The Effect of Silencing Fatty Acid Elongase 4 and 6 Genes on the Proliferation and Migration of Colorectal Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Baseline Expression of Elongase 4 and 6
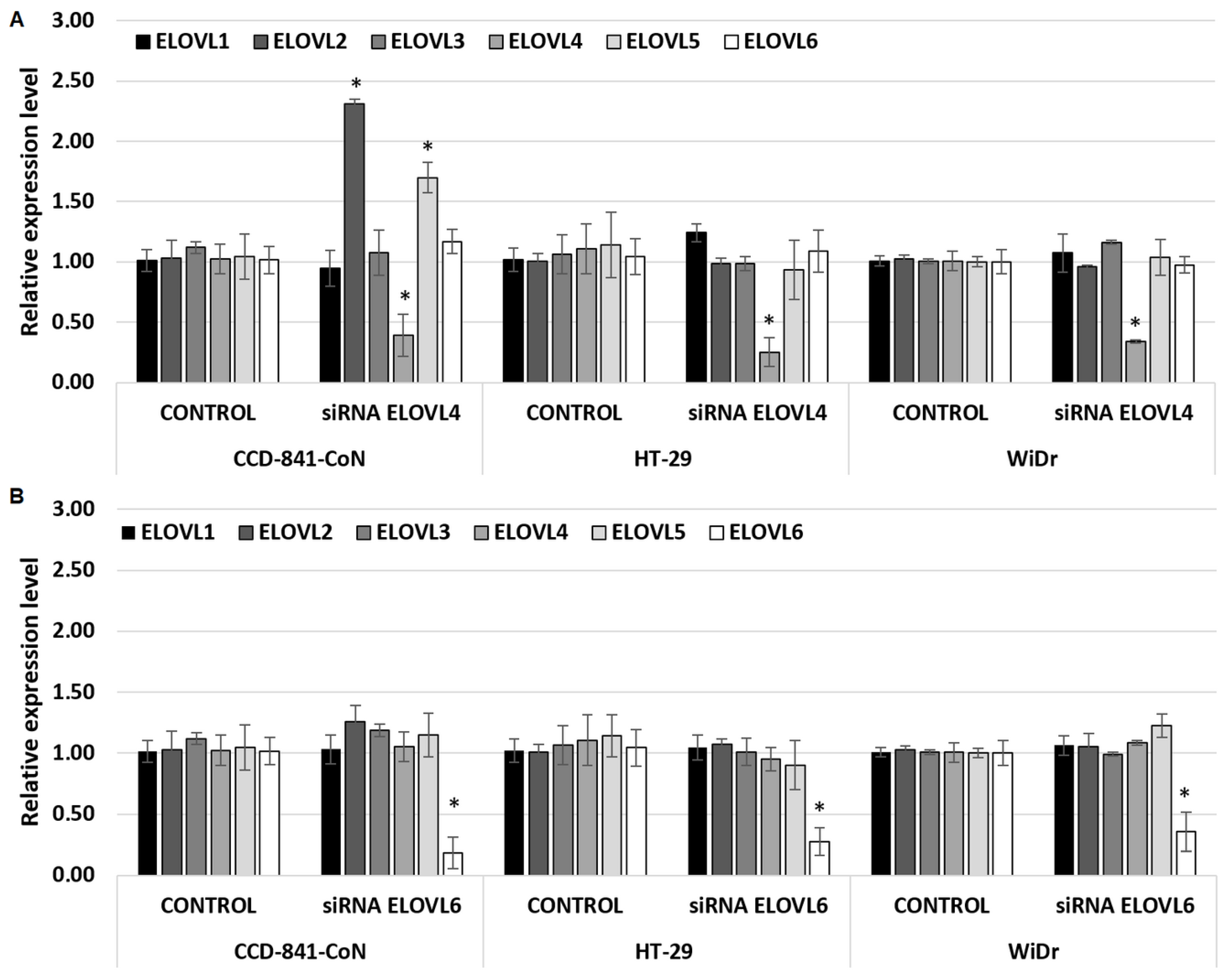
2.2. Modulation of ELOVL4 and ELOVL6 Genes Expression
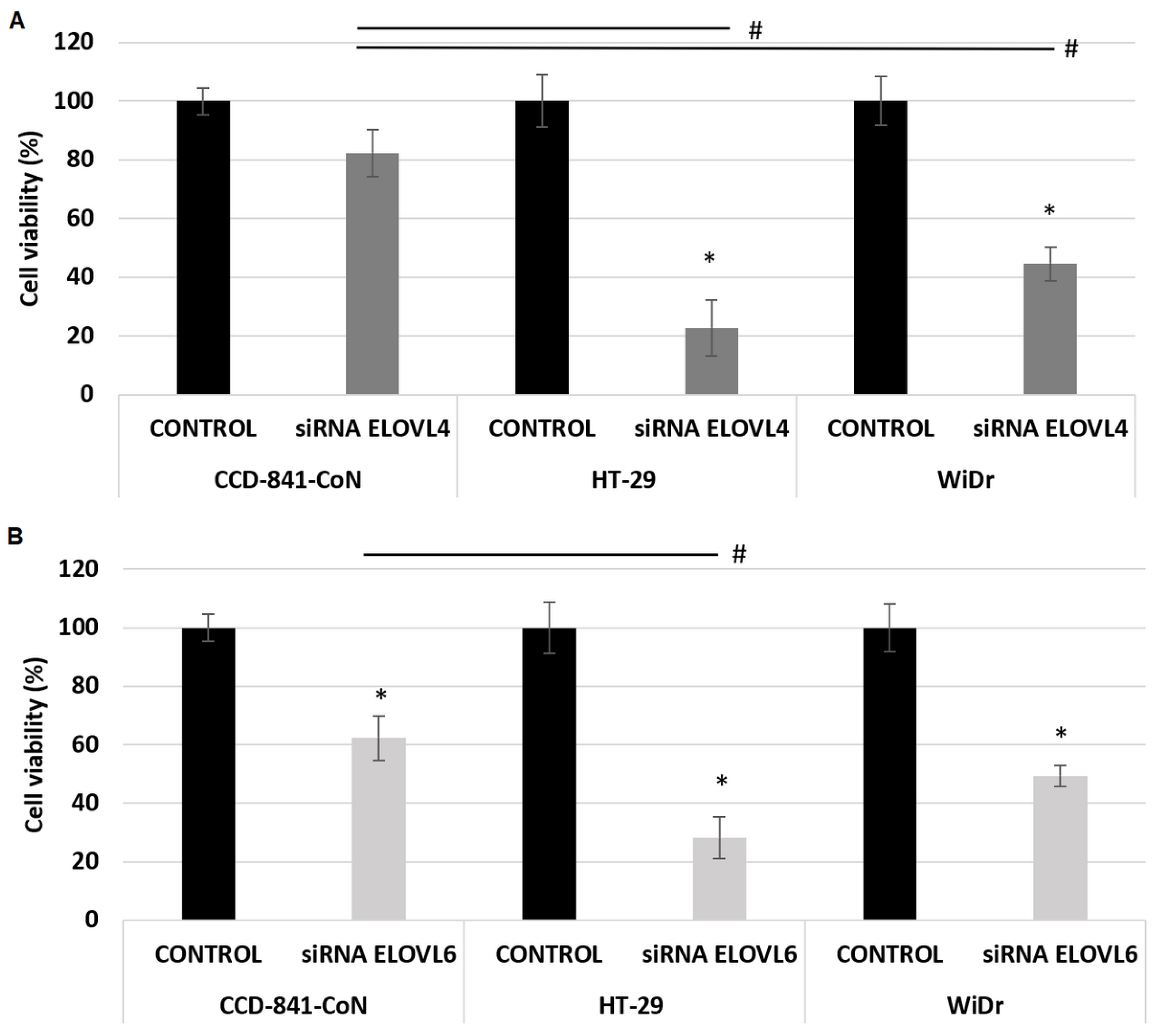
2.3. The Effect of ELOVL4 and ELOVL6 Silencing on Cell Viability
2.4. The Effect of ELOVL4 and ELOVL6 Silencing on Cell Migration
3. Discussion
4. Materials and Methods
4.1. Tissue Samples
4.2. Cell Culture
4.3. Modulation of Fatty Acid Elongases Expression with siRNA
4.4. Cell Proliferation Assay
4.5. Cell Migration Assay
4.6. RNA Extraction and Real-Time Polymerase Chain Reaction (PCR) Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, M. Colon Cancer: A Clinician’s Perspective in 2019. Gastroenterol. Res. 2020, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Malki, A.; Elruz, R.A.; Gupta, I.; Allouch, A.; Vranic, S.; Al Moustafa, A.E. Molecular mechanisms of colon cancer progression and metastasis: Recent insights and advancements. Int. J. Mol. Sci. 2021, 22, 130. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Liu, R.; Meng, Y.; Xing, D.; Xu, D.; Lu, Z. Lipid metabolism and cancer. J. Exp. Med. 2021, 218, e20201606. [Google Scholar] [CrossRef] [PubMed]
- Molendijk, J.; Robinson, H.; Djuric, Z.; Hill, M.M. Lipid mechanisms in hallmarks of cancer. Mol. Omics 2020, 16, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Salita, T.; Rustam, Y.H.; Mouradov, D.; Sieber, O.M.; Reid, G.E. Reprogrammed Lipid Metabolism and the Lipid-Associated Hallmarks of Colorectal Cancer. Cancers 2022, 14, 3714. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Nishiumi, S.; Shinohara, M.; Hatano, N.; Ikeda, A.; Yoshie, T.; Kobayashi, T.; Shiomi, Y.; Irino, Y.; Takenawa, T.; et al. Serum fatty acid profiling of colorectal cancer by gas chromatography/mass spectrometry. Biomark. Med. 2011, 5, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Yang, L.; Li, L.; Bai, Y.; Cai, C.; Liu, H. A plasma lipidomics strategy reveals perturbed lipid metabolic pathways and potential lipid biomarkers of human colorectal cancer. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1068, 41–48. [Google Scholar] [CrossRef]
- Mika, A.; Kobiela, J.; Czumaj, A.; Chmielewski, M.; Stepnowski, P.; Sledzinski, T. Hyper-Elongation in Colorectal Cancer Tissue—Cerotic Acid is a Potential Novel Serum Metabolic Marker of Colorectal Malignancies. Cell. Physiol. Biochem. 2017, 41, 722–730. [Google Scholar] [CrossRef]
- Matsuzaka, T. Role of fatty acid elongase Elovl6 in the regulation of energy metabolism and pathophysiological significance in diabetes. Diabetol. Int. 2021, 12, 68–73. [Google Scholar] [CrossRef]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef]
- Deák, F.; Anderson, R.E.; Fessler, J.L.; Sherry, D.M. Novel Cellular Functions of Very Long Chain-Fatty Acids: Insight from ELOVL4 Mutations. Front. Cell. Neurosci. 2019, 13, 428. [Google Scholar] [CrossRef] [PubMed]
- Hoxha, M.; Zappacosta, B. A review on the role of fatty acids in colorectal cancer progression. Front. Pharmacol. 2022, 13, 5277. [Google Scholar] [CrossRef] [PubMed]
- Sassa, T.; Kihara, A. Metabolism of very long-chain Fatty acids: Genes and pathophysiology. Biomol. Ther. 2014, 22, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Nagase, T.; Sasaki, T.; Nagumo, A.; Shimamura, K.; Miyamoto, Y.; Kitazawa, H.; Kanesaka, M.; Yoshimoto, R.; Aragane, K.; et al. Synthesis and evaluation of a novel indoledione class of long chain fatty acid elongase 6 (ELOVL6) inhibitors. J. Med. Chem. 2009, 52, 3142–3145. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, K.; Nagumo, A.; Miyamoto, Y.; Kitazawa, H.; Kanesaka, M.; Yoshimoto, R.; Aragane, K.; Morita, N.; Ohe, T.; Takahashi, T.; et al. Discovery and characterization of a novel potent, selective and orally active inhibitor for mammalian ELOVL6. Eur. J. Pharmacol. 2010, 630, 34–41. [Google Scholar] [CrossRef]
- Takamiya, M.; Sakurai, M.; Teranishi, F.; Ikeda, T.; Kamiyama, T.; Asai, A. Lead discovery for mammalian elongation of long chain fatty acids family 6 using a combination of high-throughput fluorescent-based assay and RapidFire mass spectrometry assay. Biochem. Biophys. Res. Commun. 2016, 480, 721–726. [Google Scholar] [CrossRef]
- de Fougerolles, A.; Vornlocher, H.P.; Maraganore, J.; Lieberman, J. Interfering with disease: A progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007, 6, 443–453. [Google Scholar] [CrossRef]
- Charbe, N.B.; Amnerkar, N.D.; Ramesh, B.; Tambuwala, M.M.; Bakshi, H.A.; Aljabali, A.A.A.; Khadse, S.C.; Satheeshkumar, R.; Satija, S.; Metha, M.; et al. Small interfering RNA for cancer treatment: Overcoming hurdles in delivery. Acta Pharm. Sin. B 2020, 10, 2075–2109. [Google Scholar] [CrossRef]
- Hattab, D.; Gazzali, A.M.; Bakhtiar, A. Clinical advances of sirna-based nanotherapeutics for cancer treatment. Pharmaceutics 2021, 13, 1009. [Google Scholar] [CrossRef]
- Schmitz, J.C.; Chen, T.-M.; Chu, E. Small Interfering Double-Stranded RNAs as Therapeutic Molecules to Restore Chemosensitivity to Thymidylate Synthase Inhibitor Compounds. Cancer Res. 2004, 64, 1431–1435. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, G.; Liu, J.; Ma, N.; Chivukula, P.; Perelman, L.; Okada, K.; Chen, Z.; Gough, D.; Yu, L. Novel biodegradable lipid nano complex for siRNA delivery significantly improving the chemosensitivity of human colon cancer stem cells to paclitaxel. J. Control Release 2009, 140, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chen, H. The role of microRNAs in colorectal cancer. J. Genet. Genom. 2010, 37, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Piccinno, E.; Scalavino, V.; Armentano, R.; Giannelli, G. miR-195-5p as Regulator of γ-Catenin and Desmosome Junctions in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 17084. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Okayama, H.; Harris, C.C. The role of microRNAs in colorectal cancer. Cancer J. 2012, 18, 244–252. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, C.; Wu, Y.; Liu, J.; Wang, T.; Wang, B. BAP31-Mediated miR-206/133b Cluster Promotes Transendothelial Migration and Metastasis of Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 16740. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Kobiela, J.; Pakiet, A.; Czumaj, A.; Sokołowska, E.; Makarewicz, W.; Chmielewski, M.; Stepnowski, P.; Marino-Gamazza, A.; Sledzinski, T. Preferential uptake of polyunsaturated fatty acids by colorectal cancer cells. Sci. Rep. 2020, 10, 1954. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknaes, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.C.G.; Eide, P.W.; Eilertsen, I.A.; Johannessen, B.; Bruun, J.; Danielsen, S.A.; Bjørnslett, M.; Meza-Zepeda, L.A.; Eknæs, M.; Lind, G.E.; et al. Multi-omics of 34 colorectal cancer cell lines—A resource for biomedical studies. Mol. Cancer 2017, 16, 116. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.-S.; Hyeon Kim, J.; Jeong, N.; Shin, Y.-K.; Jung Kim, M.; Won Park, J.; Jeong, S.-Y.; Ku, J.-L. Establishment and characterization of 18 human colorectal cancer cell lines. Cancer Res. 2020, 44, 5813–5821. [Google Scholar] [CrossRef]
- Green, C.D.; Ozguden-Akkoc, C.G.; Wang, Y.; Jump, D.B.; Olson, L.K. Role of fatty acid elongases in determination of de novo synthesized monounsaturated fatty acid species. J. Lipid Res. 2010, 51, 1871–1877. [Google Scholar] [CrossRef]
- Wang, H.; Hu, M.; Shen, Z.; Zhou, X.; Yang, S.; He, K.; Li, X.; Yan, F.; Zhao, A. A Specific microRNA Targets an Elongase of Very Long Chain Fatty Acids to Regulate Fatty Acid Composition and Mitochondrial Morphology of Skeletal Muscle Cells. Animals 2022, 12, 2274. [Google Scholar] [CrossRef] [PubMed]
- Sunaga, H.; Matsui, H.; Anjo, S.; Syamsunarno, M.R.A.A.; Koitabashi, N.; Iso, T.; Matsuzaka, T.; Shimano, H.; Yokoyama, T.; Kurabayashi, M. Elongation of Long-Chain Fatty Acid Family Member 6 (Elovl6)-Driven Fatty Acid Metabolism Regulates Vascular Smooth Muscle Cell Phenotype Through AMP-Activated Protein Kinase/Krüppel-Like Factor 4 (AMPK/KLF4) Signaling. J. Am. Heart Assoc. 2016, 5, e004014. [Google Scholar] [CrossRef] [PubMed]
- Istiqamah, N.; Matsuzaka, T.; Shimizu, M.; Motomura, K.; Ohno, H.; Hasebe, S.; Sharma, R.; Okajima, Y.; Matsuda, E.; Han, S.I.; et al. Identification of key microRNAs regulating ELOVL6 and glioblastoma tumorigenesis. BBA Adv. 2023, 3, 100078. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, G.S.; Poloznikov, A.A.; Astakhova, L.A.; Raigorodskaya, M.P.; Khesina, Z.B.; Fomicheva, K.A.; Buryak, A.K.; Alekseev, B.Y. The effect of ELOVL6 fatty acid elongase inhibition on the expression of genes associated with the metastasis of breast cancer. Russ. Chem. Bull. 2018, 67, 2307–2315. [Google Scholar] [CrossRef]
- Nitta, S.; Kandori, S.; Tanaka, K.; Sakka, S.; Siga, M.; Nagumo, Y.; Negoro, H.; Kojima, T.; Mathis, B.J.; Shimazui, T.; et al. ELOVL5-mediated fatty acid elongation promotes cellular proliferation and invasion in renal cell carcinoma. Cancer Sci. 2022, 113, 2738–2752. [Google Scholar] [CrossRef] [PubMed]
- Gonzá Lez-Bengtsson, A.; Asadi, A.; Gao, H.; Dahlman-Wright, K.; Jacobsson, A. Estrogen Enhances the Expression of the Polyunsaturated Fatty Acid Elongase Elovl2 via ERα in Breast Cancer Cells. PLoS ONE 2016, 11, e0164241. [Google Scholar] [CrossRef]
- Zadravec, D.; Tvrdik, P.; Guillou, H.; Haslam, R.; Kobayashi, T.; Napier, J.A.; Capecchi, M.R.; Jacobsson, A. ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J. Lipid Res. 2011, 52, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Torres-Gonzalez, M.; Tripathy, S.; Botolin, D.; Christian, B.; Jump, D.B. Elevated hepatic fatty acid elongase-5 activity affects multiple pathways controlling hepatic lipid and carbohydrate composition. J. Lipid Res. 2008, 49, 1538–1552. [Google Scholar] [CrossRef]
- Bond, L.M.; Miyazaki, M.; O’Neill, L.M.; Ding, F.; Ntambi, J.M. Fatty Acid Desaturation and Elongation in Mammals. In Biochemistry of Lipids, Lipoproteins and Membranes, 6th ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 185–208. ISBN 9780444634382. [Google Scholar]
- Zeng, X.; Li, S.; Liu, L.; Cai, S.; Ye, Q.; Xue, B.; Wang, X.; Zhang, S.; Chen, F.; Cai, C.; et al. Role of functional fatty acids in modulation of reproductive potential in livestock. J. Anim. Sci. Biotechnol. 2023, 14, 24. [Google Scholar] [CrossRef]
- Monroig, O.; Webb, K.; Ibarra-Castro, L.; Holt, G.J.; Tocher, D.R. Biosynthesis of long-chain polyunsaturated fatty acids in marine fish: Characterization of an Elovl4-like elongase from cobia Rachycentron canadum and activation of the pathway during early life stages. Aquaculture 2011, 312, 145–153. [Google Scholar] [CrossRef]
- Monroig, Ó.; Tocher, D.R.; Navarro, J.C. Biosynthesis of polyunsaturated fatty acids in marine invertebrates: Recent advances in molecular mechanisms. Mar. Drugs 2013, 11, 3998–4018. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Ren, T.; Li, X.; Cao, X.; Gao, J. Polyunsaturated fatty acids synthesized by freshwater fish: A new insight to the roles of elovl2 and elovl5 in vivo. Biochem. Biophys. Res. Commun. 2020, 532, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Centenera, M.M.; Scott, J.S.; Machiels, J.; Nassar, Z.D.; Miller, D.C.; Zinonos, I.; Dehairs, J.; Burvenich, I.J.G.; Zadra, G.; Chetta, P.M.; et al. ELOVL5 is a critical and targetable fatty acid elongase in prostate cancer. Cancer Res. 2021, 81, 1704–1718. [Google Scholar] [CrossRef] [PubMed]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czumaj, A.; Kobiela, J.; Mika, A.; Pappou, E.; Śledziński, T. The Effect of Silencing Fatty Acid Elongase 4 and 6 Genes on the Proliferation and Migration of Colorectal Cancer Cells. Int. J. Mol. Sci. 2023, 24, 17615. https://doi.org/10.3390/ijms242417615
Czumaj A, Kobiela J, Mika A, Pappou E, Śledziński T. The Effect of Silencing Fatty Acid Elongase 4 and 6 Genes on the Proliferation and Migration of Colorectal Cancer Cells. International Journal of Molecular Sciences. 2023; 24(24):17615. https://doi.org/10.3390/ijms242417615
Chicago/Turabian StyleCzumaj, Aleksandra, Jarosław Kobiela, Adriana Mika, Emmanouil Pappou, and Tomasz Śledziński. 2023. "The Effect of Silencing Fatty Acid Elongase 4 and 6 Genes on the Proliferation and Migration of Colorectal Cancer Cells" International Journal of Molecular Sciences 24, no. 24: 17615. https://doi.org/10.3390/ijms242417615
APA StyleCzumaj, A., Kobiela, J., Mika, A., Pappou, E., & Śledziński, T. (2023). The Effect of Silencing Fatty Acid Elongase 4 and 6 Genes on the Proliferation and Migration of Colorectal Cancer Cells. International Journal of Molecular Sciences, 24(24), 17615. https://doi.org/10.3390/ijms242417615






