Arabidopsis SEC13B Interacts with Suppressor of Frigida 4 to Repress Flowering
Abstract
:1. Introduction
2. Results
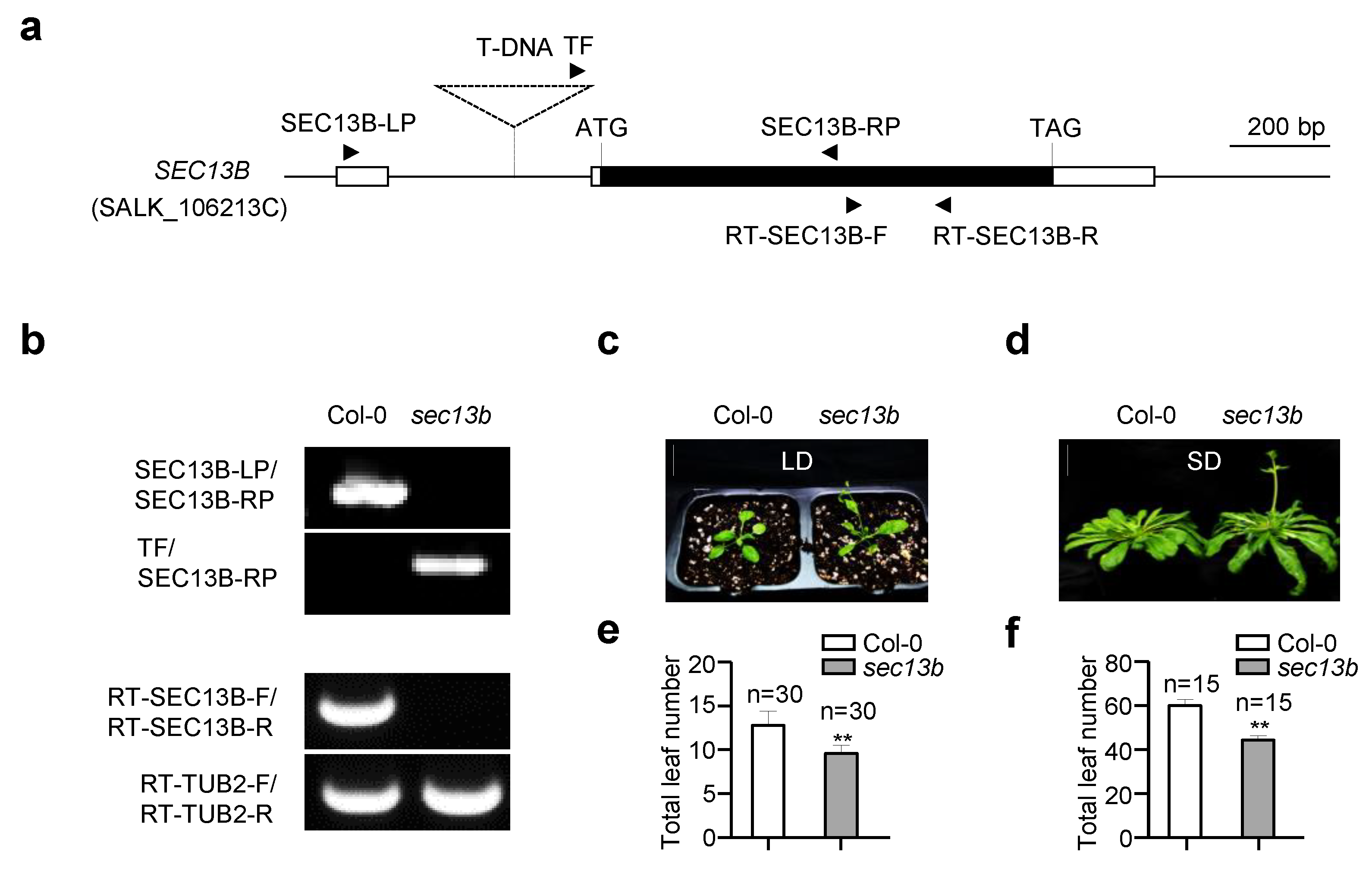
2.1. SEC13B Affects Flowering in Arabidopsis
2.2. SEC13A and SEC13B Are Expressed in the Reproductive Phase, and Their Encoding Proteins Localize in the Nucleus and ER
2.3. SEC13A and SEC13B Interact with SUF4 In Vitro and In Vivo
2.4. SEC13B and SUF4 Mutations Had to Promote Flowering in Arabidopsis
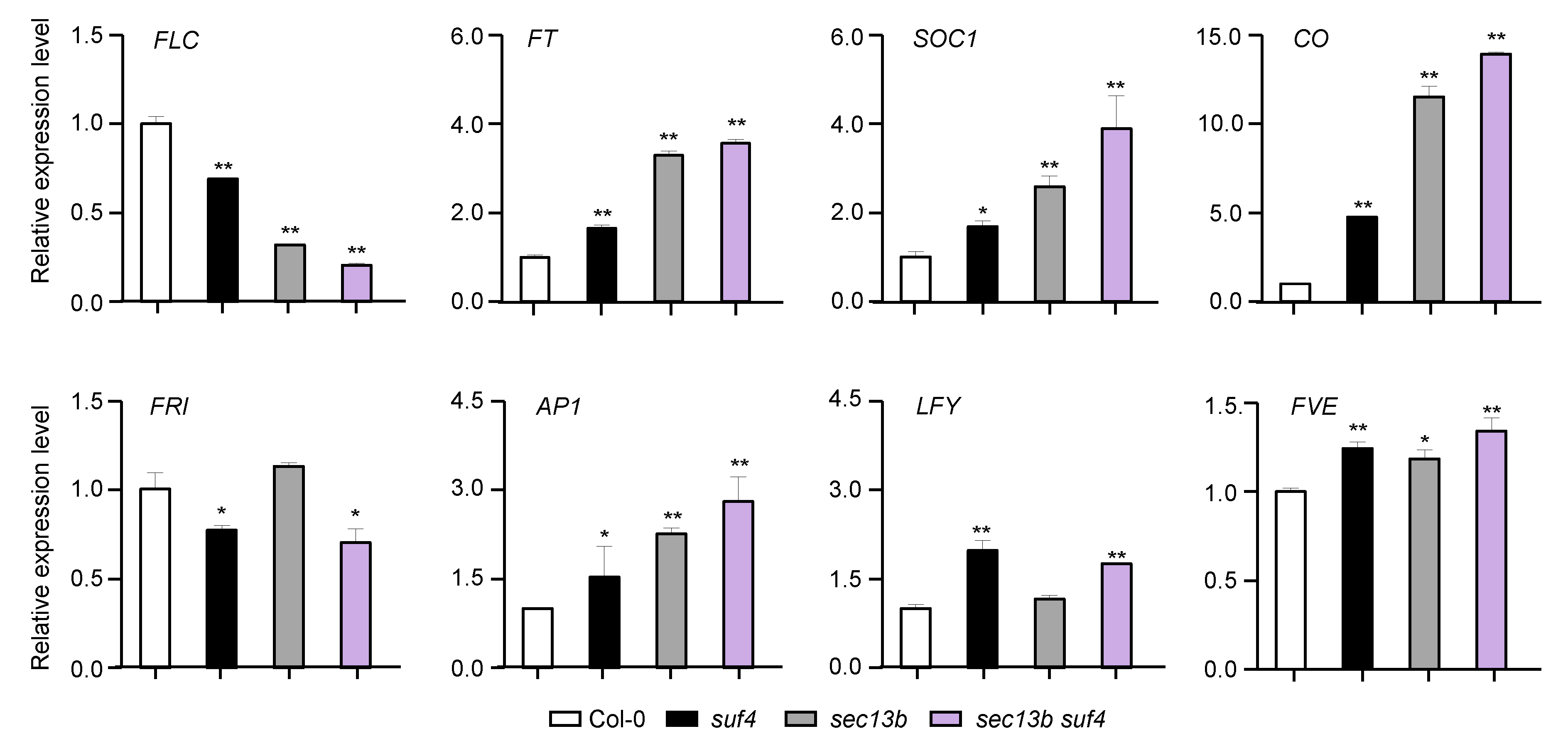
2.5. SEC13B and SUF4 Co-Regulate Flowering-Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction and Plant Transformation
4.3. qRT-PCR Assays
4.4. Yeast Two-Hybrid Assays
4.5. Pull-Down Assays
4.6. LCI Assays
4.7. Co-Immunoprecipitation
4.8. Subcellular Localization Assays
4.9. AGI Locus Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brandizzi, F. Transport from the endoplasmic reticulum to the Golgi in plants: Where are we now? Semin. Cell Dev. Biol. 2018, 80, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Goldberg, J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell 2010, 142, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Hsu, V.; Lee, S.; Yang, J. The evolving understanding of COPI vesicle formation. Nat. Rev. Mol. Cell Biol. 2009, 10, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Peotter, J.; Kasberg, W.; Pustova, I.; Audhya, A. COPII-mediated trafficking at the ER/ERGIC interface. Traffic 2019, 20, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Bassham, D.; Brandizzi, F.; Otegui, M.; Sanderfoot, A. The secretory system of Arabidopsis. Arabidopsis Book 2008, 6, e0116. [Google Scholar] [CrossRef]
- Bhattacharya, N.; O Donnell, J.; Stagg, S. The structure of the Sec13/31 COPII cage bound to Sec23. J. Mol. Biol. 2012, 420, 324–334. [Google Scholar] [CrossRef]
- Fath, S.; Mancias, J.; Bi, X.; Goldberg, J. Structure and organization of coat proteins in the COPII cage. Cell 2007, 129, 1325–1336. [Google Scholar] [CrossRef]
- Hanton, S.; Chatre, L.; Matheson, L.; Rossi, M.; Held, M.; Brandizzi, F. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol. Biol. 2008, 67, 283–294. [Google Scholar] [CrossRef]
- Miller, E.; Beilharz, T.; Malkus, P.; Lee, M.; Hamamoto, S.; Orci, L.; Schekman, R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 2003, 114, 497–509. [Google Scholar] [CrossRef]
- Stagg, S.; Gürkan, C.; Fowler, D.; Lapointe, P.; Foss, T.; Potter, C.; Carragher, B.; Balch, W. Structure of the Sec13/31 COPII coat cage. Nature 2006, 439, 234–238. [Google Scholar] [CrossRef]
- Chung, K.; Zeng, Y.; Jiang, L. COPII paralogs in plants: Functional redundancy or diversity? Trends Plant Sci. 2016, 21, 758–769. [Google Scholar] [CrossRef]
- Liang, X.; Li, S.; Gong, L.; Li, S.; Zhang, Y. COPII components Sar1b and Sar1c play distinct yet interchangeable roles in pollen development. Plant Physiol. 2020, 183, 974–985. [Google Scholar] [CrossRef]
- Zeng, Y.; Chung, K.; Li, B.; Lai, C.; Lam, S.; Wang, X.; Cui, Y.; Gao, C.; Luo, M.; Wong, K.; et al. Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 14360–14365. [Google Scholar] [CrossRef]
- Aboulela, M.; Nakagawa, T.; Oshima, A.; Nishimura, K.; Tanaka, Y. The Arabidopsis COPII components, AtSEC23A and AtSEC23D, are essential for pollen wall development and exine patterning. J. Exp. Bot. 2018, 69, 1615–1633. [Google Scholar] [CrossRef]
- Conger, R.; Chen, Y.; Fornaciari, S.; Faso, C.; Held, M.; Renna, L.; Brandizzi, F. Evidence for the involvement of the Arabidopsis SEC24A in male transmission. J. Exp. Bot. 2011, 62, 4917–4926. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nishimura, K.; Kawamukai, M.; Oshima, A.; Nakagawa, T. Redundant function of two Arabidopsis COPII components, AtSec24B and AtSec24C, is essential for male and female gametogenesis. Planta 2013, 238, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tong, M.; Zhang, A.; Liu, M.; Zhao, B.; Liu, Z.; Li, Z.; Zhu, X.; Guo, Y.; Li, R. COPII genes SEC31A/B are essential for gametogenesis and interchangeable in pollen development in Arabidopsis. Plant J. 2021, 105, 1600–1614. [Google Scholar] [CrossRef] [PubMed]
- Hino, T.; Tanaka, Y.; Kawamukai, M.; Nishimura, K.; Mano, S.; Nakagawa, T. Two Sec13p homologs, AtSec13A and AtSec13B, redundantly contribute to the formation of COPII transport vesicles in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2011, 75, 1848–1852. [Google Scholar] [CrossRef]
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops–what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, C.; Dean, C. The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 2017, 33, 555–575. [Google Scholar] [CrossRef]
- Freytes, S.; Canelo, M.; Cerdan, P. Regulation of flowering time: When and where? Curr. Opin. Plant Biol. 2021, 63, 102049. [Google Scholar] [CrossRef]
- Gu, X.; Wang, Y.; He, Y. Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol. 2013, 11, e1001649. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, X.; Yuan, W.; Schmitz, R.; He, Y. Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev. Cell 2014, 28, 727–736. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Michaels, S.; Amasino, R. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Suh, S.; Park, E.; Cho, E.; Ahn, J.; Kim, S.; Lee, J.; Kwon, Y.; Lee, I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000, 14, 2366–2376. [Google Scholar] [CrossRef] [PubMed]
- Johanson, U.; West, J.; Lister, C.; Michaels, S.; Amasino, R.; Dean, C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 2000, 290, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, J.; Hwang, H.; Kim, S.; Park, C.; Kim, S.; Lee, I. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 2011, 23, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Michaels, S. SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 2006, 133, 4699–4707. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–204. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Gu, X.; He, Y. Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 2009, 21, 1733–1746. [Google Scholar] [CrossRef]
- Kim, S.; Choi, K.; Park, C.; Hwang, H.; Lee, I. SUPPRESSOR OF FRIGIDA4, encoding a C2H2-type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell 2006, 18, 2985–2998. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, D.; He, Y. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants 2018, 4, 836–846. [Google Scholar] [CrossRef]
- Choi, J.; Hyun, Y.; Kang, M.; Yun, H.; Yun, J.; Lister, C.; Dean, C.; Amasino, R.; Noh, B.; Noh, Y.; et al. Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J. 2009, 57, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Liu, Y.; Xu, S.; Xiao, J.; Wang, B.; Deng, H.; Lu, Z.; Xu, Y.; Chong, K. Arabidopsis O-GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. EMBO J. 2018, 37, e98115. [Google Scholar] [CrossRef]
- Gibson, D.; Young, L.; Chuang, R.; Venter, J.; Hutchison, C.; Smith, H. Enzymatic assemblt of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gong, X.; Li, Y.; Li, H.; Zhang, H.; Liu, L.; Liang, D.; Yuan, W. Interaction analysis between the Arabidopsis transcription repressor VAL1 and transcription co-regulators SIN3-LIKEs (SNLs). Int. J. Mol. Sci. 2022, 23, 6987. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Milne, T.; Dou, Y.; Zhang, X.; Burlingame, A.; Roeder, R.; Brivanlou, A.; Allis, C. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005, 121, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef]
- Jung, J.; Lee, H.; Ryu, J.; Park, C. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 2016, 9, 1647–1659. [Google Scholar] [CrossRef]
- Ma, H.; Chen, J.; Zhang, Z.; Ma, L.; Yang, Z.; Zhang, Q.; Li, X.; Xiao, J.; Wang, S. MAPK kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J. 2017, 92, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.; Cho, Y.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Tian, H.; Xu, C.; Li, H.; Li, Y.; Zhang, H.; Zhang, B.; Yuan, W. Arabidopsis SEC13B Interacts with Suppressor of Frigida 4 to Repress Flowering. Int. J. Mol. Sci. 2023, 24, 17248. https://doi.org/10.3390/ijms242417248
Yang Y, Tian H, Xu C, Li H, Li Y, Zhang H, Zhang B, Yuan W. Arabidopsis SEC13B Interacts with Suppressor of Frigida 4 to Repress Flowering. International Journal of Molecular Sciences. 2023; 24(24):17248. https://doi.org/10.3390/ijms242417248
Chicago/Turabian StyleYang, Yanqi, Hao Tian, Chunxue Xu, Haitao Li, Yan Li, Haitao Zhang, Biaoming Zhang, and Wenya Yuan. 2023. "Arabidopsis SEC13B Interacts with Suppressor of Frigida 4 to Repress Flowering" International Journal of Molecular Sciences 24, no. 24: 17248. https://doi.org/10.3390/ijms242417248
APA StyleYang, Y., Tian, H., Xu, C., Li, H., Li, Y., Zhang, H., Zhang, B., & Yuan, W. (2023). Arabidopsis SEC13B Interacts with Suppressor of Frigida 4 to Repress Flowering. International Journal of Molecular Sciences, 24(24), 17248. https://doi.org/10.3390/ijms242417248






