A Genome-Wide Analysis of the Jasmonic Acid Biosynthesis Gene Families in Peanut Reveals Their Crucial Roles in Growth and Abiotic Stresses
Abstract
:1. Introduction
2. Results
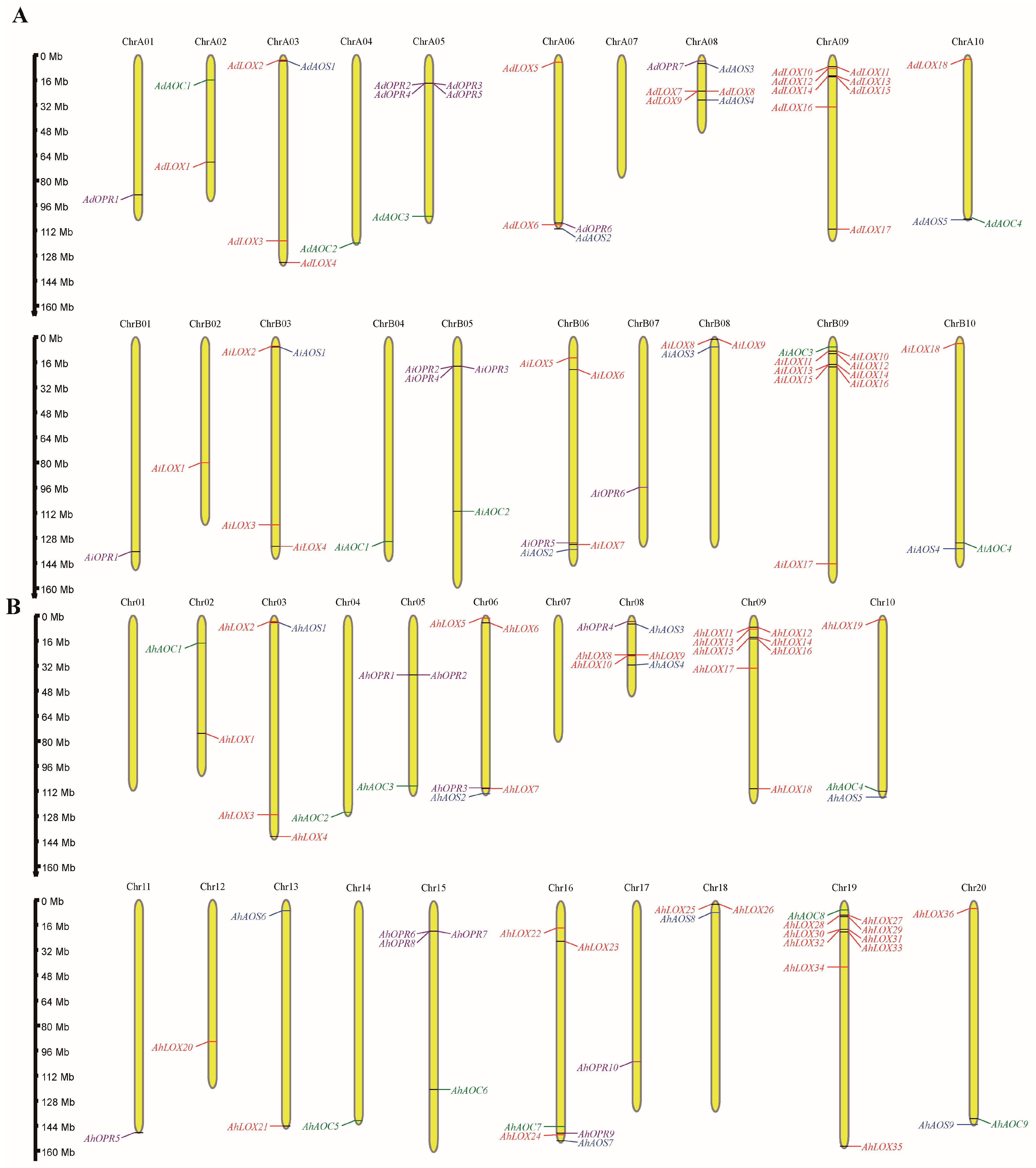
2.1. Identification and Characterization of JA Biosynthesis Gene Families in Arachis
2.2. Physicochemical Properties of JA Biosynthesis Genes
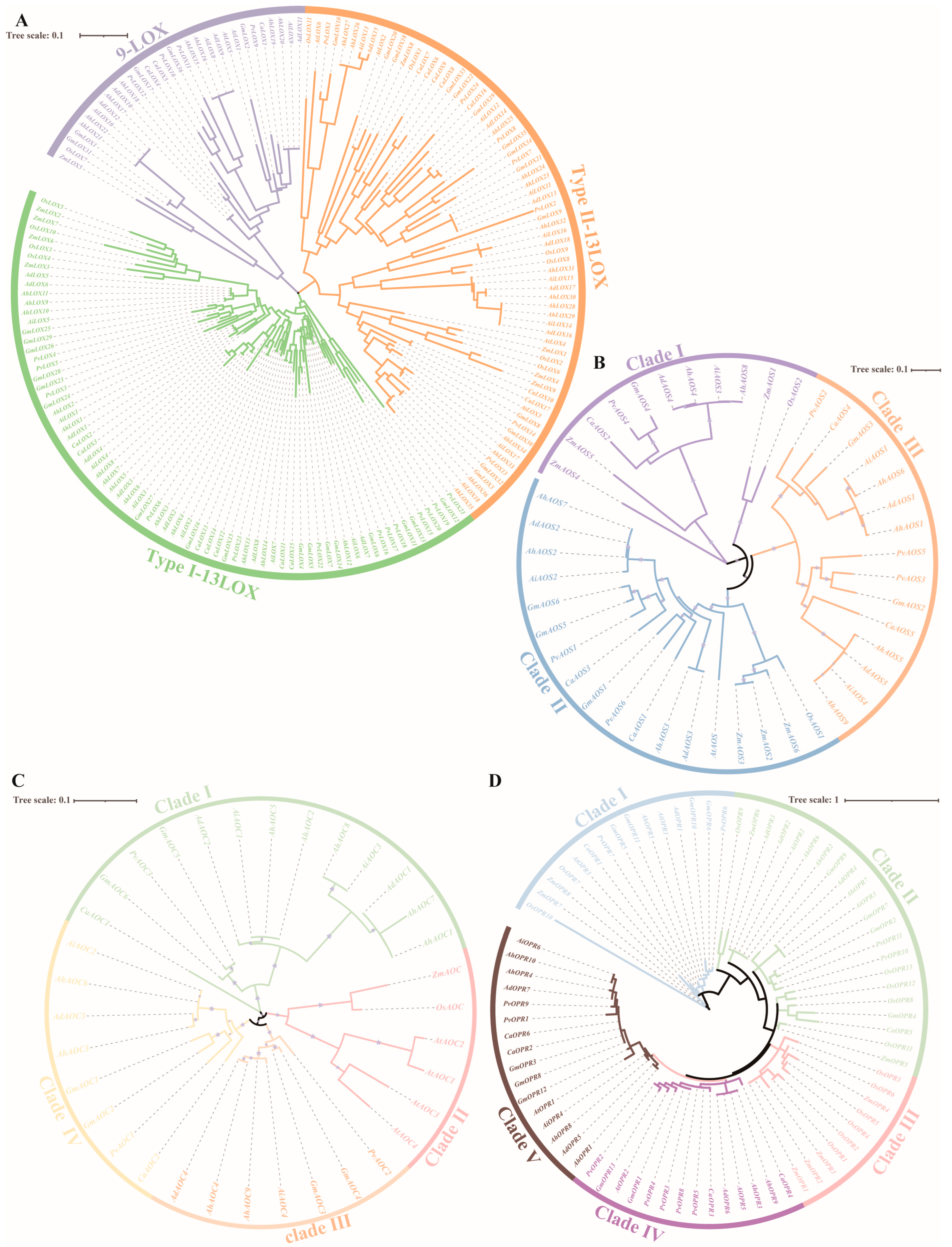
2.3. Phylogenetic Analysis of JA Biosynthesis Gene Families
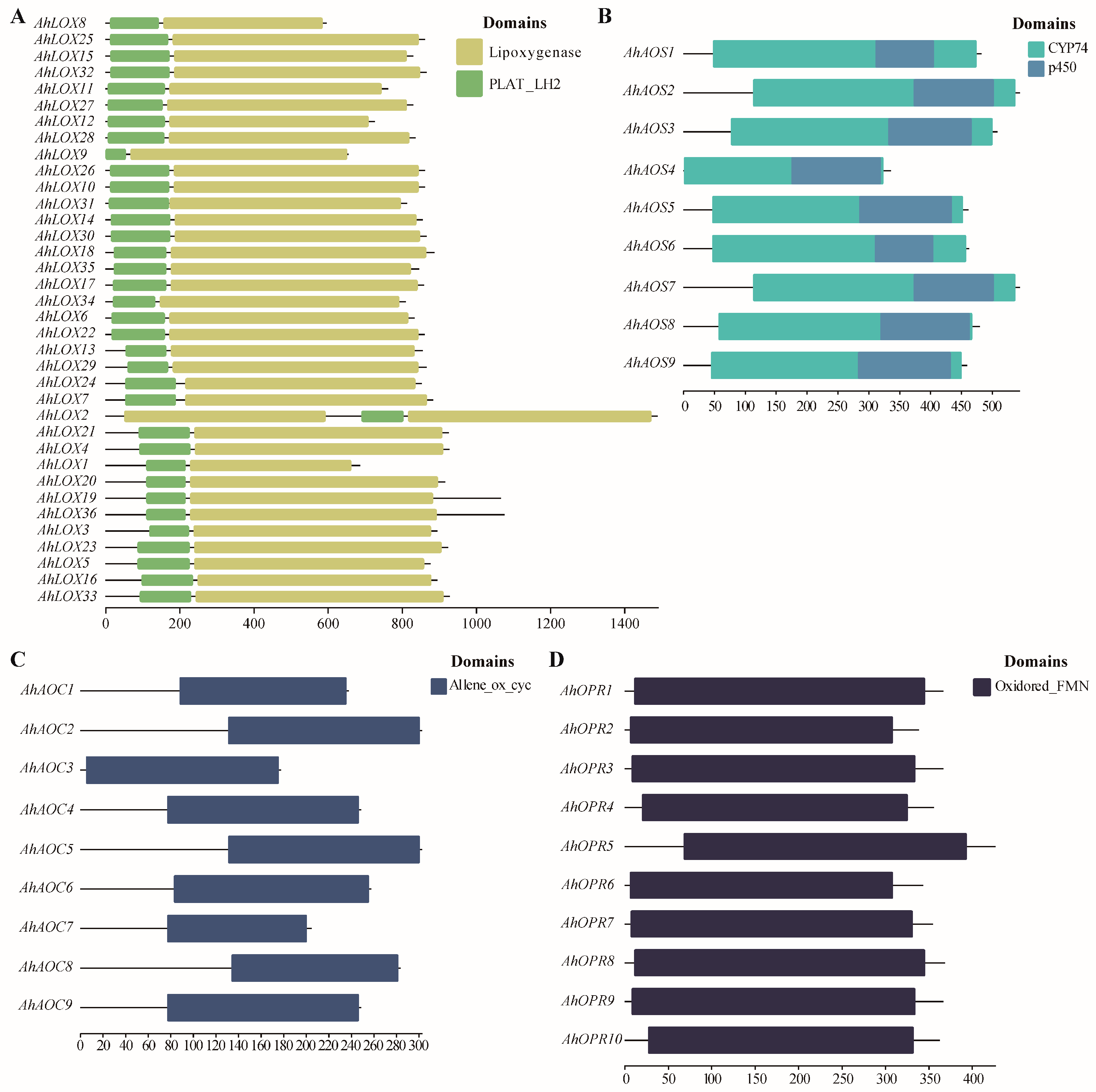
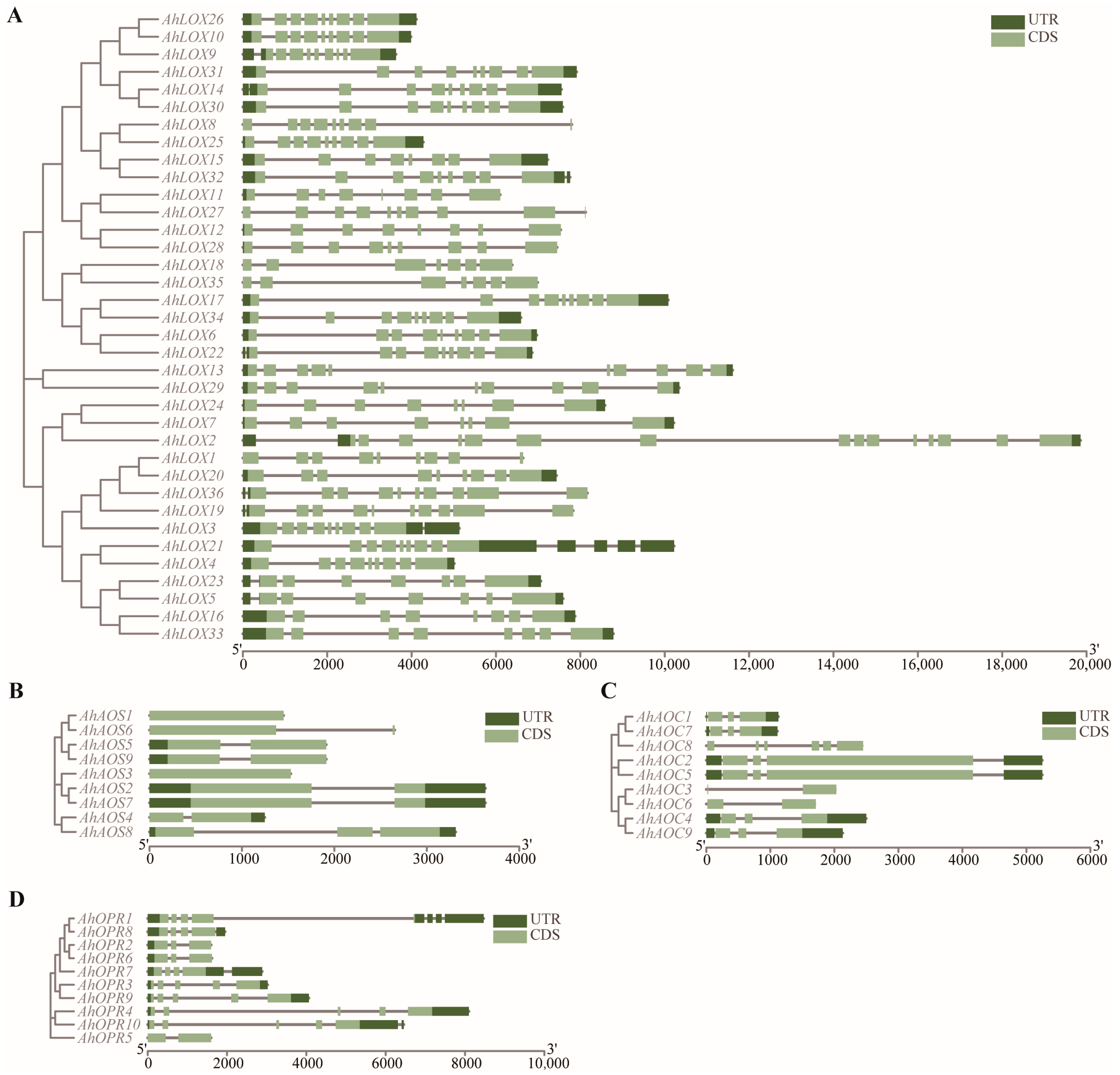
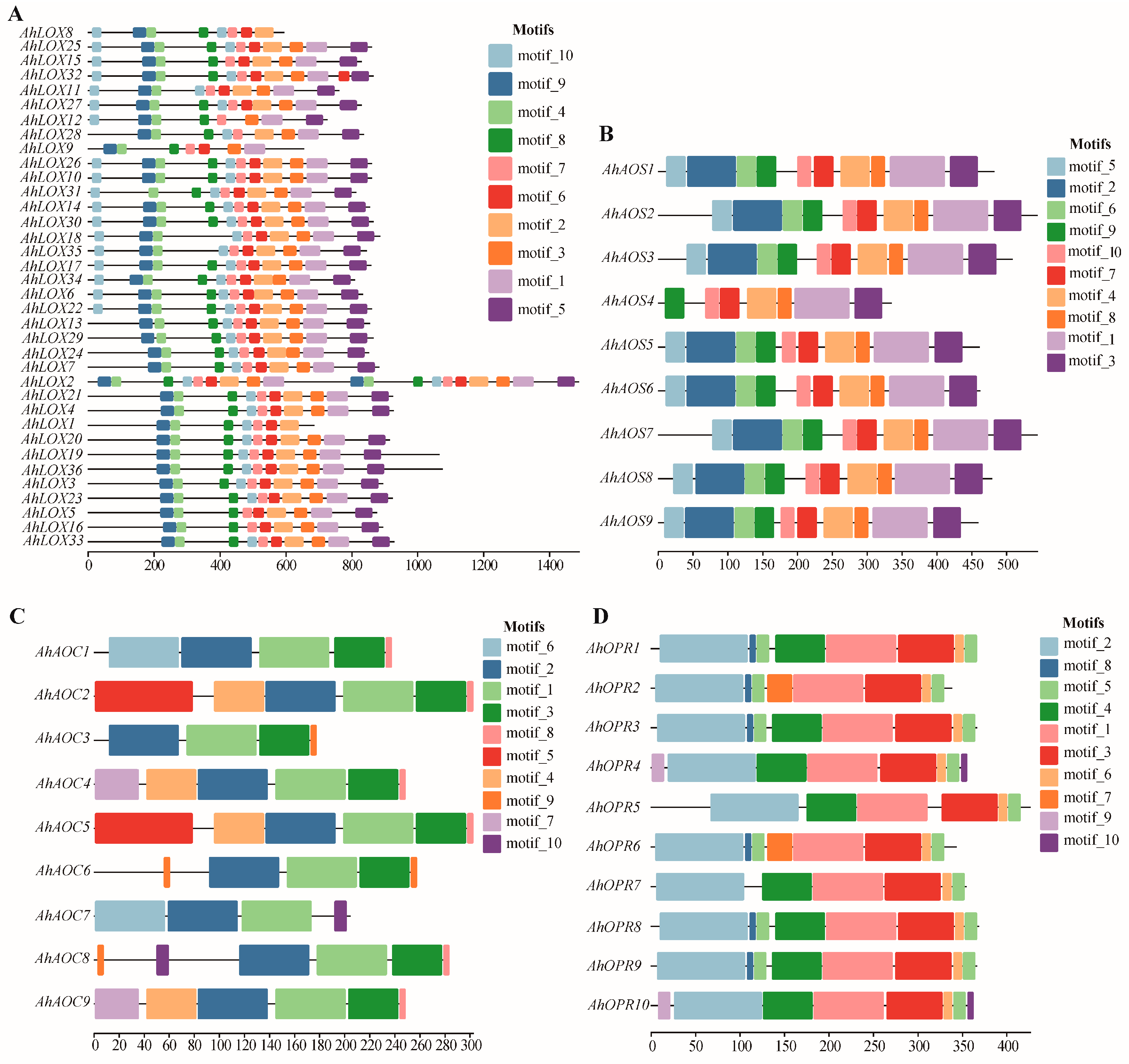
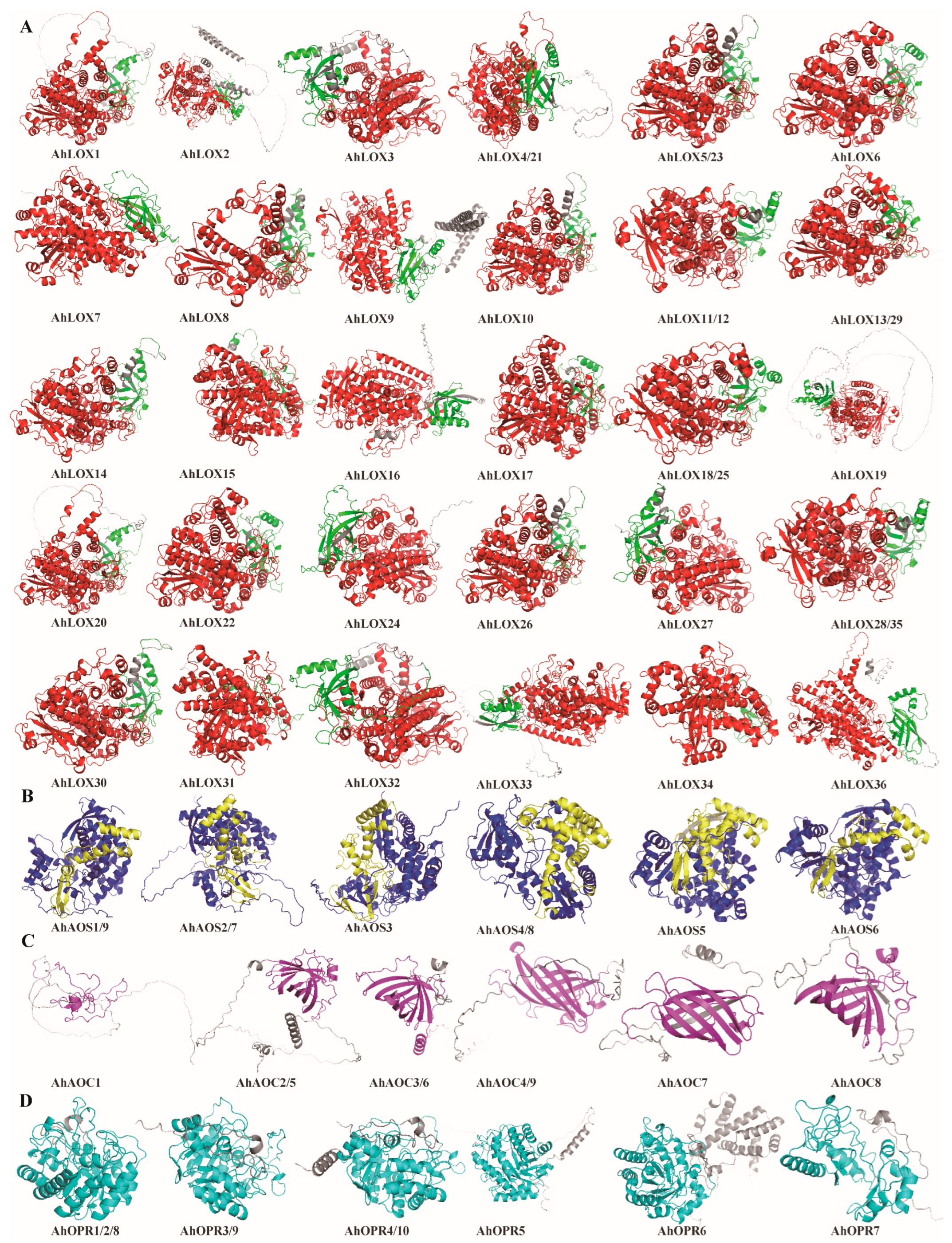
2.4. Domain Characterization, Gene Structure, Conserved Motif Distribution, and Protein Structure of JA Biosynthesis Gene Families
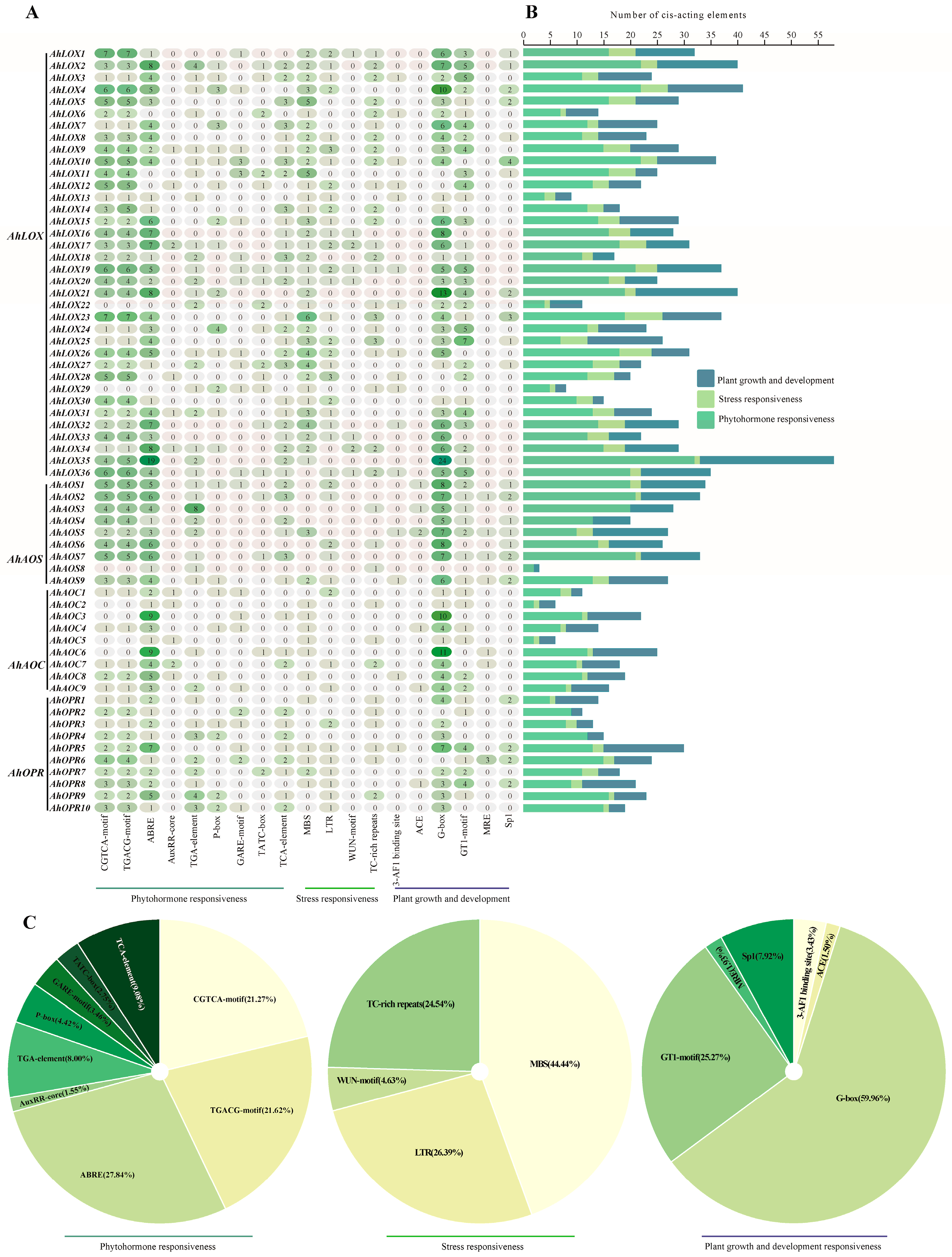
2.5. Cis-Regulatory Elements in the Promoter Regions of JA Biosynthesis Genes
2.6. Protein–Protein Interactions and miRNA Genes’ Regulatory Networks of JA Biosynthesis Gene Families
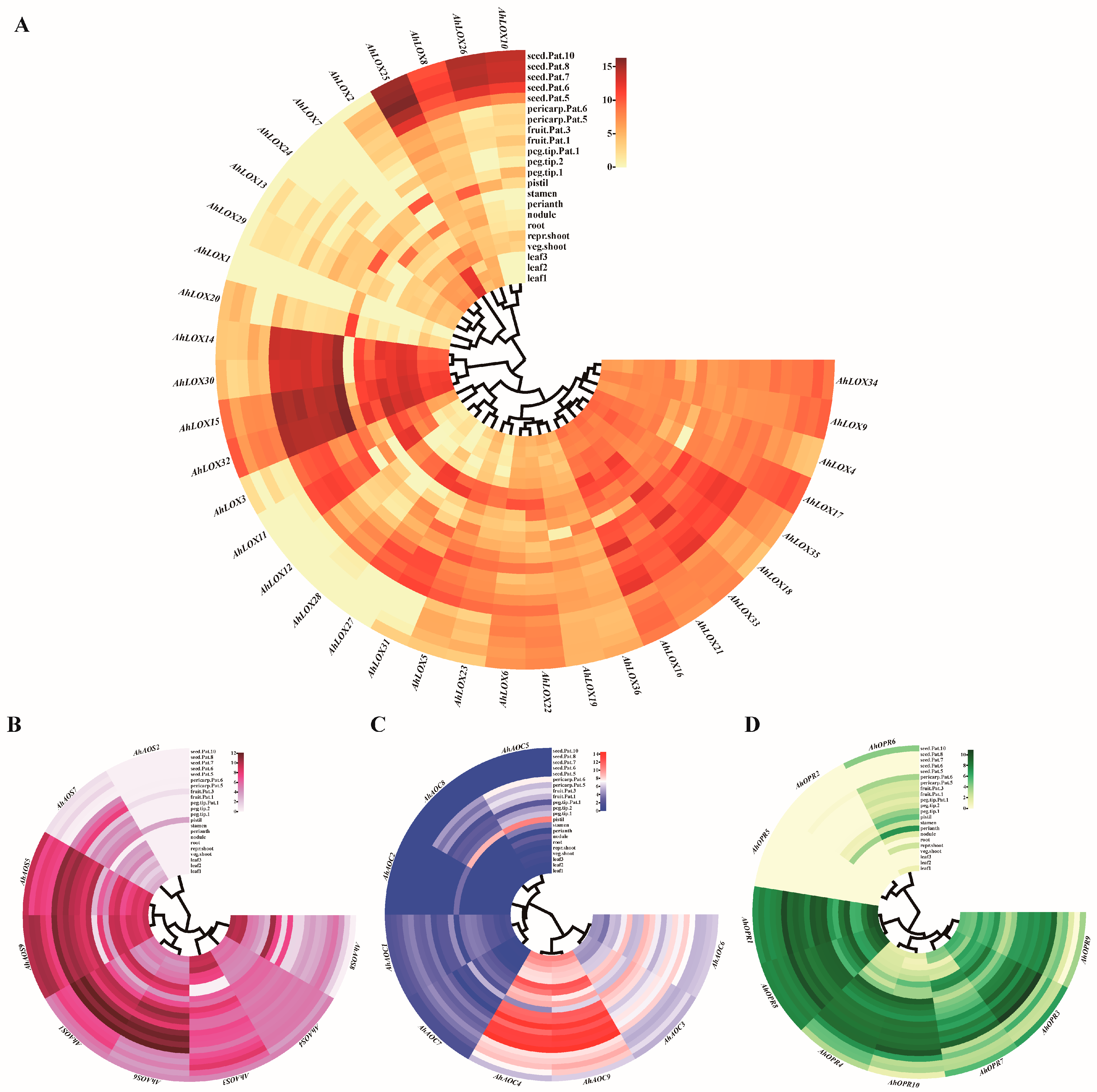
2.7. Expression Profiles of JA Biosynthesis Genes in Different Tissues
2.8. Expression Patterns of JA Biosynthesis Genes in Response to Different Abiotic Stress Treatments
3. Discussion
4. Materials and Methods
4.1. Identification of Key Enzyme Genes of JA Biosynthesis in Peanut
4.2. Gene Structure and Conserved Motifs and Protein Structures
4.3. Phylogenetic Analysis and Classification
4.4. Chromosomal Location
4.5. Cis-Regulatory Element Analysis of Promoter Region
4.6. Protein–Protein Interaction and miRNA Genes’ Regulatory Network
4.7. Tissue Expression Pattern
4.8. Plant Materials and Abiotic Stress Treatment
4.9. RNA Extraction and Gene Expression Analysis under Abiotic Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. Jasmonates: An Update on Biosynthesis, Signal Transduction and Action in Plant Stress Response, Growth and Development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Li, R.; Zhang, C.; Li, W.; Li, L.; Hu, S.; Niu, J.; Cao, X.; Wang, D.; Wang, Z. Identification and Characterization of Jasmonic Acid Biosynthetic Genes in Salvia miltiorrhiza Bunge. Int. J. Mol. Sci. 2022, 23, 9384. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Baek, K.H. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of Jasmonic Acid in Plant Regulation and Response to Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Aleman, G.H.; Scholz, S.S.; Heyer, M.; Reichelt, M.; Mithöfer, A.; Boland, W. Synthesis, Metabolism and Systemic Transport of a Fluorinated Mimic of the Endogenous Jasmonate Precursor OPC-8:0. Biochim. Biophys. Acta 2015, 1851, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, Perception, Signal Transduction and Action in Plant Stress Response, Growth and Development. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Lahari, Z.; van Boerdonk, S.; Omoboye, O.O.; Reichelt, M.; Höfte, M.; Gershenzon, J.; Gheysen, G.; Ullah, C. Strigolactone Deficiency Induces Jasmonate, Sugar and Flavonoid Phytoalexin Accumulation Enhancing Rice Defense against the Blast Fungus Pyricularia Oryzae. New Phytol. 2024, 241, 827–844. [Google Scholar] [CrossRef]
- Farmer, E.E.; Goossens, A. Jasmonates: What ALLENE OXIDE SYNTHASE Does for Plants. J. Exp. Bot. 2019, 70, 3373–3378. [Google Scholar] [CrossRef]
- Sun, T.; Cen, G.; You, C.; Lou, W.; Wang, Z.; Su, W.; Wang, W.; Li, D.; Que, Y.; Su, Y. ScAOC1, an Allene Oxide Cyclase Gene, Confers Defense Response to Biotic and Abiotic Stresses in Sugarcane. Plant Cell Rep. 2020, 39, 1785–1801. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Monte, I.; Zamarreño, A.M.; García-Mina, J.M.; Solano, R. Evolution of the Jasmonate Ligands and Their Biosynthetic Pathways. New Phytol. 2023, 238, 2236–2246. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Luo, Y.; Yu, J.; Kou, X.; Xie, L.; Deng, P.; Li, T.; Chen, C.; Ji, W.; Liu, X. Genome-Wide Identification and Characterization of Lipoxygenase Genes Related to the English Grain Aphid Infestation Response in Wheat. Planta 2023, 257, 84. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.A.T.; Dobson, E.P.; Henderson, L.C.; Barrow, C.J.; Adcock, J.L. Soy Flour as an Alternative to Purified Lipoxygenase for the Enzymatic Synthesis of Resolvin Analogues. New Biotechnol. 2018, 41, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Feussner, I.; Wasternack, C. The Lipoxygenase Pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Uehara, S.; Uno, Y.; Yamazaki, H. The Marmoset Cytochrome P450 Superfamily: Sequence/Phylogenetic Analyses, Genomic Structure, and Catalytic Function. Biochem. Pharmacol. 2020, 171, 113721. [Google Scholar] [CrossRef] [PubMed]
- Pazhamala, L.T.; Giri, J. Plant Phosphate Status Influences Root Biotic Interactions. J. Exp. Bot. 2023, 74, 2829–2844. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, H.; Shan, Z.; Hao, Q.; Zhang, C.; Yang, Z.; Zhang, X.; Yuan, S.; Qiu, D.; Chen, S.; et al. Herbivore Defense Responses and Associated Herbivore Defense Mechanism as Revealed by Comparing a Resistant Wild Soybean with a Susceptible Cultivar. Crop J. 2015, 3, 451–467. [Google Scholar] [CrossRef]
- Ziegler, J.; Stenzel, I.; Hause, B.; Maucher, H.; Hamberg, M.; Grimm, R.; Ganal, M.; Wasternack, C. Molecular Cloning of Allene Oxide Cyclase: The Enzyme Establishing the Stereochemistry of Octadecanoids and Jasmonates. J. Biol. Chem. 2000, 275, 19132–19138. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Jwa, N.S.; Agrawal, S.K.; Tamogami, S.; Iwahashi, H.; Rakwal, R. Cloning of Novel Rice Allene Oxide Cyclase (OsAOC): MRNA Expression and Comparative Analysis with Allene Oxide Synthase (OsAOS) Gene Provides Insight into the Transcriptional Regulation of Octadecanoid Pathway Biosynthetic Genes in Rice. Plant Sci. 2003, 164, 979–992. [Google Scholar] [CrossRef]
- Stenzel, I.; Hause, B.; Miersch, O.; Kurz, T.; Maucher, H.; Weichert, H.; Ziegler, J.; Feussner, I.; Wasternack, C. Jasmonate Biosynthesis and the Allene Oxide Cyclase Family of Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 895–911. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, J.; Sun, H.; Zhang, D.; Yu, D. Sequence and Expression Divergence of the AOC Gene Family in Soybean: Insights into Functional Diversity for Stress Responses. Biotechnol. Lett. 2011, 33, 1351–1359. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, W.; Zhang, N.; Ai, X.; Wang, M.; Huang, Z.; Xiao, L.; Xia, G. A Wheat Allene Oxide Cyclase Gene Enhances Salinity Tolerance via Jasmonate Signaling. Plant Physiol. 2014, 164, 1068–1076. [Google Scholar] [CrossRef]
- Borrego, E.J.; Robertson, M.; Taylor, J.; Schultzhaus, Z.; Espinoza, E.M. Oxylipin Biosynthetic Gene Families of Cannabis sativa. PLoS ONE 2023, 18, e0272893. [Google Scholar] [CrossRef]
- Wang, X.; Wu, L.; Xiang, L.; Gao, R.; Yin, Q.; Wang, M.; Liu, Z.; Leng, L.; Su, Y.; Wan, H.; et al. Promoter Variations in DBR2-like Affect Artemisinin Production in Different Chemotypes of Artemisia Annua. Hortic. Res. 2023, 10, uhad164. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, X.; Fu, Y.; Wang, H.; Yu, C.; Chu, J.; Jiang, B.; Zhu, J. Genome-Wide Identification and Expression Analysis of VQ Gene Family under Abiotic Stress in Coix lacryma-jobi L. BMC Plant Biol. 2023, 23, 327. [Google Scholar] [CrossRef]
- Schaller, F.; Weiler, E.W. Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana. Structural and functional relationship to yeast old yellow enzyme. J. Biol. Chem. 1997, 272, 28066–28072. [Google Scholar] [CrossRef]
- Li, W.; Zhou, F.; Liu, B.; Feng, D.; He, Y.; Qi, K.; Wang, H.; Wang, J. Comparative Characterization, Expression Pattern and Function Analysis of the 12-Oxo-Phytodienoic Acid Reductase Gene Family in Rice. Plant Cell Rep. 2011, 30, 981–995. [Google Scholar] [CrossRef]
- Lian, H.; Qin, C.; Shen, J.; Ahanger, M.A. Alleviation of Adverse Effects of Drought Stress on Growth and Nitrogen Metabolism in Mungbean (Vigna radiata) by Sulphur and Nitric Oxide Involves Up-Regulation of Antioxidant and Osmolyte Metabolism and Gene Expression. Plants 2023, 12, 3082. [Google Scholar] [CrossRef]
- Chauvin, A.; Caldelari, D.; Wolfender, J.L.; Farmer, E.E. Four 13-Lipoxygenases Contribute to Rapid Jasmonate Synthesis in Wounded Arabidopsis Thaliana Leaves: A Role for Lipoxygenase 6 in Responses to Long-Distance Wound Signals. New Phytol. 2013, 197, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, C.; Wang, S.; Yang, F.; Sun, Y.; Tang, J.; Luo, J.; Wu, J. Ethylene and Jasmonate Signaling Converge on Gibberellin Catabolism during Thigmomorphogenesis in Arabidopsis. Plant Physiol. 2023, 194, 758–773. [Google Scholar] [CrossRef]
- Walper, E.; Weiste, C.; Mueller, M.J.; Hamberg, M.; Dröge-Laser, W. Screen Identifying Arabidopsis Transcription Factors Involved in the Response to 9-Lipoxygenase-Derived Oxylipins. PLoS ONE 2016, 11, e0153216. [Google Scholar] [CrossRef]
- Christensen, S.A.; Huffaker, A.; Kaplan, F.; Sims, J.; Ziemann, S.; Doehlemann, G.; Ji, L.; Schmitz, R.J.; Kolomiets, M.V.; Alborn, H.T.; et al. Maize Death Acids, 9-Lipoxygenase-Derived Cyclopente(a)Nones, Display Activity as Cytotoxic Phytoalexins and Transcriptional Mediators. Proc. Natl. Acad. Sci. USA 2015, 112, 11407–11412. [Google Scholar] [CrossRef]
- Luo, H.; Xu, Z.; Li, Z.; Li, X.; Lv, J.; Ren, X.; Huang, L.; Zhou, X.; Chen, Y.; Yu, J.; et al. Development of SSR Markers and Identification of Major Quantitative Trait Loci Controlling Shelling Percentage in Cultivated Peanut (Arachis hypogaea L.). Theor. Appl. Genet. 2017, 130, 1635–1648. [Google Scholar] [CrossRef]
- Huai, D.; Zhi, C.; Wu, J.; Xue, X.; Hu, M.; Zhang, J.; Liu, N.; Huang, L.; Yan, L.; Chen, Y.; et al. Unveiling the molecular regulatory mechanisms underlying sucrose accumulation and oil reduction in peanut kernels through genetic mapping and transcriptome analysis. Plant Physiol. Biochem. 2024, 208, 108448. [Google Scholar] [CrossRef]
- Lu, J.; Liu, Y.; Zou, X.; Zhang, X.; Yu, X.; Wang, Y.; Si, T. Rotational Strip Peanut/Cotton Intercropping Improves Agricultural Production through Modulating Plant Growth, Root Exudates, and Soil Microbial Communities. Agric. Ecosyst. Environ. 2024, 359, 108767. [Google Scholar] [CrossRef]
- Lawas, L.M.F.; Erban, A.; Kopka, J.; Jagadish, S.V.K.; Zuther, E.; Hincha, D.K. Metabolic Responses of Rice Source and Sink Organs during Recovery from Combined Drought and Heat Stress in the Field. Gigascience 2019, 8, giz102. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Dong, W.; Jiao, B.; Wang, J.; Sun, L.; Li, S.; Wu, Z.; Gao, J.; Zhou, S. Genome-Wide Identification and Expression Analysis of Lipoxygenase Genes in Rose (Rosa chinensis). Genes 2023, 14, 1957. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.C.; Tate, M.; Berg-Falloure, K.M.; Christensen, S.A.; Zhang, J.; Schirawski, J.; Meeley, R.; Kolomiets, M.V. A Non-JA Producing Oxophytodienoate Reductase Functions in Salicylic Acid-Mediated Antagonism with Jasmonic Acid during Pathogen Attack. Mol. Plant Pathol. 2023, 24, 725–741. [Google Scholar] [CrossRef]
- Rani, V. Computational Methods to Dissect Cis-Regulatory Transcriptional Networks. J. Biosci. 2007, 32, 1325–1330. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A Developmental Transcriptome Map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef]
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of Jasmonic Acid in Plants: The Molecular Point of View. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef]
- Alonso, C.; Ramos-Cruz, D.; Becker, C. The Role of Plant Epigenetics in Biotic Interactions. New Phytol. 2019, 221, 731–737. [Google Scholar] [CrossRef]
- Pottosin, I.; Velarde-Buendía, A.M.; Bose, J.; Zepeda-Jazo, I.; Shabala, S.; Dobrovinskaya, O. Cross-Talk between Reactive Oxygen Species and Polyamines in Regulation of Ion Transport across the Plasma Membrane: Implications for Plant Adaptive Responses. J. Exp. Bot. 2014, 65, 1271–1283. [Google Scholar] [CrossRef]
- Gupta, A.; Hisano, H.; Hojo, Y.; Matsuura, T.; Ikeda, Y.; Mori, I.C.; Senthil-Kumar, M. Global Profiling of Phytohormone Dynamics during Combined Drought and Pathogen Stress in Arabidopsis Thaliana Reveals ABA and JA as Major Regulators. Sci. Rep. 2017, 7, 4017. [Google Scholar] [CrossRef]
- Dhakarey, R.; Raorane, M.L.; Treumann, A.; Peethambaran, P.K.; Schendel, R.R.; Sahi, V.P.; Hause, B.; Bunzel, M.; Henry, A.; Kohli, A.; et al. Physiological and Proteomic Analysis of the Rice Mutant Cpm2 Suggests a Negative Regulatory Role of Jasmonic Acid in Drought Tolerance. Front. Plant Sci. 2017, 8, 1903. [Google Scholar] [CrossRef]
- Dumin, W.; Rostas, M.; Winefield, C. Identification and Functional Characterisation of an Allene Oxide Synthase from Grapevine (Vitis vinifera L. Sauvignon Blanc). Mol. Biol. Rep. 2018, 45, 263–277. [Google Scholar] [CrossRef]
- Mou, Y.; Liu, Y.; Tian, S.; Guo, Q.; Wang, C.; Wen, S. Genome-Wide Identification and Characterization of the OPR Gene Family in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2019, 20, 1914. [Google Scholar] [CrossRef]
- Guang, Y.; Luo, S.; Ahammed, G.J.; Xiao, X.; Li, J.; Zhou, Y.; Yang, Y. The OPR Gene Family in Watermelon: Genome-Wide Identification and Expression Profiling under Hormone Treatments and Root-Knot Nematode Infection. Plant Biol. 2021, 23, 80–88. [Google Scholar] [CrossRef]
- Nie, W.F.; Chen, Y.; Tao, J.; Li, Y.; Liu, J.; Zhou, Y.; Yang, Y. Identification of the 12-Oxophytoeienoic Acid Reductase (OPR) Gene Family in Pepper (Capsicum annuum L.) and Functional Characterization of CaOPR6 in Pepper Fruit Development and Stress Response. Genome 2022, 65, 537–545. [Google Scholar] [CrossRef]
- Zhang, J.; Ng, C.; Jiang, Y.; Wang, X.; Wang, S.; Wang, S. Genome-Wide Identification and Analysis of LOX Genes in Soybean Cultivar “Zhonghuang 13”. Front. Genet. 2022, 13, 1020554. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Z.; Guo, C.; Zhao, X.; Li, Z.; Mou, Y.; Sun, Q.; Wang, J.; Yuan, C.; Li, C.; et al. Hsf Transcription Factor Gene Family in Peanut (Arachis hypogaea L.): Genome-Wide Characterization and Expression Analysis under Drought and Salt Stresses. Front. Plant Sci. 2023, 14, 1214732. [Google Scholar] [CrossRef]
- Laudert, D.; Weiler, E.W. Allene Oxide Synthase: A Major Control Point in Arabidopsis Thaliana Octadecanoid Signalling. Plant J. 1998, 15, 675–684. [Google Scholar] [CrossRef]
- Bannenberg, G.; Martínez, M.; Hamberg, M.; Castresana, C. Diversity of the Enzymatic Activity in the Lipoxygenase Gene Family of Arabidopsis Thaliana. Lipids 2009, 44, 85–95. [Google Scholar] [CrossRef]
- Stenzel, I.; Otto, M.; Delker, C.; Kirmse, N.; Schmidt, D.; Miersch, O.; Hause, B.; Wasternack, C. ALLENE OXIDE CYCLASE (AOC) Gene Family Members of Arabidopsis Thal Thaliana: Tissue-and Organ-Specific Promoter Activities and in Vivo Heteromerization. J. Exp. Bot. 2012, 63, 6125–6138. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, L.; Chi, J.; Li, R.; Han, Y.; Cui, F.; Peng, Z.; Wan, S.; Li, G. Genome-Wide Identification and Expression of SAUR Gene Family in Peanut (Arachis hypogaea L.) and Functional Identification of AhSAUR3 in Drought Tolerance. BMC Plant Biol. 2022, 22, 178. [Google Scholar] [CrossRef]
- Li, Q.; Gao, L.; Yu, F.; Lü, S.; Yang, P. Evolution and Diversification of CaM/CML Gene Family in Green Plants. Plant Physiol. Biochem. 2023, 202, 107922. [Google Scholar] [CrossRef]
- Hu, J.; Liu, T.; Huo, H.; Liu, S.; Liu, M.; Liu, C.; Zhao, M.; Wang, K.; Wang, Y.; Zhang, M. Genome-Wide Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the Trihelix Transcription Factor Family and Gene Expression Analysis under MeJA Treatment in Panax Ginseng. BMC Plant Biol. 2023, 23, 376. [Google Scholar] [CrossRef]
- Liu, X.; Li, L.; Zhang, B.; Zeng, L.; Li, L. AhHDA1-Mediated AhGLK1 Promoted Chlorophyll Synthesis and Photosynthesis Regulates Recovery Growth of Peanut Leaves after Water Stress. Plant Sci. 2020, 294, 110461. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H. Bin Plant-MPLoc: A Top-down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, Y.; Wu, T.T.; Zhang, G.L.; Yin, H.; Chen, W.; Wang, S.; Chang, F.; Gou, J.Y. Cloning of Wheat Keto-Acyl Thiolase 2B Reveals a Role of Jasmonic Acid in Grain Weight Determination. Nat. Commun. 2020, 11, 6266. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A User-Friendly Online Tool for Drawing Genetic Maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Ai, X.; Li, C.; Wang, S.; Zhang, N.; Ren, J.; Wang, J.; Zhong, C.; Zhao, X.; Zhang, H.; et al. A Genome-Wide Analysis of the Jasmonic Acid Biosynthesis Gene Families in Peanut Reveals Their Crucial Roles in Growth and Abiotic Stresses. Int. J. Mol. Sci. 2024, 25, 7054. https://doi.org/10.3390/ijms25137054
Ma X, Ai X, Li C, Wang S, Zhang N, Ren J, Wang J, Zhong C, Zhao X, Zhang H, et al. A Genome-Wide Analysis of the Jasmonic Acid Biosynthesis Gene Families in Peanut Reveals Their Crucial Roles in Growth and Abiotic Stresses. International Journal of Molecular Sciences. 2024; 25(13):7054. https://doi.org/10.3390/ijms25137054
Chicago/Turabian StyleMa, Xinlei, Xin Ai, Chenghua Li, Shiyu Wang, Nan Zhang, Jingyao Ren, Jing Wang, Chao Zhong, Xinhua Zhao, He Zhang, and et al. 2024. "A Genome-Wide Analysis of the Jasmonic Acid Biosynthesis Gene Families in Peanut Reveals Their Crucial Roles in Growth and Abiotic Stresses" International Journal of Molecular Sciences 25, no. 13: 7054. https://doi.org/10.3390/ijms25137054
APA StyleMa, X., Ai, X., Li, C., Wang, S., Zhang, N., Ren, J., Wang, J., Zhong, C., Zhao, X., Zhang, H., & Yu, H. (2024). A Genome-Wide Analysis of the Jasmonic Acid Biosynthesis Gene Families in Peanut Reveals Their Crucial Roles in Growth and Abiotic Stresses. International Journal of Molecular Sciences, 25(13), 7054. https://doi.org/10.3390/ijms25137054





