Unique Substrate Recognition and Sodium–Substrate Binding Stoichiometry in a Bacterial Serotonin Transporter, TuriSERT
Abstract
1. Introduction
2. Results
2.1. TuriSERT Expressed in E. coli Cells Is Functional for APP+ Transport
2.2. Ionic Requirement for APP+ Uptake by TuriSERT
2.3. Mutation of Asp262 to Asparagine Renders TuriSERT Cl−-Dependent
2.4. Mutations of the Na1 Site Do Not Alter Na+ Binding Dependence of APP+ Transport
2.5. Identification of 5-HT Binding Site in TuriSERT
2.6. Effect of Antidepressant Drugs on APP+ Transport
3. Discussion
4. Materials and Methods
4.1. Expression Plasmids
4.2. Expression of TuriSERT
4.3. Homology Modeling and Molecular Docking
4.4. APP+ Uptake Assay
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torres, G.E.; Gainetdinov, R.R.; Caron, M.G. Plasma membrane monoamine transporters: Structure, regulation and function. Nat. Rev. Neurosci. 2003, 4, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, A.S.; Andersen, J.; Jorgensen, T.N.; Sorensen, L.; Eriksen, J.; Loland, C.J.; Stromgaard, K.; Gether, U. SLC6 neurotransmitter transporters: Structure, function, and regulation. Pharmacol. Rev. 2011, 63, 585–640. [Google Scholar] [CrossRef] [PubMed]
- Broer, S.; Gether, U. The solute carrier 6 family of transporters. Br. J. Pharmacol. 2012, 167, 256–278. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.H.; Bahar, I. Monoamine transporters: Structure, intrinsic dynamics and allosteric regulation. Nat. Struct. Mol. Biol. 2019, 26, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Gether, U.; Andersen, P.H.; Larsson, O.M.; Schousboe, A. Neurotransmitter transporters: Molecular function of important drug targets. Trends Pharmacol. Sci. 2006, 27, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Kurian, M.A.; Gissen, P.; Smith, M.; Heales, S., Jr.; Clayton, P.T. The monoamine neurotransmitter disorders: An expanding range of neurological syndromes. Lancet Neurol. 2011, 10, 721–733. [Google Scholar] [CrossRef]
- Rudnick, G.; Kramer, R.; Blakely, R.D.; Murphy, D.L.; Verrey, F. The SLC6 transporters: Perspectives on structure, functions, regulation, and models for transporter dysfunction. Pflügers Arch. Eur. J. Physiol. 2014, 466, 25–42. [Google Scholar] [CrossRef]
- Navratna, V.; Gouaux, E. Insights into the mechanism and pharmacology of neurotransmitter sodium symporters. Curr. Opin. Struct. Biol. 2019, 54, 161–170. [Google Scholar] [CrossRef]
- Howell, L.L.; Kimmel, H.L. Monoamine transporters and psychostimulant addiction. Biochem. Pharmacol. 2008, 75, 196–217. [Google Scholar] [CrossRef]
- Yamashita, A.; Singh, S.K.; Kawate, T.; Jin, Y.; Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 2005, 437, 215–223. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Rudnick, G. The cytoplasmic substrate permeation pathway of serotonin transporter. J. Biol. Chem. 2006, 281, 36213–36220. [Google Scholar] [CrossRef] [PubMed]
- Forrest, L.R.; Zhang, Y.W.; Jacobs, M.T.; Gesmonde, J.; Xie, L.; Honig, B.H.; Rudnick, G. Mechanism for alternating access in neurotransmitter transporters. Proc. Natl. Acad. Sci. USA 2008, 105, 10338–10343. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, G. Cytoplasmic permeation pathway of neurotransmitter transporters. Biochemistry 2011, 50, 7462–7475. [Google Scholar] [CrossRef]
- Penmatsa, A.; Wang, K.H.; Gouaux, E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 2013, 503, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Penmatsa, A.; Wang, K.H.; Gouaux, E. X-ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine. Nat. Struct. Mol. Biol. 2015, 22, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.A.; Green, E.M.; Gouaux, E. X-ray structures and mechanism of the human serotonin transporter. Nature 2016, 532, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.A.; Yang, D.; Zhao, Z.; Wen, P.C.; Yoshioka, C.; Tajkhorshid, E.; Gouaux, E. Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport. Nature 2019, 569, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Gouaux, E. Illumination of serotonin transporter mechanism and role of the allosteric site. Sci. Adv. 2021, 7, eabl3587. [Google Scholar] [CrossRef]
- Barker, E.L.; Moore, K.R.; Rakhshan, F.; Blakely, R.D. Transmembrane domain I contributes to the permeation pathway for serotonin and ions in the serotonin transporter. J. Neurosci. 1999, 19, 4705–4717. [Google Scholar] [CrossRef]
- Singh, S.K.; Piscitelli, C.L.; Yamashita, A.; Gouaux, E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science 2008, 322, 1655–1661. [Google Scholar] [CrossRef]
- Malinauskaite, L.; Quick, M.; Reinhard, L.; Lyons, J.A.; Yano, H.; Javitch, J.A.; Nissen, P. A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat. Struct. Mol. Biol. 2014, 21, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Shahsavar, A.; Stohler, P.; Bourenkov, G.; Zimmermann, I.; Siegrist, M.; Guba, W.; Pinard, E.; Sinning, S.; Seeger, M.A.; Schneider, T.R.; et al. Structural insights into the inhibition of glycine reuptake. Nature 2021, 591, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Motiwala, Z.; Aduri, N.G.; Shaye, H.; Han, G.W.; Lam, J.H.; Katritch, V.; Cherezov, V.; Gati, C. Structural basis of GABA reuptake inhibition. Nature 2022, 606, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Noskov, S.Y. The role of local hydration and hydrogen-bonding dynamics in ion and solute release from ion-coupled secondary transporters. Biochemistry 2011, 50, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, H.; Gouaux, E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature 2012, 481, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, G. How do transporters couple solute movements? Mol. Membr. Biol. 2013, 30, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Tavoulari, S.; Margheritis, E.; Nagarajan, A.; DeWitt, D.C.; Zhang, Y.W.; Rosado, E.; Ravera, S.; Rhoades, E.; Forrest, L.R.; Rudnick, G. Two Na+ sites control conformational change in a neurotransmitter transporter homolog. J. Biol. Chem. 2016, 291, 1456–1471. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Tavoulari, S.; Sinning, S.; Aleksandrova, A.A.; Forrest, L.R.; Rudnick, G. Structural elements required for coupling ion and substrate transport in the neurotransmitter transporter homolog LeuT. Proc. Natl. Acad. Sci. USA 2018, 115, E8854–E8862. [Google Scholar] [CrossRef]
- Nelson, N. The family of Na+/Cl− neurotransmitter transporters. J. Neurochem. 1998, 71, 1785–1803. [Google Scholar] [CrossRef]
- Androutsellis-Theotokis, A.; Goldberg, N.R.; Ueda, K.; Beppu, T.; Beckman, M.L.; Das, S.; Javitch, J.A.; Rudnick, G. Characterization of a functional bacterial homologue of sodium-dependent neurotransmitter transporters. J. Biol. Chem. 2003, 278, 12703–12709. [Google Scholar] [CrossRef]
- Tavoulari, S.; Rizwan, A.N.; Forrest, L.R.; Rudnick, G. Reconstructing a chloride-binding site in a bacterial neurotransmitter transporter homologue. J. Biol. Chem. 2011, 286, 2834–2842. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.; Gray, J.A.; Roth, B.L. The expanded biology of serotonin. Annu. Rev. Med. 2009, 60, 355–366. [Google Scholar] [CrossRef]
- Layunta, E.; Buey, B.; Mesonero, J.E.; Latorre, E. Crosstalk between intestinal serotonergic system and pattern recognition receptors on the microbiota-gut-brain axis. Front. Endocrinol. 2021, 12, 748254. [Google Scholar] [CrossRef] [PubMed]
- Mohammad-Zadeh, L.F.; Moses, L.; Gwaltney-Brant, S.M. Serotonin: A review. J. Vet. Pharmacol. Ther. 2008, 31, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Mawe, G.M.; Hoffman, J.M. Serotonin signalling in the gut--functions, dysfunctions and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 473–486. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Fung, T.C.; Vuong, H.E.; Luna, C.D.G.; Pronovost, G.N.; Aleksandrova, A.A.; Riley, N.G.; Vavilina, A.; McGinn, J.; Rendon, T.; Forrest, L.R.; et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol. 2019, 4, 2064–2073. [Google Scholar] [CrossRef]
- Forrest, L.R.; Tavoulari, S.; Zhang, Y.W.; Rudnick, G.; Honig, B. Identification of a chloride ion binding site in Na+/Cl--dependent transporters. Proc. Natl. Acad. Sci. USA 2007, 104, 12761–12766. [Google Scholar] [CrossRef]
- Zomot, E.; Bendahan, A.; Quick, M.; Zhao, Y.; Javitch, J.A.; Kanner, B.I. Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature 2007, 449, 726–730. [Google Scholar] [CrossRef]
- Barker, E.L.; Perlman, M.A.; Adkins, E.M.; Houlihan, W.J.; Pristupa, Z.B.; Niznik, H.B.; Blakely, R.D. High affinity recognition of serotonin transporter antagonists defined by species-scanning mutagenesis. An aromatic residue in transmembrane domain I dictates species-selective recognition of citalopram and mazindol. J. Biol. Chem. 1998, 273, 19459–19468. [Google Scholar] [CrossRef]
- Henry, L.K.; Field, J.R.; Adkins, E.M.; Parnas, M.L.; Vaughan, R.A.; Zou, M.F.; Newman, A.H.; Blakely, R.D. Tyr-95 and Ile-172 in transmembrane segments 1 and 3 of human serotonin transporters interact to establish high affinity recognition of antidepressants. J. Biol. Chem. 2006, 281, 2012–2023. [Google Scholar] [CrossRef] [PubMed]
- Walline, C.C.; Nichols, D.E.; Carroll, F.I.; Barker, E.L. Comparative molecular field analysis using selectivity fields reveals residues in the third transmembrane helix of the serotonin transporter associated with substrate and antagonist recognition. J. Pharmacol. Exp. Ther. 2008, 325, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Taboureau, O.; Hansen, K.B.; Olsen, L.; Egebjerg, J.; Stromgaard, K.; Kristensen, A.S. Location of the antidepressant binding site in the serotonin transporter: Importance of Ser-438 in recognition of citalopram and tricyclic antidepressants. J. Biol. Chem. 2009, 284, 10276–10284. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Olsen, L.; Hansen, K.B.; Taboureau, O.; Jorgensen, F.S.; Jorgensen, A.M.; Bang-Andersen, B.; Egebjerg, J.; Stromgaard, K.; Kristensen, A.S. Mutational mapping and modeling of the binding site for (S)-citalopram in the human serotonin transporter. J. Biol. Chem. 2010, 285, 2051–2063. [Google Scholar] [CrossRef]
- Coleman, J.A.; Gouaux, E. Structural basis for recognition of diverse antidepressants by the human serotonin transporter. Nat. Struct. Mol. Biol. 2018, 25, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Quick, M.; Yano, H.; Goldberg, N.R.; Duan, L.; Beuming, T.; Shi, L.; Weinstein, H.; Javitch, J.A. State-dependent conformations of the translocation pathway in the tyrosine transporter Tyt1, a novel neurotransmitter:sodium symporter from Fusobacterium nucleatum. J. Biol. Chem. 2006, 281, 26444–26454. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Quick, M.; Shi, L.; Mehler, E.L.; Weinstein, H.; Javitch, J.A. Substrate-dependent proton antiport in neurotransmitter:sodium symporters. Nat. Chem. Biol. 2010, 6, 109–116. [Google Scholar] [CrossRef][Green Version]
- Talvenheimo, J.; Fishkes, H.; Nelson, P.J.; Rudnick, G. The serotonin transporter-imipramine “receptor”. J. Biol. Chem. 1983, 258, 6115–6119. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Protein. Sci. 2016, 86, 5–6. [Google Scholar] [CrossRef]
- Olivella, M.; Gonzalez, A.; Pardo, L.; Deupi, X. Relation between sequence and structure in membrane proteins. Bioinformatics 2013, 29, 1589–1592. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Solis, E., Jr.; Zdravkovic, I.; Tomlinson, I.D.; Noskov, S.Y.; Rosenthal, S.J.; De Felice, L.J. 4-(4-(dimethylamino)phenyl)-1-methylpyridinium (APP+) is a fluorescent substrate for the human serotonin transporter. J. Biol. Chem. 2012, 287, 8852–8863. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, Q.; Zhang, Y.W. Determining Ligand and Ion-Induced Conformational Changes in Serotonin Transporter with Its Fluorescent Substrates. Int. J. Mol. Sci. 2022, 23, 10919. [Google Scholar] [CrossRef]
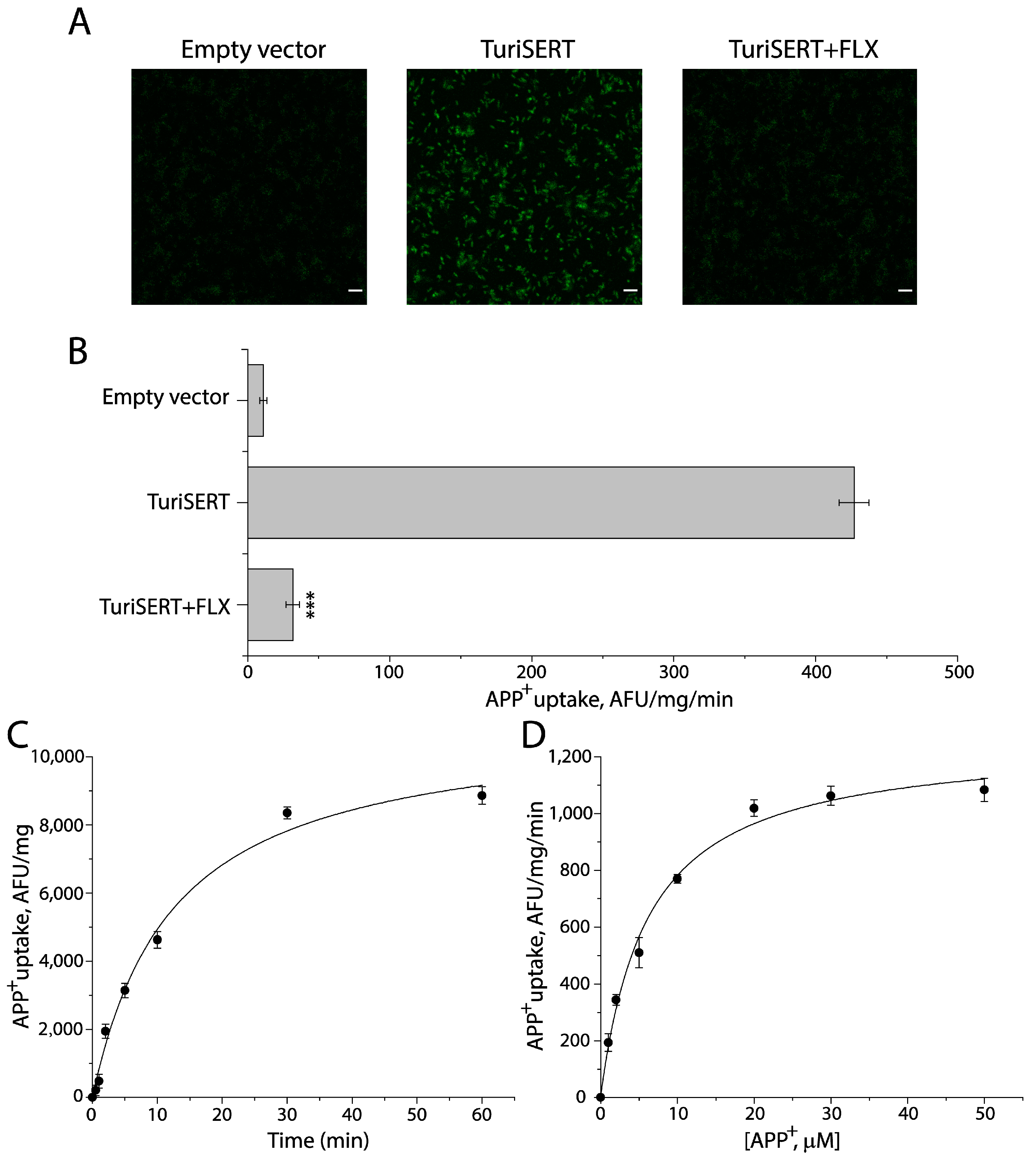
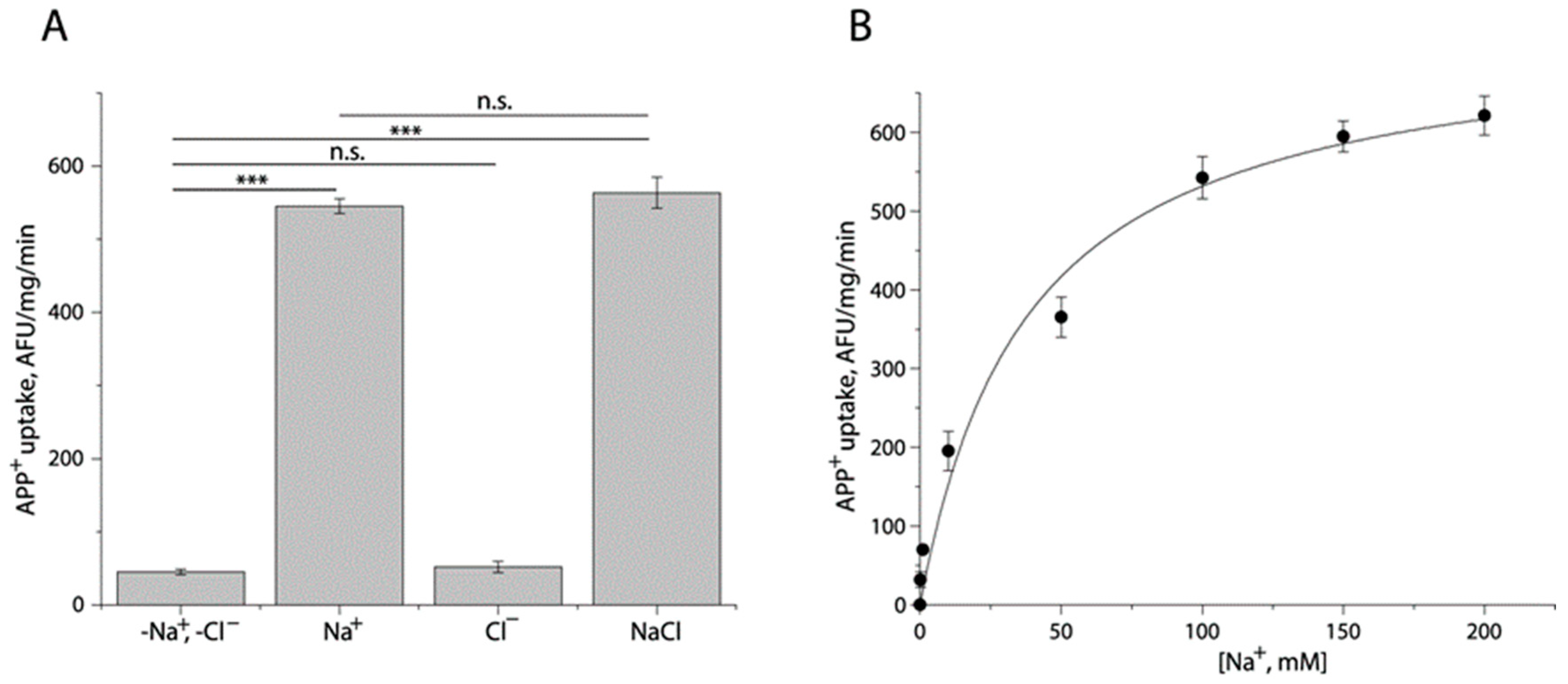

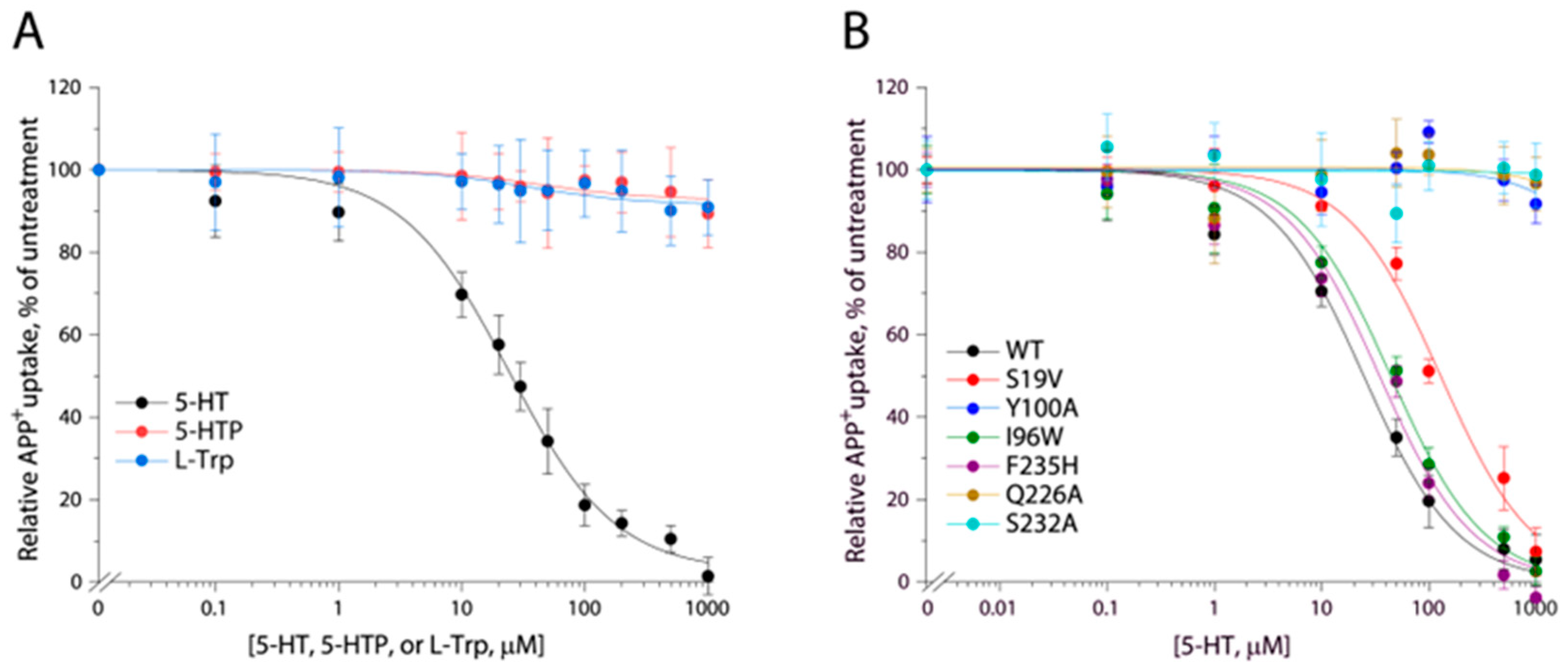

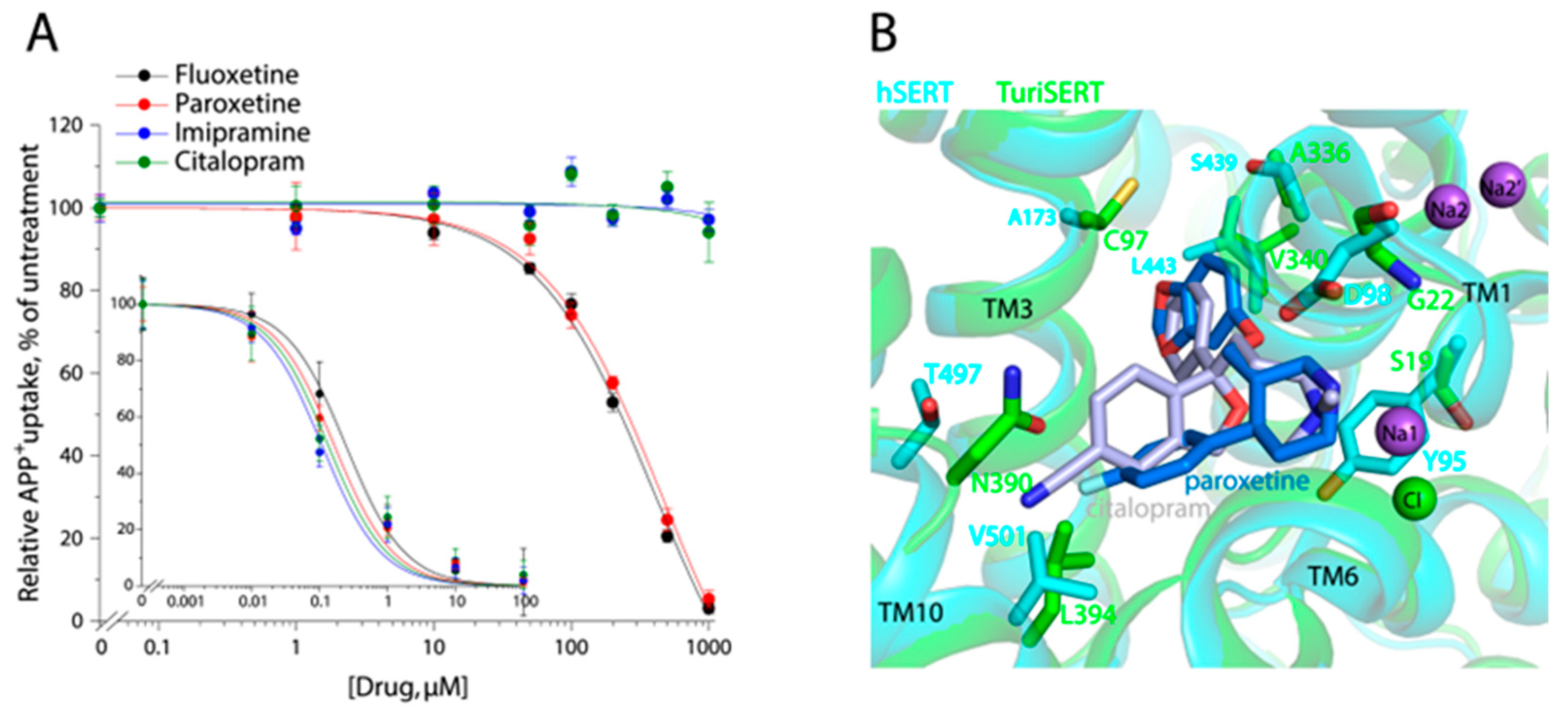
| Binding Site | Km for APP+ (μM) | Vmax (AFU/mg/min) | Km for Na+ (μM) | Ki for 5-HT (μM) | |
|---|---|---|---|---|---|
| WT | 6.12 ± 0.27 | 1285 ± 14 | 33.33 ± 1.32 | 26.13 ± 2.21 | |
| Na1 or Cl− | N25S | 12.78 ± 1.22 * | 857.4 ± 46.08 * | 42.41 ± 0.92 * | 36.25 ± 0.72 |
| S230A | 13.66 ± 0.60 * | 902.5 ± 17.81 * | 37.49 ± 0.81 | 37.82 ± 1.47 | |
| D262N | 14.61 ± 1.43 * | 841.4 ± 22.3 * | 21.56 ± 0.82 * | 65.03 ± 7.53 * | |
| Na2 | S335A | NF | NF | NF | NF |
| 5-HT | S19Y | 10.47 ± 0.40 * | 1205 ± 53.42 | ND | 30.72 ± 1.78 |
| G22D | 10.29 ± 0.54 * | 593.2 ± 59.4 * | 32.33 ± 0.94 | 15.63 ± 0.83 | |
| C97A | 9.93 ± 1.16 | 1195 ± 7.75 | ND | 30.77 ± 1.63 | |
| A336T | 9.23 ± 1.12 | 1431 ± 145.1 | ND | 37.43 ± 1.42 | |
| S19V | 18.22 ± 0.44 ** | 1127 ± 23.99 | ND | 124.32 ± 3.31 ** | |
| Y100A | 19.17 ± 1.99 ** | 773.2 ± 89.66 * | ND | >1000 | |
| I96W | 9.12 ± 1.52 | 1294 ± 175.9 | ND | 41.21 ± 2.16 | |
| F235H | 12.71 ± 0.35 * | 1051 ± 9.82 | ND | 33.77 ± 1.89 | |
| Q226A | 22.32 ± 0.51 ** | 1292 ± 51.22 | ND | >1000 | |
| S232A | 16.24 ± 0.64 ** | 1163 ± 34.43 | ND | >1000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Zhang, X.; Chen, S.; Liu, H.; Zhang, Y.-W. Unique Substrate Recognition and Sodium–Substrate Binding Stoichiometry in a Bacterial Serotonin Transporter, TuriSERT. Int. J. Mol. Sci. 2023, 24, 17112. https://doi.org/10.3390/ijms242317112
Li M, Zhang X, Chen S, Liu H, Zhang Y-W. Unique Substrate Recognition and Sodium–Substrate Binding Stoichiometry in a Bacterial Serotonin Transporter, TuriSERT. International Journal of Molecular Sciences. 2023; 24(23):17112. https://doi.org/10.3390/ijms242317112
Chicago/Turabian StyleLi, Mu, Xintong Zhang, Sixiang Chen, Hanhe Liu, and Yuan-Wei Zhang. 2023. "Unique Substrate Recognition and Sodium–Substrate Binding Stoichiometry in a Bacterial Serotonin Transporter, TuriSERT" International Journal of Molecular Sciences 24, no. 23: 17112. https://doi.org/10.3390/ijms242317112
APA StyleLi, M., Zhang, X., Chen, S., Liu, H., & Zhang, Y.-W. (2023). Unique Substrate Recognition and Sodium–Substrate Binding Stoichiometry in a Bacterial Serotonin Transporter, TuriSERT. International Journal of Molecular Sciences, 24(23), 17112. https://doi.org/10.3390/ijms242317112







