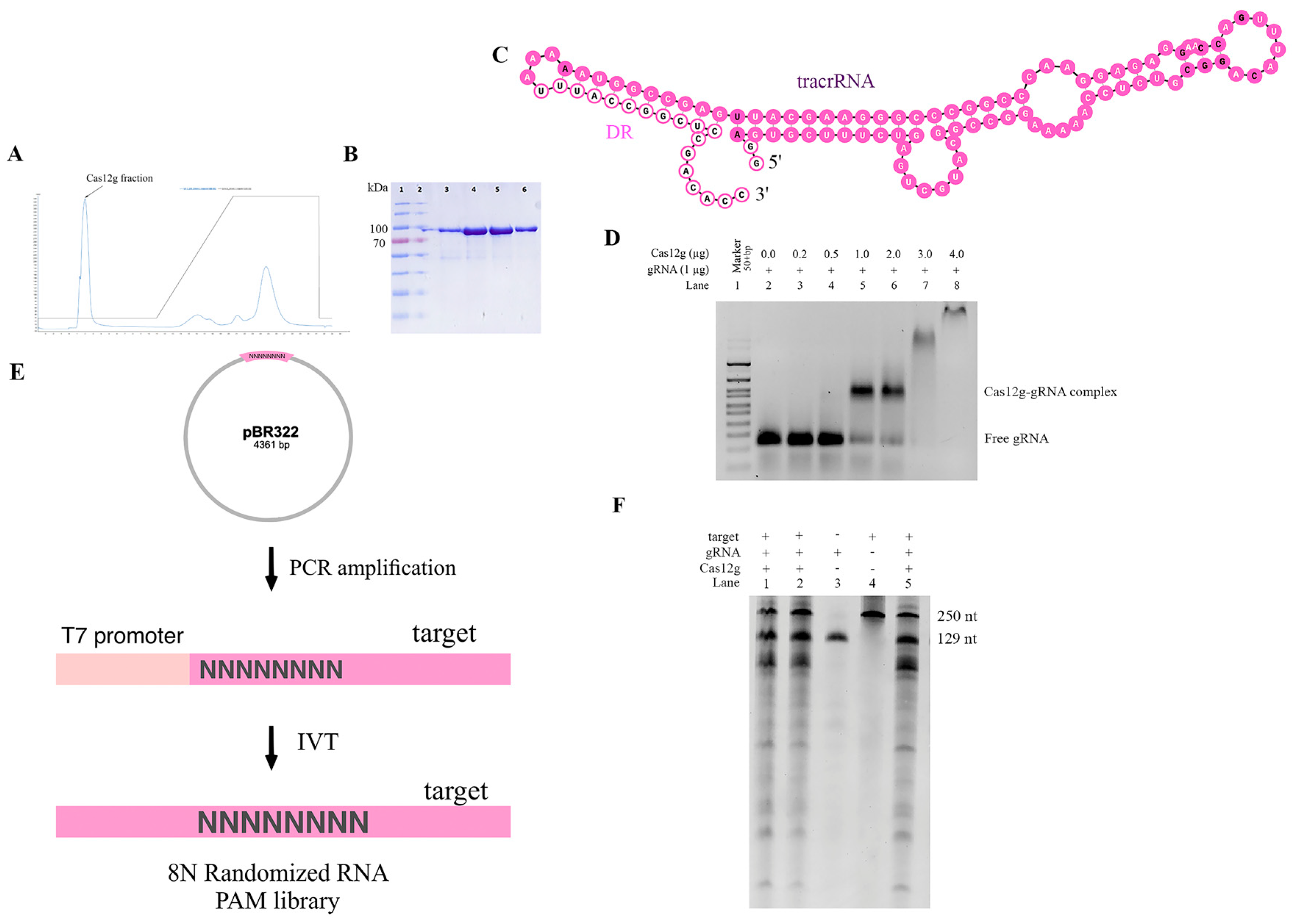
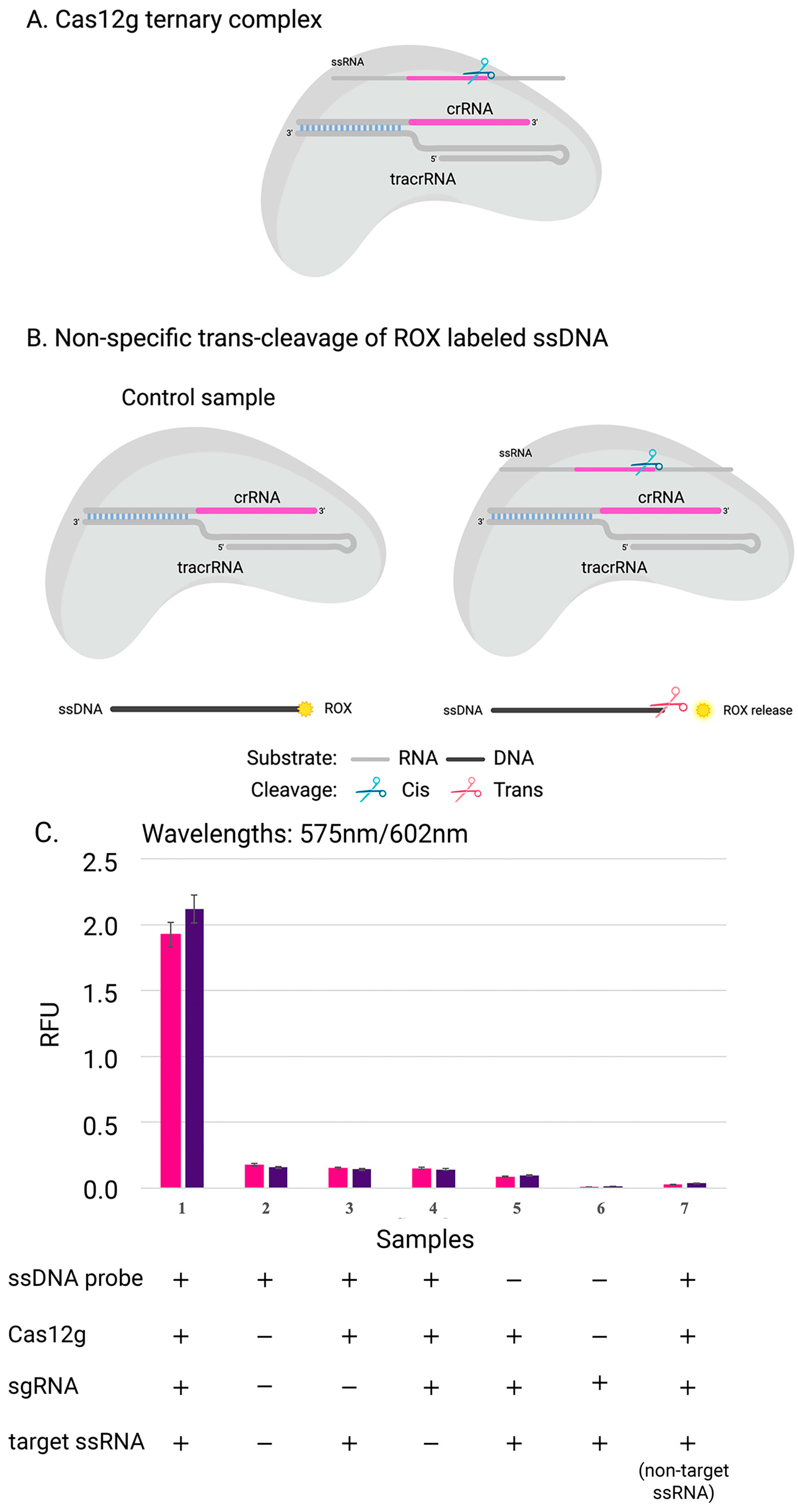
2.3. In Vitro Cas12g Characterization
3. Materials and Methods
3.1. CRISPR-Cas System Identification and Bioinformatics Analysis
3.2. Cas12g Structure Analysis and Visualization
3.3. Phylogenetics
3.4. Bacterial Strains and Growth Conditions
3.5. Plasmids Construction
3.5.1. Recombinant Plasmid Cloning
3.5.2. PAM Library Construction
3.5.3. gRNA Encoding Plasmid Cloning
3.6. Expression and Purification of Cas12g Proteins
3.7. In Vitro RNA Preparation
3.8. Electrophoretic Mobility Shift Assay (EMSA)
3.9. In Vitro Cleavage Assay
3.10. Collateral Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas System and Its Role in Phage-Bacteria Interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Karginov, F.V.; Hannon, G.J. The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. Mol. Cell 2010, 37, 7–19. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. CRISPR-Cas: An Adaptive Immunity System in Prokaryotes. F1000 Biol. Rep. 2009, 1, 95. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Kunin, V.; Hugenholtz, P. CRISPR—A Widespread System That Provides Acquired Resistance against Phages in Bacteria and Archaea. Nat. Rev. Microbiol. 2008, 6, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary Classification of CRISPR–Cas Systems: A Burst of Class 2 and Derived Variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Pinilla-Redondo, R.; Mayo-Muñoz, D.; Russel, J.; Garrett, R.A.; Randau, L.; Sørensen, S.J.; Shah, S.A. Type IV CRISPR–Cas Systems Are Highly Diverse and Involved in Competition between Plasmids. Nucleic Acids Res. 2020, 48, 2000–2012. [Google Scholar] [CrossRef]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR Technologies in Research and Beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, H.; Xiao, R.; Han, R.; Chang, L. Cryo-EM Structure of the RNA-Guided Ribonuclease Cas12g. Nat. Chem. Biol. 2021, 17, 387–393. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Wang, J.Y.; Doudna, J.A. CRISPR Technology: A Decade of Genome Editing Is Only the Beginning. Science 2023, 379, eadd8643. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and Classification of the CRISPR–Cas Systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.A.; Utkina, I.; Wolf, Y.I.; Makarova, K.S.; Severinov, K.V.; Koonin, E.V. CRISPR Arrays Away from Cas Genes. CRISPR J. 2020, 3, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Khanzadi, M.N.; Khan, A.A. CRISPR/Cas9: Nature’s Gift to Prokaryotes and an Auspicious Tool in Genome Editing. J. Basic Microbiol. 2020, 60, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Horvath, P. A Decade of Discovery: CRISPR Functions and Applications. Nat. Microbiol. 2017, 2, 17092. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Platt, R.J.; Zhang, F. Therapeutic Genome Editing: Prospects and Challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas Guides the Future of Genetic Engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An Updated Evolutionary Classification of CRISPR–Cas Systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Gunitseva, N.; Evteeva, M.; Borisova, A.; Patrushev, M.; Subach, F. RNA-Dependent RNA Targeting by CRISPR-Cas Systems: Characterizations and Applications. Int. J. Mol. Sci. 2023, 24, 6894. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, Z.; Chen, J.; Lin, J.; Lu, Q.; Ye, Y.; Zhang, H.; Zhang, B.; Ouyang, S. Structural Transitions upon Guide RNA Binding and Their Importance in Cas12g-Mediated RNA Cleavage. PLOS Genet. 2023, 19, e1010930. [Google Scholar] [CrossRef] [PubMed]
- van Beljouw, S.P.B.; Sanders, J.; Rodríguez-Molina, A.; Brouns, S.J.J. RNA-Targeting CRISPR–Cas Systems. Nat. Rev. Microbiol. 2023, 21, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Morelli, K.H.; Wu, Q.; Gosztyla, M.L.; Liu, H.; Yao, M.; Zhang, C.; Chen, J.; Marina, R.J.; Lee, K.; Jones, K.L.; et al. An RNA-Targeting CRISPR–Cas13d System Alleviates Disease-Related Phenotypes in Huntington’s Disease Models. Nat. Neurosci. 2023, 26, 27–38. [Google Scholar] [CrossRef]
- Cao, C.; Li, A.; Xu, C.; Wu, B.; Liu, J.; Liu, Y. Enhancement of Protein Translation by CRISPR/dCasRx Coupled with SINEB2 Repeat of Noncoding RNAs. Nucleic Acids Res. 2023, 51, 33. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Cheng, Q.-X.; Liu, J.-K.; Nie, X.-Q.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a Has Both Cis- and Trans-Cleavage Activities on Single-Stranded DNA. Cell Res. 2018, 28, 491–493. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Yan, W.X.; Hunnewell, P.; Alfonse, L.E.; Carte, J.M.; Keston-Smith, E.; Sothiselvam, S.; Garrity, A.J.; Chong, S.; Makarova, K.S.; Koonin, E.V.; et al. Functionally Diverse Type V CRISPR-Cas Systems. Science 2019, 363, 88–91. [Google Scholar] [CrossRef]
- Toshchakov, S.V.; Izotova, A.O.; Vinogradova, E.N.; Kachmazov, G.S.; Tuaeva, A.Y.; Abaev, V.T.; Evteeva, M.A.; Gunitseva, N.M.; Korzhenkov, A.A.; Elcheninov, A.G.; et al. Culture-Independent Survey of Thermophilic Microbial Communities of the North Caucasus. Biology 2021, 10, 1352. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The Conserved Domain Database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Garner, M.M.; Revzin, A. The Use of Gel Electrophoresis to Detect and Study Nucleic Acid—Protein Interactions. Trends Biochem. Sci. 1986, 11, 395–396. [Google Scholar] [CrossRef]
- Fried, M.G. Measurement of protein-DNA Interaction Parameters by Electrophoresis Mobility Shift Assay. Electrophoresis 1989, 10, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.; Prentki, P.; Chandler, M. Use of Gel Retardation to Analyze Protein-Nucleic Acid Interactions. Microbiol. Rev. 1992, 56, 509–528. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 Is a Single-Component Programmable RNA-Guided RNA-Targeting CRISPR Effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP—A Flexible Pipeline for Genome-Resolved Metagenomic Data Analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory Friendly Classification with the Genome Taxonomy Database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Vasilev, R.; Gunitseva, N.; Shebanova, R.; Korzhenkov, A.; Vlaskina, A.; Evteeva, M.; Polushkina, I.; Nikitchina, N.; Toshchakov, S.; Kamenski, P.; et al. Targeted Modification of Mammalian DNA by a Novel Type V Cas12a Endonuclease from Ruminococcus Bromii. Int. J. Mol. Sci. 2022, 23, 9289. [Google Scholar] [CrossRef] [PubMed]
- Kadam, U.S.; Cho, Y.; Park, T.Y.; Hong, J.C. Aptamer-Based CRISPR-Cas Powered Diagnostics of Diverse Biomarkers and Small Molecule Targets. Appl. Biol. Chem. 2023, 66, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, Z.; Xiao, R.; Chang, L. Mechanisms for Target Recognition and Cleavage by the Cas12i RNA-Guided Endonuclease. Nat. Struct. Mol. Biol. 2020, 27, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Stella, S.; Mesa, P.; Thomsen, J.; Paul, B.; Alcón, P.; Jensen, S.B.; Saligram, B.; Moses, M.E.; Hatzakis, N.S.; Montoya, G. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity. Cell 2018, 175, 1856–1871.e21. [Google Scholar] [CrossRef]
- Yang, H.; Gao, P.; Rajashankar, K.R.; Patel, D.J. PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell 2016, 167, 1814–1828.e12. [Google Scholar] [CrossRef]
- Pausch, P.; Soczek, K.M.; Herbst, D.A.; Tsuchida, C.A.; Al-Shayeb, B.; Banfield, J.F.; Nogales, E.; Doudna, J.A. DNA Interference States of the Hypercompact CRISPR–CasΦ Effector. Nat. Struct. Mol. Biol. 2021, 28, 652–661. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gunitseva, N.; Evteeva, M.; Korzhenkov, A.; Patrushev, M. A New RNA-Dependent Cas12g Nuclease. Int. J. Mol. Sci. 2023, 24, 17105. https://doi.org/10.3390/ijms242317105
Gunitseva N, Evteeva M, Korzhenkov A, Patrushev M. A New RNA-Dependent Cas12g Nuclease. International Journal of Molecular Sciences. 2023; 24(23):17105. https://doi.org/10.3390/ijms242317105
Chicago/Turabian StyleGunitseva, Natalia, Martha Evteeva, Aleksei Korzhenkov, and Maxim Patrushev. 2023. "A New RNA-Dependent Cas12g Nuclease" International Journal of Molecular Sciences 24, no. 23: 17105. https://doi.org/10.3390/ijms242317105
APA StyleGunitseva, N., Evteeva, M., Korzhenkov, A., & Patrushev, M. (2023). A New RNA-Dependent Cas12g Nuclease. International Journal of Molecular Sciences, 24(23), 17105. https://doi.org/10.3390/ijms242317105





