Heat Shock Transcription Factor GhHSFB2a Is Crucial for Cotton Resistance to Verticillium dahliae
Abstract
1. Introduction
2. Results
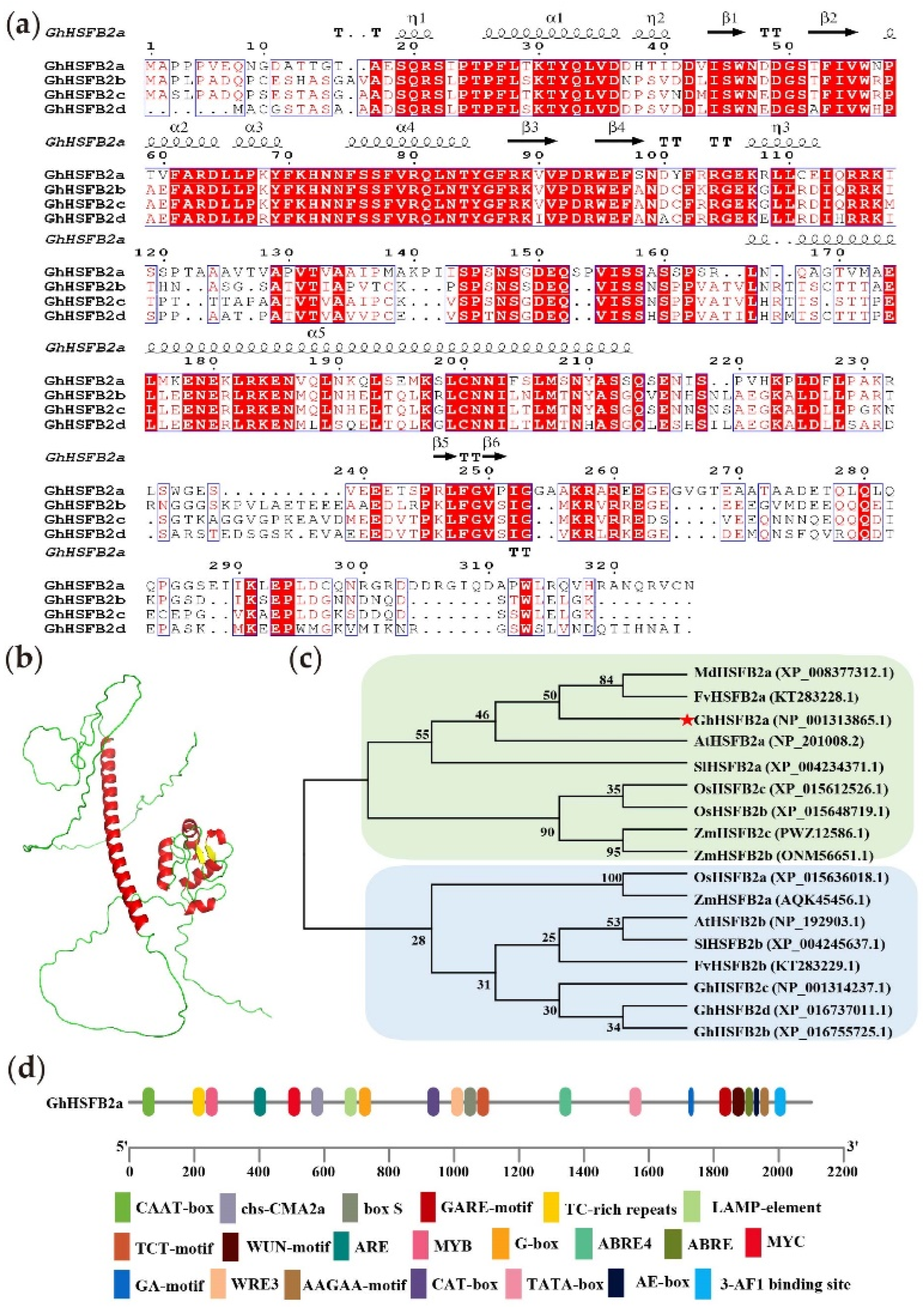
2.1. Bioinformatics Analysis of GhHSFB2a
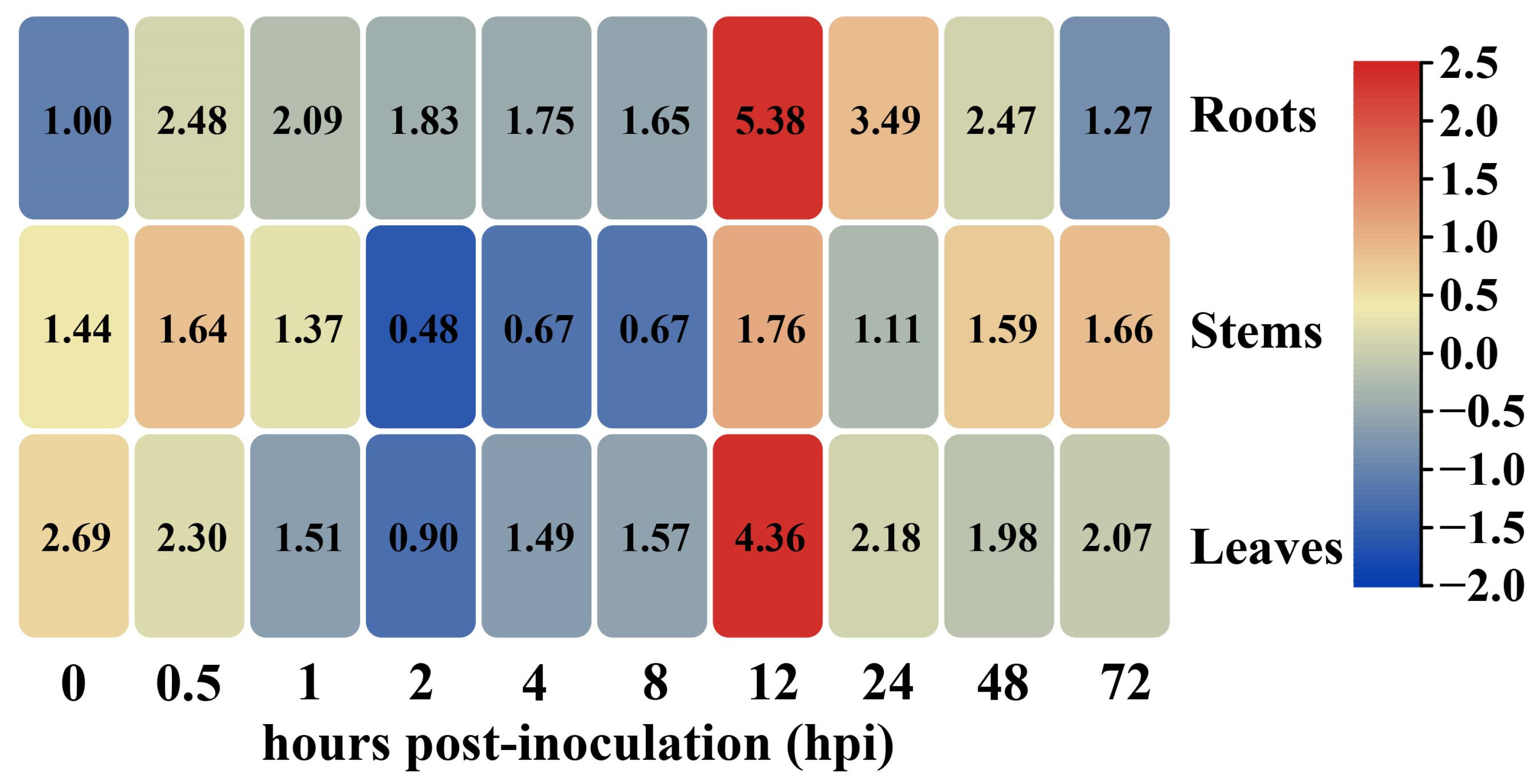
2.2. Transcriptional Levels of GhHSFB2a Were Upregulated under V. dahliae Stress
2.3. GhHSFB2a Is Localized in the Nucleus
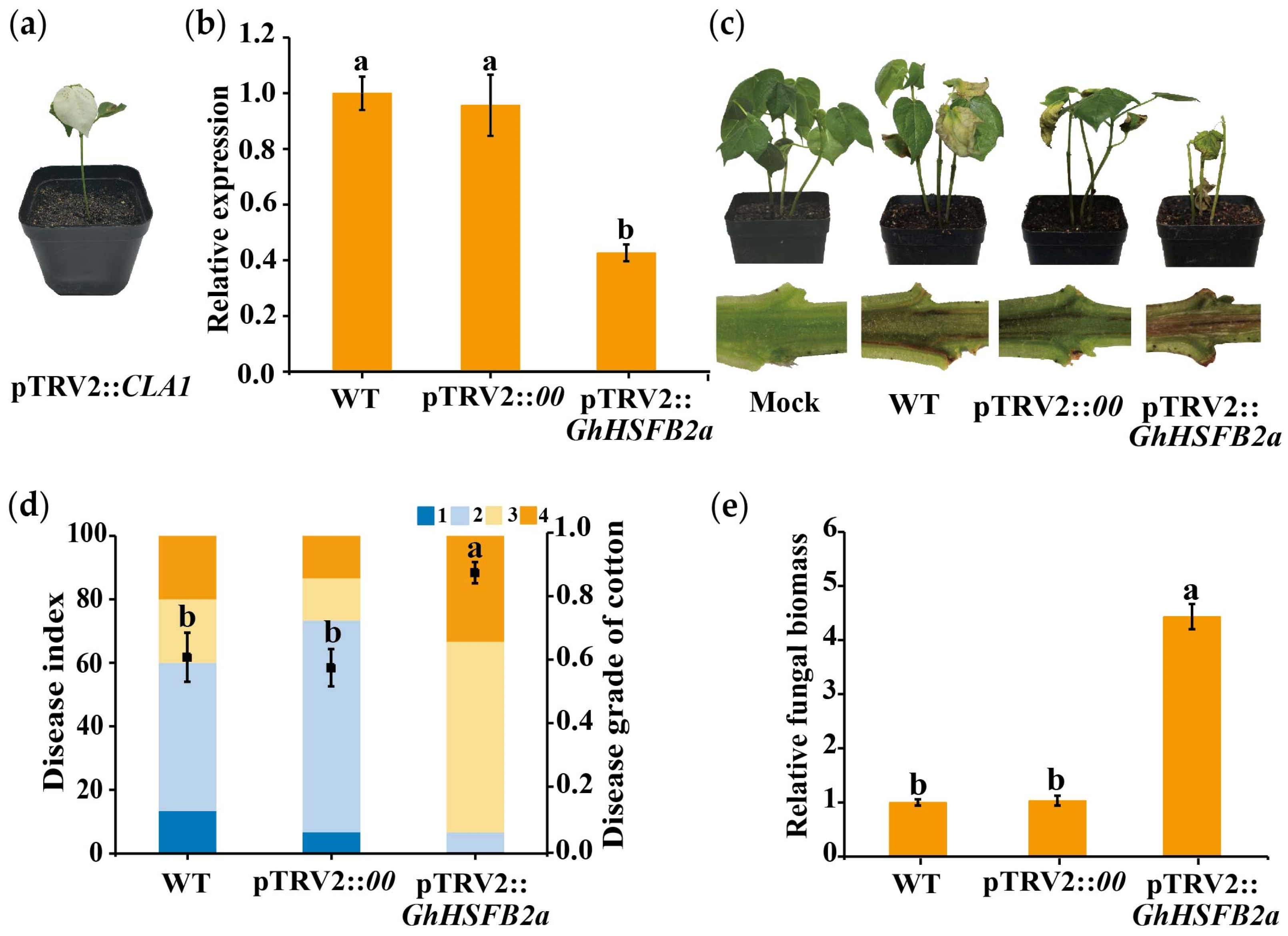
2.4. Knockdown of GhHSFB2a Increases the Susceptibility of Cotton to V. dahliae
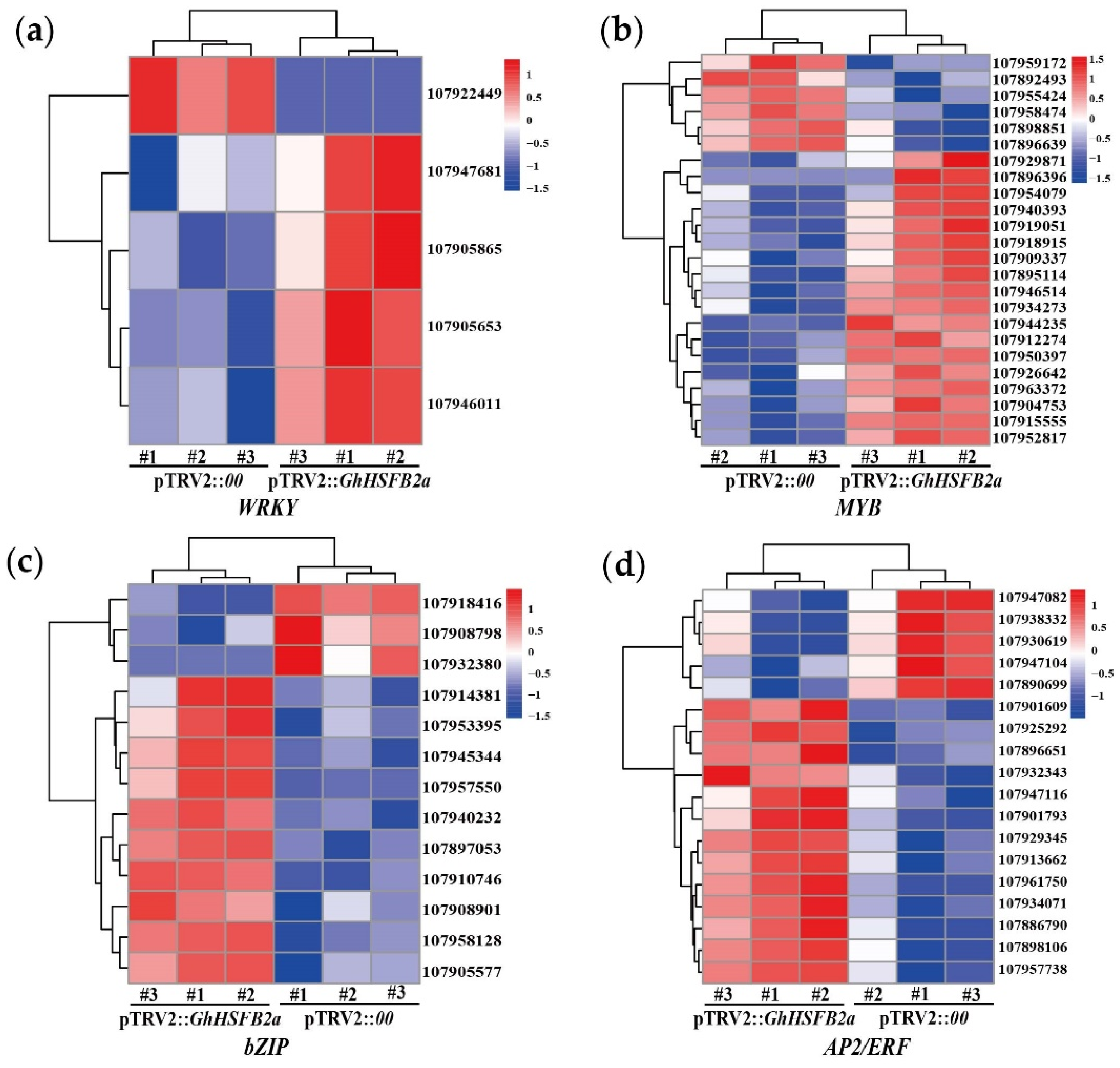
2.5. GhHSFB2a Regulates Resistance via Plant Hormones and Secondary Metabolites Pathways
2.6. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Protein-Protein Interaction (PPI) Analyses Revealed GhHSFB2a Functions
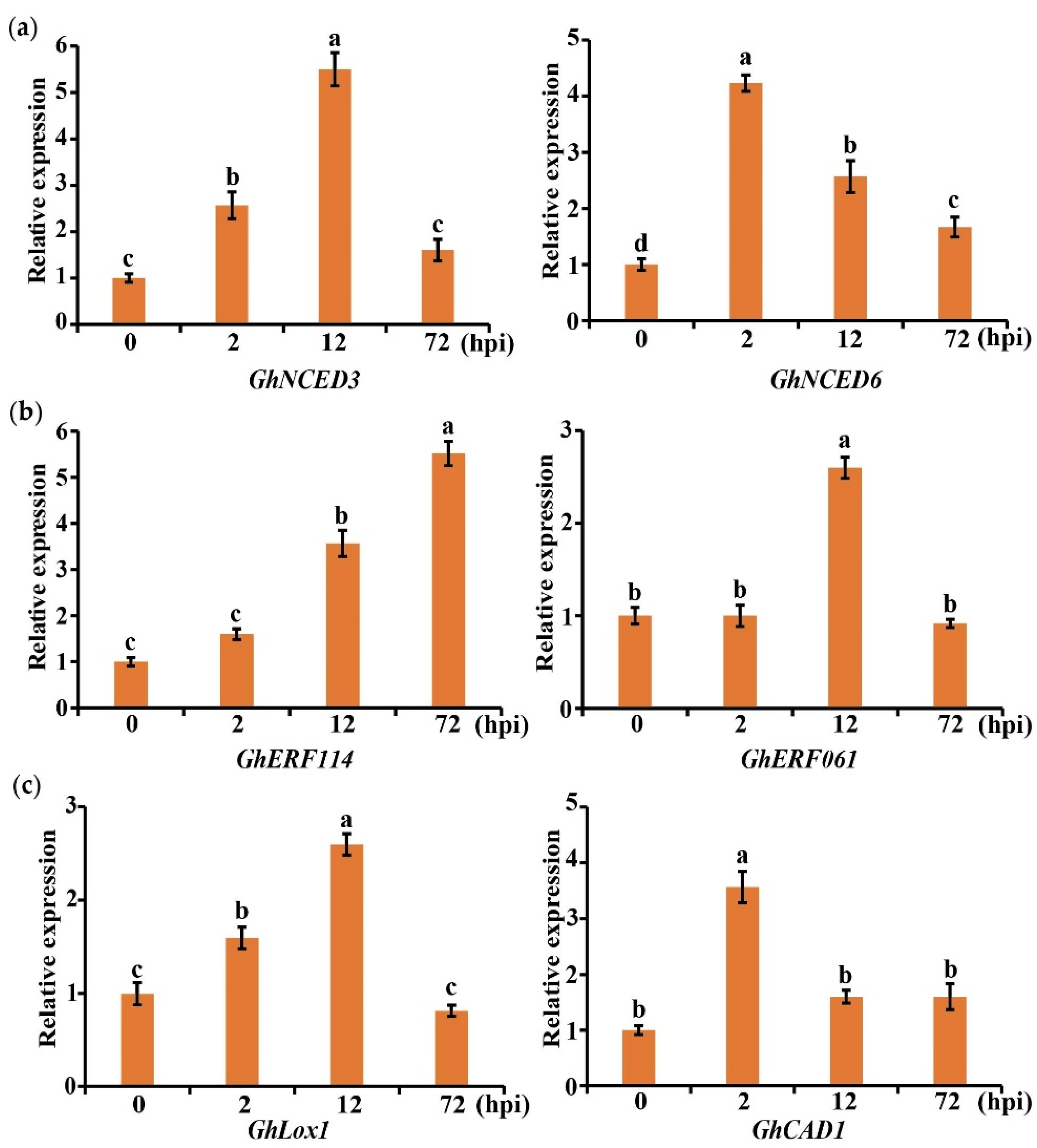
2.7. Gene Expression Related to Plant Hormone and Secondary Metabolites in Cotton Was Induced by V. dahliae
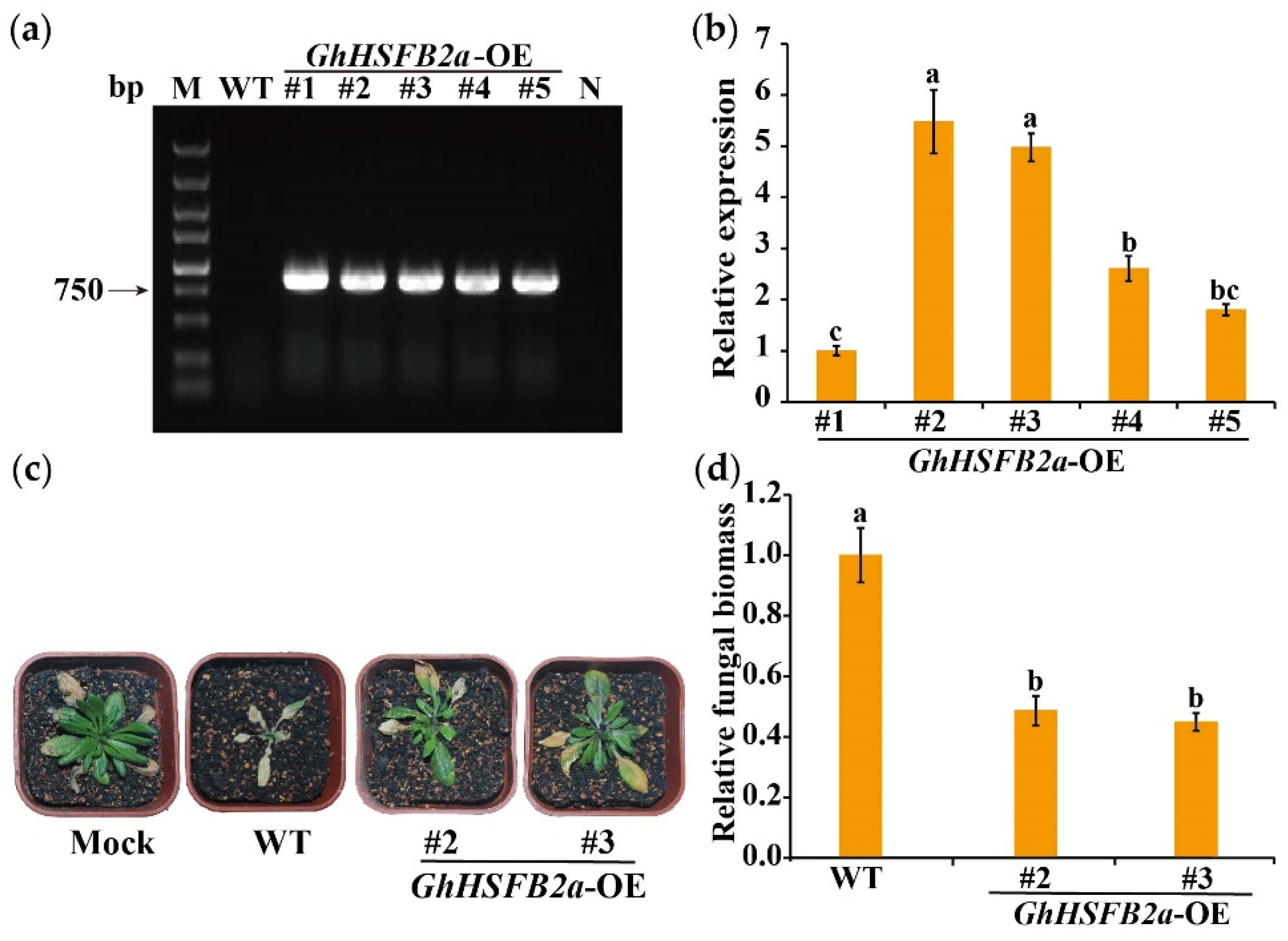
2.8. Overexpression of GhHSFB2a Enhanced V. dahliae Resistance in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Analysis
4.2. Growth of Plant Material and Pathogen Cultures
4.3. Gene Expression Analysis
4.4. Subcellular Localization and Confocal Microscopy
4.5. VIGS and RNA-seq Analysis
4.6. A. thaliana Transformation and Molecular Analysis
4.7. Pathogenicity Assay and Determination of Fungal Biomass
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, Z.; Achar, P.N.; Benkang, G. Vegetative compatibility groupings of Verticillium dahliae from cotton in mainland China. Eur. J. Plant Pathol. 1998, 104, 871–876. [Google Scholar] [CrossRef]
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Deketelaere, S.; Tyvaert, L.; França, S.C.; Höfte, M. Desirable traits of a good biocontrol agent against Verticillium wilt. Front. Microbiol. 2017, 8, 1186–1208. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, J.; Xu, J.; Zhao, L.; Wu, Q.; Xiao, S. Quantitative trait locus mapping and candidate gene analysis for Verticillium wilt resistance using Gossypium barbadense chromosomal segment introgressed line. Front. Plant Sci. 2018, 9, 682–698. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, C.; Wu, S.; Zhang, X.; Tang, J.; Jian, G.; Jiao, G.; Li, F.; Chu, C. Significant improvement of cotton Verticillium wilt resistance by manipulating the expression of gastrodia antifungal proteins. Mol. Plant 2016, 9, 1436–1439. [Google Scholar] [CrossRef]
- Amorim, L.L.B.; da Fonseca Dos Santos, R.; Neto, J.P.B.; Guida-Santos, M.; Crovella, S.; Benko-Iseppon, A.M. Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 2017, 18, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114–126. [Google Scholar] [CrossRef]
- Wang, J.; Sun, N.; Deng, T.; Zhang, L.; Zuo, K. Genome-Wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genom. 2014, 15, 961–979. [Google Scholar] [CrossRef]
- Sakurai, H.; Enoki, Y. Novel aspects of heat shock factors: DNA recognition, chromatin modulation and gene expression. FEBS J. 2010, 277, 4140–4149. [Google Scholar] [CrossRef]
- Bharti, K.; Von Koskull-Döring, P.; Bharti, S.; Kumar, P.; Tintschl-Körbitzer, A.; Treuter, E.; Nover, L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. BBA-Gene Struct. Expr. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Xue, G.P.; Drenth, J.; McIntyre, C.L. TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 2015, 66, 1025–1039. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, Z.S.; Li, P.; Yang, L.; Wei, Y.; Chen, M.; Li, L.; Zhang, G.; Ma, Y. Overexpression of TaHSF3 in transgenic Arabidopsis enhances tolerance to extreme temperatures. Plant Mol. Biol. Rep. 2013, 31, 688–697. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, L.; Wang, A.; Xu, X.; Li, J. Ectopic overexpression of SlHsfA3, a heat stress transcription factor from tomato, confers increased thermotolerance and salt hypersensitivity in germination in transgenic Arabidopsis. PLoS One 2013, 8, e54880–e54895. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Ye, C.; Lü, H.; Chen, X.; Chai, G.; Chen, J.; Wang, C. Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J. Plant Res. 2006, 119, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Shao, K.; Huang, Y.; Lei, Y.; Tan, L.; Chan, Z. Natural variation analysis of perennial ryegrass in response to abiotic stress highlights LpHSFC1b as a positive regulator of heat stress. Environ. Exp. Bot. 2020, 179, 104192–104203. [Google Scholar] [CrossRef]
- Zhuang, L.; Cao, W.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Characterization and functional analysis of FaHsfC1b from Festuca arundinacea conferring heat tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. [Google Scholar] [CrossRef]
- Schmidt, R.; Schippers, J.H.; Welker, A.; Mieulet, D.; Guiderdoni, E.; Mueller-Roeber, B. Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB PLANTS 2012, 2012, pls011–pls027. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.T.; Zhang, K.; Zhao, F.L.; Li, Y.J.; Zheng, Y.; Wang, Y.J.; Wen, Y.Q. Identification and expression analysis of heat shock transcription factors in the wild Chinese grapevine (Vitis pseudoreticulata). Plant Physiol. Biochem. 2016, 99, 1–10. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Jia, H.X.; Li, J.B.; Huang, J.; Lu, M.Z.; Hu, J.J. The heat shock factor gene family in Salix suchowensis: A genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 2015, 6, 748–761. [Google Scholar] [CrossRef]
- Huang, Y.; Li, M.Y.; Wang, F.; Xu, Z.S.; Huang, W.; Wang, G.L.; Ma, J.; Xiong, A.S. Heat shock factors in carrot: Genome-Wide identification, classification, and expression profiles response to abiotic stress. Mol. Biol. Rep. 2015, 42, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Ran, J.; Zou, J.; Zhou, X.; Liu, A.; Zhang, X.; Peng, Y.; Tang, N.; Luo, G.; Chen, X. Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Rep. 2013, 32, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Busch, W.; Birke, H.; Kemmerling, B.; Nurnberger, T.; Schoffl, F. Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol. Plant 2009, 2, 152–165. [Google Scholar] [CrossRef]
- Singh, A.; Mittal, D.; Lavania, D.; Agarwal, M.; Mishra, R.C.; Grover, A. OsHsfA2c and OsHsfB4b are involved in the transcriptional regulation of cytoplasmic OsClpB (Hsp100) gene in rice (Oryza sativa L.). Cell Stress Chaperon. 2012, 17, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, L.; Zhang, Q.; Ma, C.; Su, X.; Cheng, H.; Guo, H. AmCBF1 transcription factor regulates plant architecture by repressing GhPP2C1 or GhPP2C2 in Gossypium hirsutum. Front. Plant Sci. 2022, 13, 914206–914219. [Google Scholar] [CrossRef]
- Kessler, A.; Halitschke, R.; Baldwin, I.T. Silencing the jasmonate cascade: Induced plant defenses and insect populations. Science 2004, 305, 665–668. [Google Scholar] [CrossRef]
- Kessler, A.; Baldwin, I.T. Defensive function of herbivore-induced plant volatile emissions in nature. Science 2001, 291, 2141–2144. [Google Scholar] [CrossRef]
- Pick, T.; Jaskiewicz, M.; Peterhänsel, C.; Conrath, U. Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol. 2012, 159, 52–55. [Google Scholar] [CrossRef]
- Ashraf, M.F.; Yang, S.; Wu, R.; Wang, Y.; Hussain, A.; Noman, A.; Khan, M.I.; Liu, Z.; Qiu, A.; Guan, D.; et al. Capsicum annuum HsfB2a positively regulates the response to Ralstonia solanacearum infection or high temperature and high humidity forming transcriptional cascade with CaWRKY6 and CaWRKY40. Plant Cell Physiol. 2018, 59, 2608–2623. [Google Scholar] [CrossRef]
- Liao, W.Y.; Lin, L.F.; Jheng, J.L.; Wang, C.C.; Yang, J.H.; Chou, M.L. Identification of heat shock transcription factor genes involved in thermotolerance of octoploid cultivated strawberry. Int. J. Mol. Sci. 2016, 17, 2130. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.T.; Wei, W.; Li, Y.J.; Zhang, K.; Gao, Y.R.; Zhao, F.L.; Feng, J.Y. Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 2015, 6, 736–751. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Schiff, M.; Liu, Y.; Dinesh-Kumar, S.P. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 2006, 142, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xie, Y.; Liu, G.; Lin, D.; He, C.; Shi, H. Molecular identification of GAPDHs in cassava highlights the antagonism of MeGAPCs and MeATG8s in plant disease resistance against cassava bacterial blight. Plant Mol. Biol. 2018, 97, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, N.; Wang, K.; Xian, Q.; Dong, J.; Qi, X.; Chen, X. Chitinase chi 2 positively regulates cucumber resistance against Fusarium oxysporum f. sp. cucumerinum. Genes 2021, 13, 62. [Google Scholar] [CrossRef]
- Dong, S.; Dong, X.; Han, X.; Zhang, F.; Zhu, Y.; Xin, X.; Wang, Y.; Hu, Y.; Yuan, D.; Wang, J.; et al. OsPDCD5 negatively regulates plant architecture and grain yield in rice. Proc. Natl. Acad. Sci. USA 2021, 118, e2018799118–e2018799128. [Google Scholar] [CrossRef]
- Dang, F.; Wang, Y.; She, J.; Lei, Y.; Liu, Z.; Eulgem, T.; Lai, Y.; Lin, J.; Yu, L.; Lei, D.; et al. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 2014, 150, 397–411. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Liu, Q.; Yan, S.; Huang, W.; Yang, J.; Dong, J.; Zhang, S.; Zhao, J.; Yang, T.; Mao, X.; Zhu, X.; et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 2018, 98, 289–302. [Google Scholar] [CrossRef]
- Gao, Y.; Li, S.J.; Zhang, S.W.; Feng, T.; Zhang, Z.Y.; Luo, S.J.; Mao, H.Y.; Borkovich, K.A.; Ouyang, S.Q. SlymiR482e-3p mediates tomato wilt disease by modulating ethylene response pathway. Plant Biotechnol. J. 2021, 19, 17–19. [Google Scholar] [CrossRef]
- Sumayo, M.S.; Kwon, D.K.; Ghim, S.Y. Linoleic acid-induced expression of defense genes and enzymes in tobacco. J. Plant Physiol. 2014, 171, 1757–1762. [Google Scholar] [CrossRef]
- Su, X.; Wu, S.; Liu, L.; Lu, G.; Liu, H.; Jin, X.; Wang, Y.; Guo, H.; Wang, C.; Cheng, H. Potential antagonistic bacteria against Verticillium dahliae isolated from artificially infested nursery. Cells 2021, 10, 3588. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Lu, G.; Li, X.; Rehman, L.; Liu, W.; Sun, G.; Guo, H.; Wang, G.; Cheng, H. Host-induced gene silencing of an adenylate kinase gene involved in fungal energy metabolism improves plant resistance to Verticillium dahliae. Biomolecules 2020, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Su, X.; Lu, G.; Sun, G.; Zhang, Z.; Guo, H.; Guo, N.; Cheng, H. VdOGDH is involved in energy metabolism and required for virulence of Verticillium dahliae. Curr. Genet. 2020, 66, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.P.; Shen, J.L.; Li, W.J.; Wu, N.; Chen, C.; Hou, Y.X. Evolutionary and characteristic analysis of RING-DUF1117 E3 ubiquitin ligase genes in Gossypium discerning the role of GhRDUF4D in Verticillium dahliae resistance. Biomolecules 2021, 11, 1145. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Wang, Q.; Zhu, L.; Guo, H.; Cheng, H.; Su, X. Heat Shock Transcription Factor GhHSFB2a Is Crucial for Cotton Resistance to Verticillium dahliae. Int. J. Mol. Sci. 2023, 24, 1845. https://doi.org/10.3390/ijms24031845
Liu L, Wang Q, Zhu L, Guo H, Cheng H, Su X. Heat Shock Transcription Factor GhHSFB2a Is Crucial for Cotton Resistance to Verticillium dahliae. International Journal of Molecular Sciences. 2023; 24(3):1845. https://doi.org/10.3390/ijms24031845
Chicago/Turabian StyleLiu, Lu, Qi Wang, Linfeng Zhu, Huiming Guo, Hongmei Cheng, and Xiaofeng Su. 2023. "Heat Shock Transcription Factor GhHSFB2a Is Crucial for Cotton Resistance to Verticillium dahliae" International Journal of Molecular Sciences 24, no. 3: 1845. https://doi.org/10.3390/ijms24031845
APA StyleLiu, L., Wang, Q., Zhu, L., Guo, H., Cheng, H., & Su, X. (2023). Heat Shock Transcription Factor GhHSFB2a Is Crucial for Cotton Resistance to Verticillium dahliae. International Journal of Molecular Sciences, 24(3), 1845. https://doi.org/10.3390/ijms24031845






