The Isolation, Identification and Immobilization Method of Three Novel Enzymes with Diosgenin-Producing Activity Derived from an Aspergillus flavus
Abstract
:1. Introduction
2. Results and Discussion
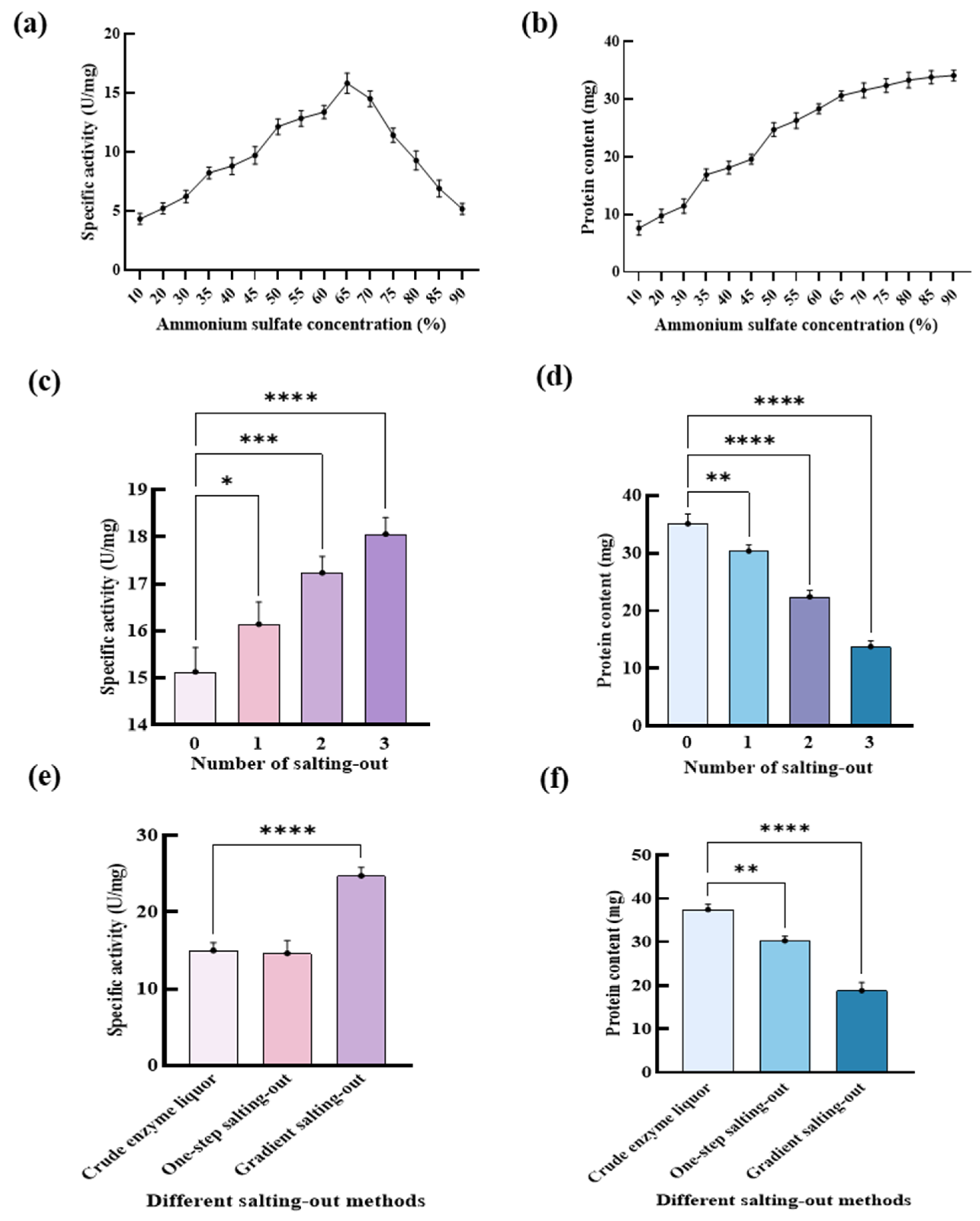
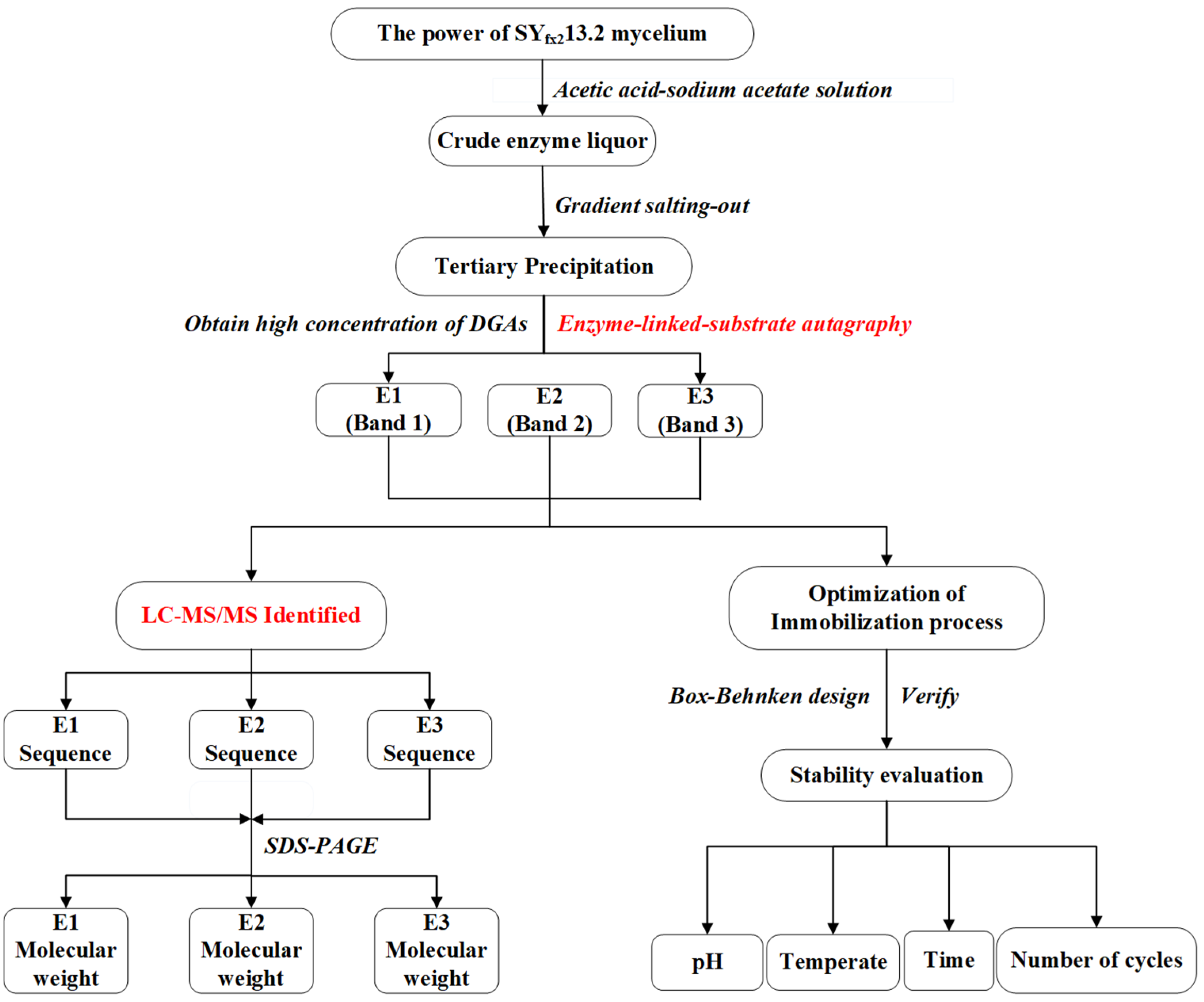
2.1. Isolation of Dioscin-Glycosidases by Fractional Precipitation with Ammonium Sulphate
2.1.1. Effects of Ammonium Sulfate Concentrations on the Specific Activity of DGAs
2.1.2. Effects of the Number of Salting-Out on the Specific Activity of DGAs
2.1.3. Effects of Gradient Ammonium Sulfate on the Specific Activity of DGAs
2.2. The Activity Verification and Identification of the Isolated DGAs
2.3. The Immobilization Optimization of the DGAs
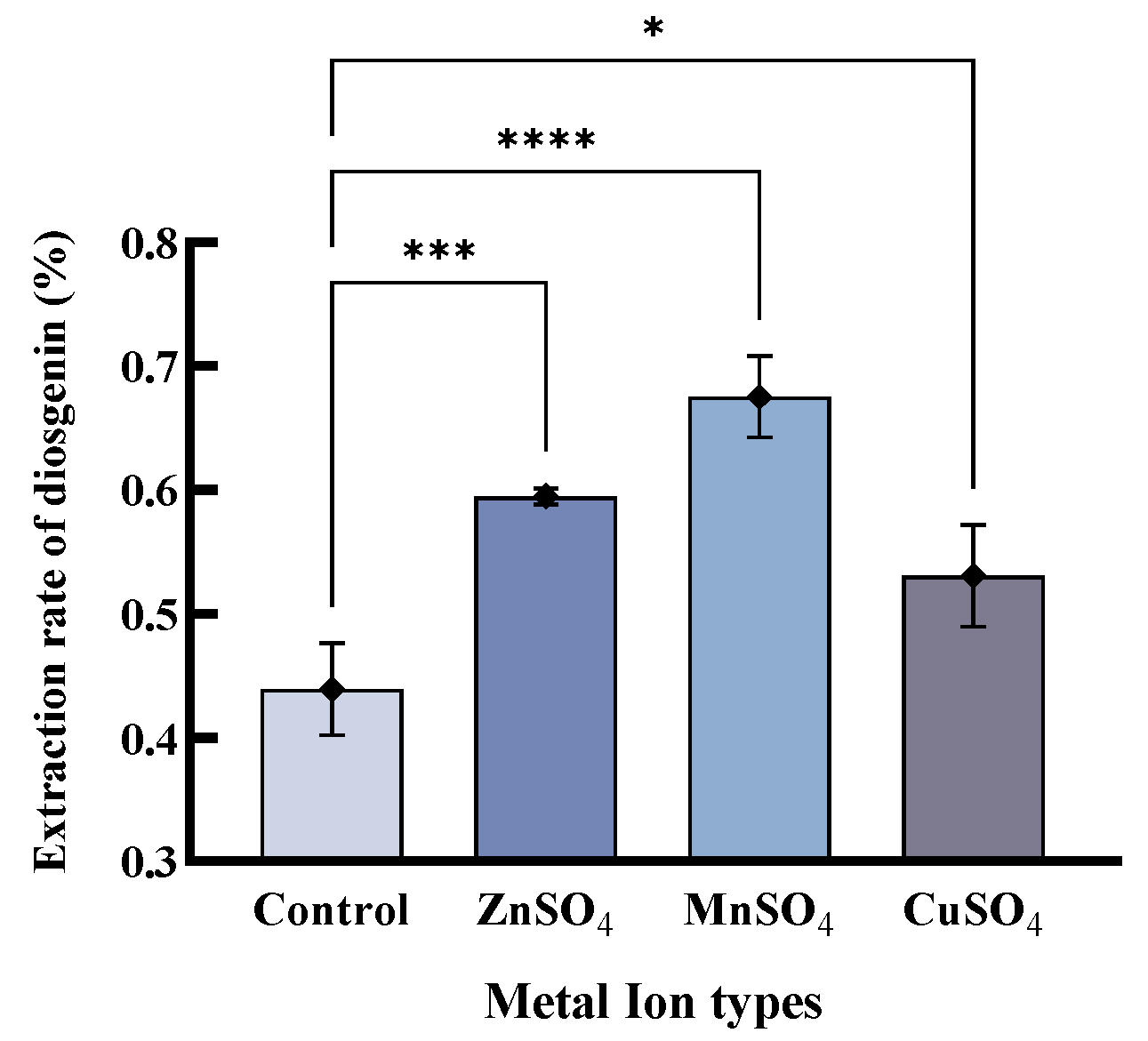
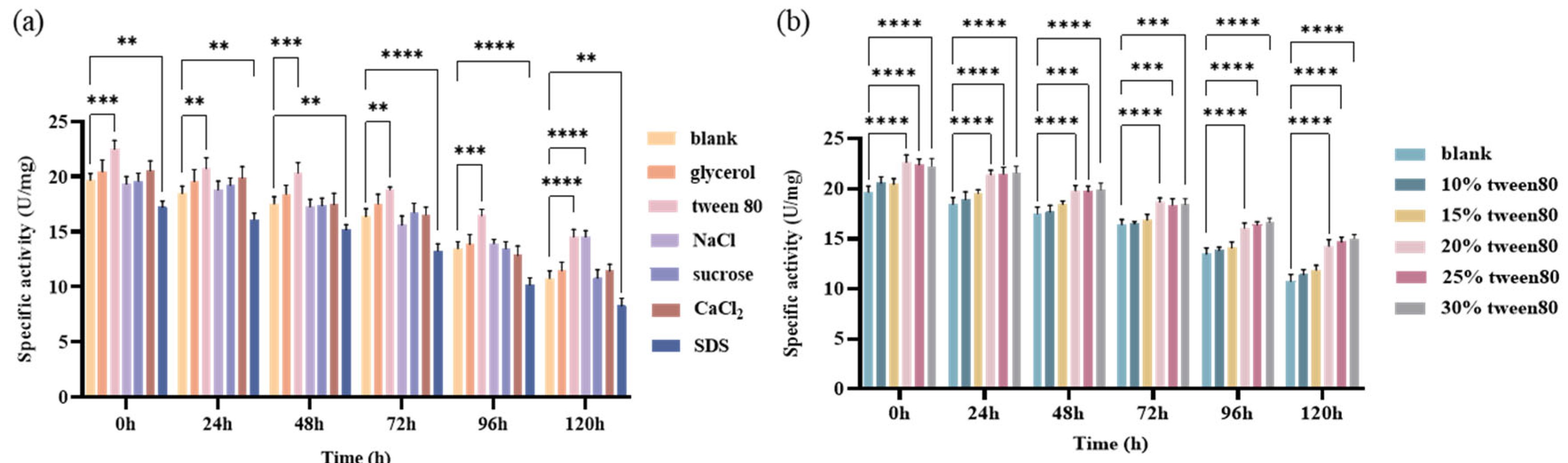
2.3.1. Screening and Optimization of Enzyme Protectants
2.3.2. The Immobilization Optimization Using the Box–Behnken Design
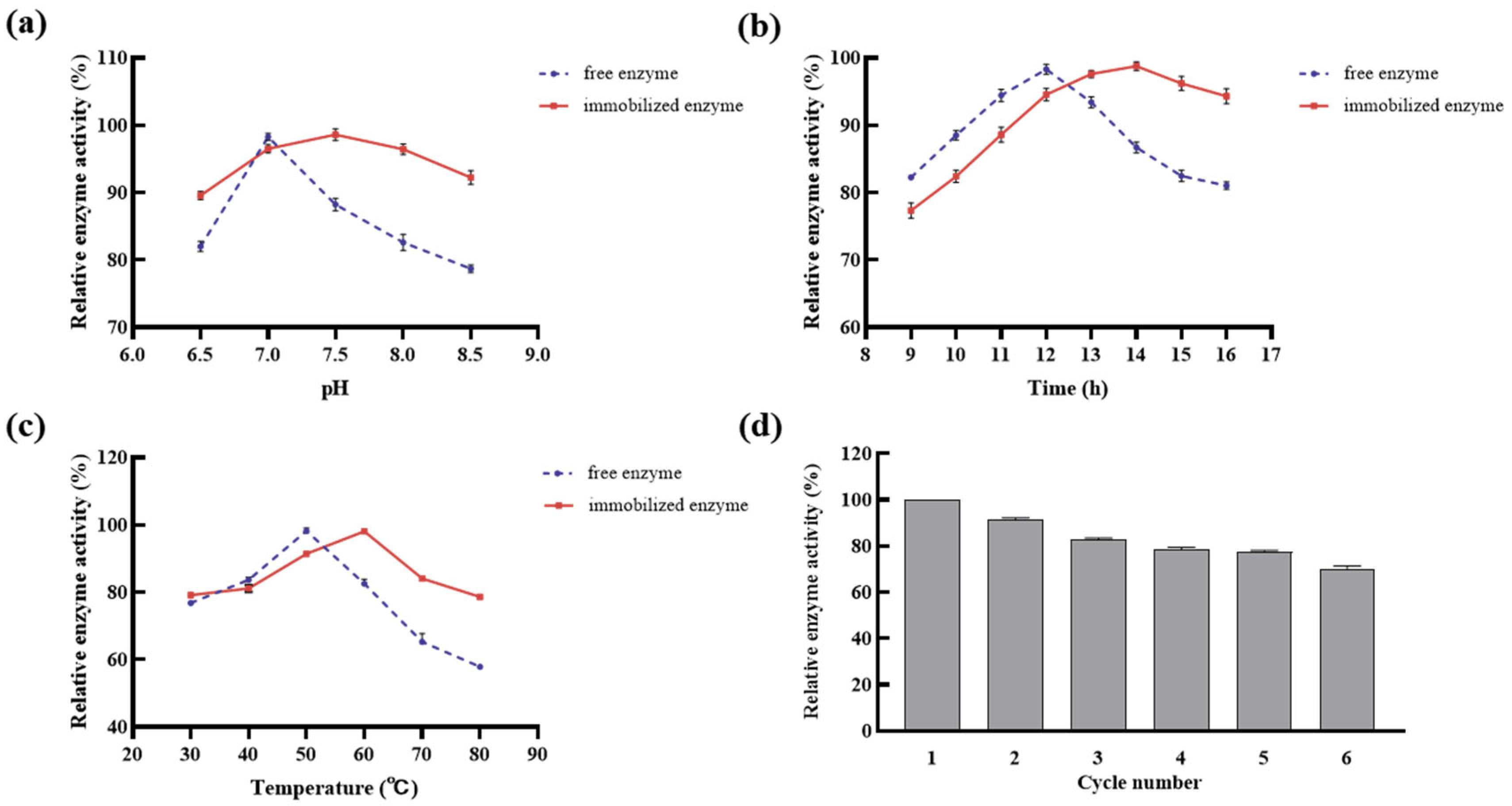
2.3.3. Stability Test of Immobilized DGAs
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Primary Culture and Liquid Fermentation of the Strain
3.3. Preparation of Crude Enzyme Solution
3.4. Isolation and Optimization of DGAs
3.5. Identification of DGAs
3.6. HPLC/MS-MS Identification of DGAs
3.7. Analysis of DGAs Activity
3.8. Screening and Optimization of Protective Agents for Enzyme Immobilization
3.9. Preparation Conditions and Optimization of Immobilized DGAs
3.10. Stability Test of Immobilized DGAs
3.10.1. Effect of pH Values on the Activities of the Immobilized DGAs
3.10.2. Effect of Temperatures on the Activities of the Immobilized DGAs
3.10.3. Effect of the Enzymatic Time on the Activities of the Immobilized DGAs
3.10.4. Recycling of Immobilized DGAs
3.11. Recovery of Immobilized DGAs Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mohammadi, M.; Mashayekh, T.; Rashidi-Monfared, S.; Ebrahimi, A.; Abedini, D. New insights into diosgenin biosynthesis pathway and its regulation in Trigonella foenum-graecum L. Phytochem. Anal. 2020, 31, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jin, M.; Tadesse, N.; Dang, J.; Zhou, T.; Zhang, H.; Wang, S.; Guo, Z.; Ito, Y. Dioscorea zingiberensis C. H. Wright: An overview on its traditional use, phytochemistry, pharmacology, clinical applications, quality control, and toxicity. J. Ethnopharmacol. 2018, 220, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Ito, Y.; Liang, J.R.; Liu, J.L.; He, J.; Sun, W.J. Therapeutic effects of total steroid saponin extracts from the rhizome of Dioscorea zingiberensis C.H.Wright in Freund’s complete adjuvant induced arthritis in rats. Int. Immunopharmacol. 2014, 23, 407–416. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Chen, H.; Li, G.; Zhu, H.; Gao, Y.; Zhang, L.; Sun, J. Diosgenin inhibits the migration of human breast cancer MDA-MB-231 cells by suppressing Vav2 activity. Phytomedicine 2014, 21, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Huang, H.Z.; Zhao, Y.; Xiong, C.-Q.; Yu, L.Y.; Ma, B.-P. Conversion of furostanol saponins into spirostanol saponins improves the yield of diosgenin from Dioscorea zingiberensis by acid hydrolysis. RSC Adv. 2015, 5, 4831–4837. [Google Scholar] [CrossRef]
- Pan, C.; Zhao, Y.; Liu, G.; Dou, G.; Ru, Z.; Zhu, K. Development and demonstration of a cleaner process to produce diosgenin from Dioscorea zingiberensis based on physical separation. J. Clean. Prod. 2014, 76, 161–166. [Google Scholar] [CrossRef]
- Wang, P.; Ma, C.; Chen, S.; Zhu, S.; Lou, Z.; Wang, H. Conversion of steroid saponins into diosgenin by catalytic hydrolysis using acid-functionalized ionic liquid under microwave irradiation. J. Clean. Prod. 2014, 79, 265–270. [Google Scholar] [CrossRef]
- Zhao, W.-Y.; Hao, H.-Q.; Xiao, W.; Zhu, Q.-S. Subcritical water extraction of Diosgenin from Dioscorea nipponica Makino and its antioxidant activity. IOP Conf. Ser. Earth Environ. Sci. 2020, 559, 012004. [Google Scholar] [CrossRef]
- Shen, B.; Yu, X.; Jiang, W.; Yuan, H.; Zhao, M.; Zhou, H.; Pan, Z. Green Conversion of Saponins to Diosgenin in an Alcoholysis System Catalyzed by Solid Acid Derived from Phosphorus Tailings. ACS Omega 2021, 6, 5423–5435. [Google Scholar] [CrossRef]
- Shen, B.; Zhang, F.; Zhao, M.; Pan, Z.; Cheng, Q.; Zhou, H. Synthesis and characterization of magnetic solid acid Fe3O4@PEI@SO3H and application for the production of diosgenin by alcoholysis of turmeric saponins. Mol. Catal. 2021, 511, 111751. [Google Scholar] [CrossRef]
- Yuan, H.; Yu, X.; Jiang, W.; Shen, B.; Zhang, F.; Pan, Z.; Zhou, H. Polymer-based solid acid catalyst for the green production of diosgenin. J. Appl. Polym. Sci. 2021, 139, 51596. [Google Scholar] [CrossRef]
- Liu, L.; Dong, Y.-S.; Qi, S.-S.; Wang, H.; Xiu, Z.-L. Biotransformation of steriodal saponins in Dioscorea zingiberensis CH Wright to diosgenin by Trichoderma harzianum. Appl. Microbiol. Biotechnol. 2010, 85, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Liu, L. Microwave-assisted three-liquid-phase extraction of diosgenin and steroidal saponins from fermentation broth of Dioscorea zingiberensis CH Wright. Solvent Extr. Res. Dev. Jpn. 2016, 23, 101–114. [Google Scholar] [CrossRef]
- Shu, G.; Wang, Z.; Chen, H. Screening and identification of probiotic Lactobacillus for the production of diosgenin from Dioscorea zingiberensis Wright by biotransformation. Biotechnol. Biotechnol. Equip. 2017, 31, 1026–1032. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, H.; Qiu, M.; Zhu, T.; Ni, J. Investigation on the mechanisms for biotransformation of saponins to diosgenin. World J. Microbiol. Biotechnol. 2014, 30, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Guo, Y.; Xue, P.; Xue, Z.; Zhang, Y.; Zhang, H.; Ito, Y.; Dou, J.; Guo, Z. Research progress of diosgenin extraction from Dioscorea zingiberensis C. H. Wright: Inspiration of novel method with environmental protection and efficient characteristics. Steroids 2023, 192, 109181. [Google Scholar] [CrossRef]
- Xiang, H.; Zhang, T.; Pang, X.; Wei, Y.; Liu, H.; Zhang, Y.; Ma, B.; Yu, L. Isolation of endophytic fungi from Dioscorea zingiberensis C. H. Wright and application for diosgenin production by solid-state fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 5519–5532. [Google Scholar] [CrossRef]
- Liu, W.; Huang, W.; Sun, W.; Zhu, Y.; Ni, J. Production of diosgenin from yellow ginger (Dioscorea zingiberensis CH Wright) saponins by commercial cellulase. World J. Microbiol. Biotechnol. 2010, 26, 1171–1180. [Google Scholar] [CrossRef]
- Zhang, J.; Li, H.; Lu, Z.; Shi, J.; Xu, Z. Screening and condition optimization of a strain for efficiently biotransformation of saponins in Dioscorea zingiberensis into diosgenin. Sheng Wu Gong Cheng Xue Bao = Chin. J. Biotechnol. 2013, 29, 848–852. [Google Scholar]
- Wei, M.; Bai, Y.; Ao, M.; Jin, W.; Yu, P.; Zhu, M.; Yu, L. Novel method utilizing microbial treatment for cleaner production of diosgenin from Dioscorea zingiberensis CH Wright (DZW). Bioresour. Technol. 2013, 146, 549–555. [Google Scholar] [CrossRef]
- Huang, W.; Zhao, H.; Ni, J.; Zuo, H.; Qiu, L.; Li, H.; Li, H. The best utilization of D. zingiberensis CH Wright by an eco-friendly process. Bioresour. Technol. 2008, 99, 7407–7411. [Google Scholar] [CrossRef] [PubMed]
- Mou, F.; Tian, J.; Li, Y.; Han, S.; Shang, R.; Song, Y.; Feng, S.; Zhang, Y.; Cao, R.; Qin, B. A Method for Improving Microbial Conversion of Diosgenin and Separation and Identification of the Product. Fermentation 2023, 9, 70. [Google Scholar] [CrossRef]
- Mosayebi, M.; Salehi, Z.; Doosthosseini, H.; Tishbi, P.; Kawase, Y. Amine, thiol, and octyl functionalization of GO-Fe3O4 nanocomposites to enhance immobilization of lipase for transesterification. Renew. Energy 2020, 154, 569–580. [Google Scholar] [CrossRef]
- Guo, H.; Lei, B.; Yu, J.; Chen, Y.; Qian, J. Immobilization of lipase by dialdehyde cellulose crosslinked magnetic nanoparticles. Int. J. Biol. Macromol. 2021, 185, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Shahedi, M.; Habibi, Z.; Yousefi, M.; Brask, J.; Mohammadi, M. Improvement of biodiesel production from palm oil by co-immobilization of Thermomyces lanuginosa lipase and Candida antarctica lipase B: Optimization using response surface methodology. Int. J. Biol. Macromol. 2021, 170, 490–502. [Google Scholar] [CrossRef]
- Yang, X.; Chen, Y.; Yao, S.; Qian, J.; Guo, H.; Cai, X. Preparation of immobilized lipase on magnetic nanoparticles dialdehyde starch. Carbohydr. Polym. 2019, 218, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Mobini-Dehkordi, M.; Javan, F.A. Application of alpha-amylase in biotechnology. J. Biol. Today World 2012, 1, 39–50. [Google Scholar] [CrossRef]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Virgen-Ortíz, J.J.; dos Santos, J.C.S.; Berenguer-Murcia, Á.; Alcantara, A.R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports: Immobilization mechanism, advantages, problems, and solutions. Biotechnol. Adv. 2019, 37, 746–770. [Google Scholar] [CrossRef]
- Facin, B.R.; Melchiors, M.S.; Valério, A.; Oliveira, J.V.; Oliveira, D.d. Driving Immobilized Lipases as Biocatalysts: 10 Years State of the Art and Future Prospects. Ind. Eng. Chem. Res. 2019, 58, 5358–5378. [Google Scholar] [CrossRef]
- Qin, Y. 7—Seaweed Hydrocolloids as Thickening, Gelling, and Emulsifying Agents in Functional Food Products. In Bioactive Seaweeds for Food Applications; Qin, Y., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 135–152. [Google Scholar] [CrossRef]
- Tsai, C.-T.; Meyer, A.S. Enzymatic Cellulose Hydrolysis: Enzyme Reusability and Visualization of β-Glucosidase Immobilized in Calcium Alginate. Molecules 2014, 19, 19390–19406. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pacheco, P.; García-Béjar, B.; Briones Pérez, A.; Arévalo-Villena, M. Free and Immobilised β-Glucosidases in Oenology: Biotechnological Characterisation and Its Effect on Enhancement of Wine Aroma. Front. Microbiol. 2021, 12, 723815. [Google Scholar] [CrossRef] [PubMed]
- Reda, F.M.; Hassan, N.S.; El-Moghazy, A.-N. Decolorization of synthetic dyes by free and immobilized laccases from newly isolated strain Brevibacterium halotolerans N11 (KY883983). Biocatal. Agric. Biotechnol. 2018, 15, 138–145. [Google Scholar] [CrossRef]
- Braccini, I.; Pérez, S. Molecular basis of Ca2+-induced gelation in alginates and pectins: The egg-box model revisited. Biomacromolecules 2001, 2, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.C.; Rodrigues, J.A.R.; Moran, P.J.S.; Valença, G.P.; Nunhez, J.R. Effect of immobilized cells in calcium alginate beads in alcoholic fermentation. AMB Express 2013, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Taqieddin, E.; Amiji, M. Enzyme immobilization in novel alginate–chitosan core-shell microcapsules. Biomaterials 2004, 25, 1937–1945. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, H.; Wang, J.; Xing, L. Preparation of active corn peptides from zein through double enzymes immobilized with calcium alginate–chitosan beads. Process Biochem. 2014, 49, 1682–1690. [Google Scholar] [CrossRef]
- Babaki, M.; Yousefi, M.; Habibi, Z.; Mohammadi, M.; Yousefi, P.; Mohammadi, J.; Brask, J. Enzymatic production of biodiesel using lipases immobilized on silica nanoparticles as highly reusable biocatalysts: Effect of water, t-butanol and blue silica gel contents. Renew. Energy 2016, 91, 196–206. [Google Scholar] [CrossRef]
- Abd Manan, F.M.; Attan, N.; Zakaria, Z.; Mahat, N.A.; Abdul Wahab, R. Insight into the Rhizomucor miehei lipase supported on chitosan-chitin nanowhiskers assisted esterification of eugenol to eugenyl benzoate. J. Biotechnol. 2018, 280, 19–30. [Google Scholar] [CrossRef]
- Drozd, R.; Rakoczy, R.; Wasak, A.; Junka, A.; Fijałkowski, K. The application of magnetically modified bacterial cellulose for immobilization of laccase. Int. J. Biol. Macromol. 2018, 108, 462–470. [Google Scholar] [CrossRef]
- Purwanto, M.G.M. The Role and Efficiency of Ammonium Sulphate Precipitation in Purification Process of Papain Crude Extract. Procedia Chem. 2016, 18, 127–131. [Google Scholar] [CrossRef]
- Fukasawa, K.M.; Hata, T.; Ono, Y.; Hirose, J. Metal preferences of zinc-binding motif on metalloproteases. J. Amino Acids 2011, 2011, 574816. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.; Gutiérrez, M.S.; González, A.M.; Gárate-Castro, C.; Sepúlveda, D.; Barahona, S.; Baeza, M.; Cifuentes, V.; Alcaíno, J. Metallopeptidase Stp1 activates the transcription factor Sre1 in the carotenogenic yeast Xanthophyllomyces dendrorhous [S]. J. Lipid Res. 2020, 61, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Martin-Cuadrado, A.B.; Fontaine, T.; Esteban, P.F.; del Dedo, J.E.; de Medina-Redondo, M.; del Rey, F.; Latge, J.P.; de Aldana, C.R. Characterization of the endo-beta-1,3-glucanase activity of S. cerevisiae Eng2 and other members of the GH81 family. Fungal Genet. Biol. 2008, 45, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Hong, T.Y.; Meng, M. Biochemical characterization and antifungal activity of an endo-1,3-β-glucanase of Paenibacillus sp. isolated from garden soil. Appl. Microbiol. Biotechnol. 2003, 61, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Arima, J.; Uesugi, Y.; Iwabuchi, M.; Hatanaka, T. Streptomyces aminopeptidase P: Biochemical characterization and insight into the roles of its N-terminal domain. Protein Eng. Des. Sel. 2008, 21, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Matsushita-Morita, M.; Tada, S.; Suzuki, S.; Hattori, R.; Kusumoto, K.I. Enzymatic characterization of a novel Xaa-Pro aminopeptidase XpmA from Aspergillus oryzae expressed in Escherichia coli. J. Biosci. Bioeng. 2017, 124, 534–541. [Google Scholar] [CrossRef]
- Chauvaux, S.; Souchon, H.; Alzari, P.M.; Chariot, P.; Béguin, P. Structural and Functional Analysis of the Metal-binding Sites of Clostridium thermocellum Endoglucanase CelD (*). J. Biol. Chem. 1995, 270, 9757–9762. [Google Scholar] [CrossRef]
- Ye, T.; Song, W.; Zhang, J.-J.; An, M.; Feng, S.; Yan, S.; Li, J. Identification and functional characterization of DzS3GT, a cytoplasmic glycosyltransferase catalyzing biosynthesis of diosgenin 3-O-glucoside in Dioscorea zingiberensis. Plant Cell Tissue Organ Cult. (PCTOC) 2017, 129, 399–410. [Google Scholar] [CrossRef]
- Li, T.; Wang, N.; Li, S.; Zhao, Q.; Guo, M.; Zhang, C. Optimization of covalent immobilization of pectinase on sodium alginate support. Biotechnol. Lett. 2007, 29, 1413–1416. [Google Scholar] [CrossRef]
- Li, L.-J.; Xia, W.-J.; Ma, G.-P.; Chen, Y.-L.; Ma, Y.-Y. A study on the enzymatic properties and reuse of cellulase immobilized with carbon nanotubes and sodium alginate. AMB Express 2019, 9, 112. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L. Horseradish peroxidase immobilized on multi-walled carbon nanotubes/cordierite composite carrier and diesel sewage treatment. JB Univ. Chem. Technol. 2015, 30, 15–34. [Google Scholar]
- Zambari, I.F.; Hafid, S.R.A.; Muhamad, N.A. Optimisation of Protease Purification from Christia vespertilionis Leaves and Its Anti-Inflammatory Activity. Sains Malays. 2023, 52, 1371–1381. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991, 199, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.S.; Walia, A.K.; Kennedy, J.F. Purification and characterization of a mitogenic lectin from Penicillium duclauxii. Int. J. Biol. Macromol. 2018, 116, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chi, Y.; Cheng, Y.; Zhao, Y. Physicochemical properties, in vitro digestibility and antioxidant activity of dry-heated egg white protein. Food Chem. 2018, 246, 18–25. [Google Scholar] [CrossRef]
- Idris, A.; Suzana, W. Effect of sodium alginate concentration, bead diameter, initial pH and temperature on lactic acid production from pineapple waste using immobilized Lactobacillus delbrueckii. Process Biochem. 2006, 41, 1117–1123. [Google Scholar] [CrossRef]
- Tavano, O.L.; Fernandez-Lafuente, R.; Goulart, A.J.; Monti, R. Optimization of the immobilization of sweet potato amylase using glutaraldehyde-agarose support. Characterization of the immobilized enzyme. Process Biochem. 2013, 48, 1054–1058. [Google Scholar] [CrossRef]
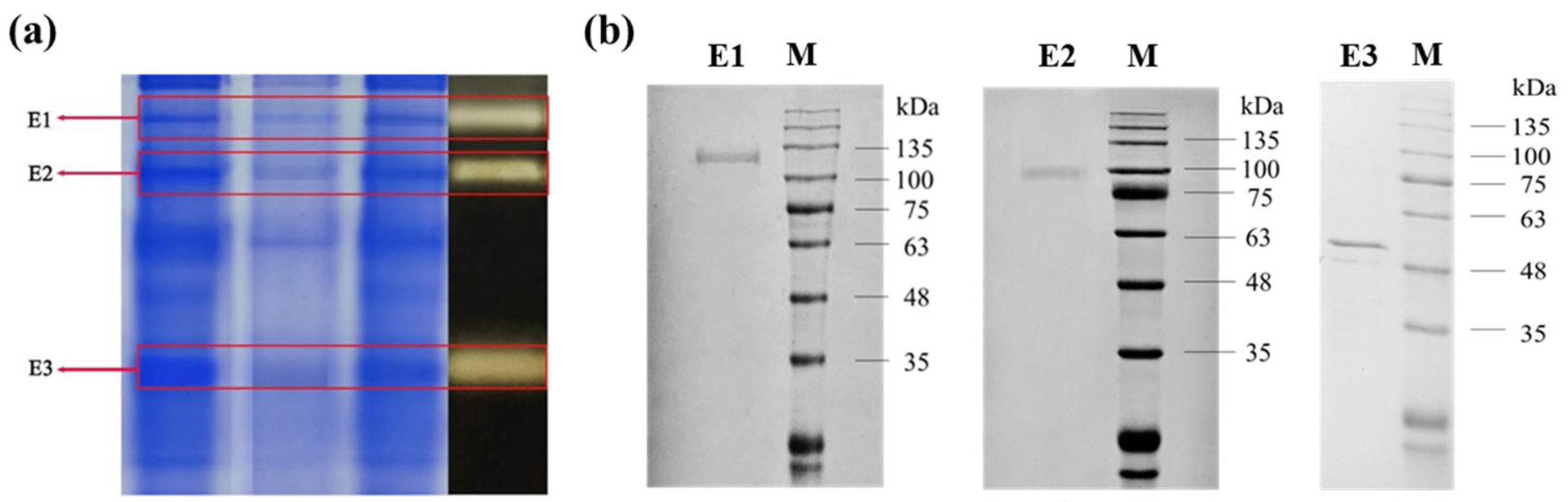

| Variable | E1 | E2 | E3 |
|---|---|---|---|
| Similar protein | A-pheromone processing metallopeptidase Ste23 | glucan endo-1,3-beta-D-glucosidase | Xaa-Pro aminopeptidase |
| Regulation gene ID | 64846173 | 64845841 | 64853314 |
| score | 323.31 | 310.4 | 323.31 |
| Unique peptides | 34 | 27 | 23 |
| Sequence coverage (%) | 35.8 | 38.2 | 61.5 |
| Number | Factor | ||||
|---|---|---|---|---|---|
| A | B | C | D | Enzyme Activity Recovery Rate (%) | |
| 1 | 0 | 0 | 0 | 0 | 40.31 |
| 2 | −1 | 0 | 0 | −1 | 32.34 |
| 3 | 0 | 0 | 0 | 0 | 39.08 |
| 4 | 0 | 1 | −1 | 0 | 31.68 |
| 5 | 1 | 1 | 0 | 0 | 38.7 |
| 6 | 1 | 0 | 0 | 1 | 38.89 |
| 7 | 1 | 0 | −1 | 0 | 34.26 |
| 8 | 1 | −1 | 0 | 0 | 30.2 |
| 9 | −1 | −1 | 0 | 0 | 30.4 |
| 10 | 0 | 1 | 0 | 1 | 38.65 |
| 11 | 0 | 1 | 1 | 0 | 37.21 |
| 12 | −1 | 0 | 1 | 0 | 29.7 |
| 13 | −1 | 0 | −1 | 0 | 30.65 |
| 14 | 0 | 1 | 0 | −1 | 34.73 |
| 15 | 0 | −1 | 0 | 1 | 28.6 |
| 16 | 1 | 0 | 1 | 0 | 37.66 |
| 17 | −1 | 1 | 0 | 0 | 28.76 |
| 18 | 0 | −1 | −1 | 0 | 34.61 |
| 19 | −1 | 0 | 0 | 1 | 27.01 |
| 20 | 0 | 0 | 0 | 0 | 39.05 |
| 21 | 0 | 0 | −1 | 1 | 32.67 |
| 22 | 0 | 0 | 0 | 0 | 38.5 |
| 23 | 0 | 0 | 1 | 1 | 36.31 |
| 24 | 0 | −1 | 1 | 0 | 29.2 |
| 25 | 0 | 0 | 0 | 0 | 39.67 |
| 26 | 1 | 0 | 0 | −1 | 35.67 |
| 27 | 0 | 0 | −1 | −1 | 36.71 |
| 28 | 0 | 0 | 1 | −1 | 36.06 |
| 29 | 0 | −1 | 0 | −1 | 34.88 |
| Source | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 431.80 | 14 | 30.84 | 39.12 | <0.0001 |
| A | 111.14 | 1 | 111.14 | 140.98 | <0.0001 |
| B | 39.75 | 1 | 39.75 | 50.42 | <0.0001 |
| C | 2.58 | 1 | 2.58 | 3.27 | 0.0922 |
| D | 5.69 | 1 | 5.69 | 7.21 | 0.0178 |
| AB | 25.70 | 1 | 25.70 | 32.61 | <0.0001 |
| AC | 4.73 | 1 | 4.73 | 6.00 | 0.0281 |
| AD | 18.28 | 1 | 18.28 | 23.18 | 0.0003 |
| BC | 29.92 | 1 | 29.92 | 37.95 | <0.0001 |
| BD | 26.01 | 1 | 26.01 | 32.99 | <0.0001 |
| CD | 4.60 | 1 | 4.60 | 5.84 | 0.0299 |
| A2 | 100.98 | 1 | 100.98 | 128.09 | <0.0001 |
| B2 | 80.51 | 1 | 80.51 | 102.13 | <0.0001 |
| C2 | 36.91 | 1 | 36.91 | 46.83 | <0.0001 |
| D2 | 17.89 | 1 | 17.89 | 22.69 | 0.0003 |
| Residual | 11.04 | 14 | 0.7883 | ||
| Lack of Fit | 9.13 | 10 | 0.9131 | 1.92 | 0.2777 |
| Error | 1.91 | 4 | 0.4764 | ||
| Total | 442.84 | 28 |
| Variable | Unit | Code Level-Value | Factor Value |
|---|---|---|---|
| Sodium alginate concentration | % | −1, 0, 1 | 2.5; 3; 3.5 |
| CaCl2 concentration | % | −1, 0, 1 | 2.5; 3; 3.5 |
| Immobilization time | h | −1, 0, 1 | 0.5; 1; 1.5 |
| pH | - | −1, 0, 1 | 7.0; 8; 9.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, S.; Pan, L.; Li, Q.; Zhang, Y.; Mou, F.; Liu, Z.; Zhang, Y.; Duan, L.; Qin, B.; Hu, Z. The Isolation, Identification and Immobilization Method of Three Novel Enzymes with Diosgenin-Producing Activity Derived from an Aspergillus flavus. Int. J. Mol. Sci. 2023, 24, 17611. https://doi.org/10.3390/ijms242417611
Feng S, Pan L, Li Q, Zhang Y, Mou F, Liu Z, Zhang Y, Duan L, Qin B, Hu Z. The Isolation, Identification and Immobilization Method of Three Novel Enzymes with Diosgenin-Producing Activity Derived from an Aspergillus flavus. International Journal of Molecular Sciences. 2023; 24(24):17611. https://doi.org/10.3390/ijms242417611
Chicago/Turabian StyleFeng, Shirong, Lintao Pan, Quanshun Li, Yi Zhang, Fangyuan Mou, Zhao Liu, Yuanyuan Zhang, Longfei Duan, Baofu Qin, and Zhongqiu Hu. 2023. "The Isolation, Identification and Immobilization Method of Three Novel Enzymes with Diosgenin-Producing Activity Derived from an Aspergillus flavus" International Journal of Molecular Sciences 24, no. 24: 17611. https://doi.org/10.3390/ijms242417611





