Abstract
Schizophrenia (SZ) is a serious mental disorder that is typically treated with antipsychotic medication. Treatment-resistant schizophrenia (TRS) is the condition where symptoms remain after pharmacological intervention, resulting in long-lasting functional and social impairments. As the identification and treatment of a TRS patient requires previous failed treatments, early mechanisms of detection are needed in order to quicken the access to effective therapy, as well as improve treatment adherence. In this study, we aim to find a microRNA (miRNA) signature for TRS, as well as to shed some light on the molecular pathways potentially involved in this severe condition. To do this, we compared the blood miRNAs of schizophrenia patients that respond to medication and TRS patients, thus obtaining a 16-miRNA TRS profile. Then, we assessed the ability of this signature to separate responders and TRS patients using hierarchical clustering, observing that most of them are grouped correctly (~70% accuracy). We also conducted a network, pathway analysis, and bibliography search to spot molecular pathways potentially altered in TRS. We found that the response to stress seems to be a key factor in TRS and that proteins p53, SIRT1, MDM2, and TRIM28 could be the potential mediators of such responses. Finally, we suggest a molecular pathway potentially regulated by the miRNAs of the TRS profile.
1. Introduction
Schizophrenia (SZ) is a heterogeneous psychiatric disorder that affects around 1% of the world’s population [1], and it is characterised by chronic psychotic symptoms and psychosocial impairment. Since the development of chlorpromazine in the 1950s, treatment for SZ is based on antipsychotic medications; however, about 20 to 50 percent of patients do not experience an improvement in their symptoms [2,3,4,5], which is known as treatment-resistant schizophrenia (TRS). The clinical definition of TRS is based on the failure of response to at least two sequential non-clozapine antipsychotic trials of sufficient dose, duration, and adherence [6]. Patients with TRS have higher rates of substance abuse, early cognitive decline, and higher rates of suicide ideation [7,8,9,10]. As pharmacological and therapeutic approaches differ between SZ and TRS [11,12,13,14], early identification of the resistance condition is essential for a rapid and effective pharmacological intervention, limiting damage to the patient and their environment and improving the adherence to treatment [15]. As a consequence, in the last two decades, an increasing number of studies have made great efforts to find molecular mechanisms behind TRS and spot biomarkers for clinical use. However, the difficult access to neural tissues, the subtle definition of TRS, and the lack of standardization in the bioinformatic methodologies hindered such endeavors [15,16,17,18,19].
In the last decade, micro-RNAs (miRNAs) emerged as promising biomarkers for mental conditions [16,20]. These molecules are small non-coding RNAs involved in the post-transcriptional regulation of gene expression. Their presence in almost all biological processes makes them easy to sample, whereas their susceptibility to everyday events, such as sleep, stress, or medications, gives them potential as biomarkers [21]. In this study, we employed NGS sequencing to characterise the complete blood miRNAome of schizophrenia patients differing in their response to antipsychotic treatment. To carry out the bioinformatics analyses, first we employed the highly replicable pipeline “myBrain-Seq v0.1.0” [22,23] for raw data preprocessing and differential expression analysis for defining a miRNA profile associated with the TRS condition (TRS profile). Then, we performed a hierarchical clustering analysis for exploring the sample grouping based on the TRS profile, using the R package hclust [24], and a functional analysis to explore some potential biological implications, using Cytoscape v3.9.1 [25] and StringApp v2.0.0 [26] tools. Our aim is twofold: first, to find a miRNA signature involved in TRS and second, to suggest its molecular roles and biological context. As a result, we found 16 miRNAs potentially involved in poor response to antipsychotic medication. Using this 16-miRNA signature, we were able to accurately classify more than two-thirds of our SZ and TRS samples and propose a molecular pathway in which these miRNAs could be involved, thus offering an interesting direction in which future research on TRS could be oriented.
2. Results
2.1. Profile of 16 miRNAs for Antipsychotic Resistance
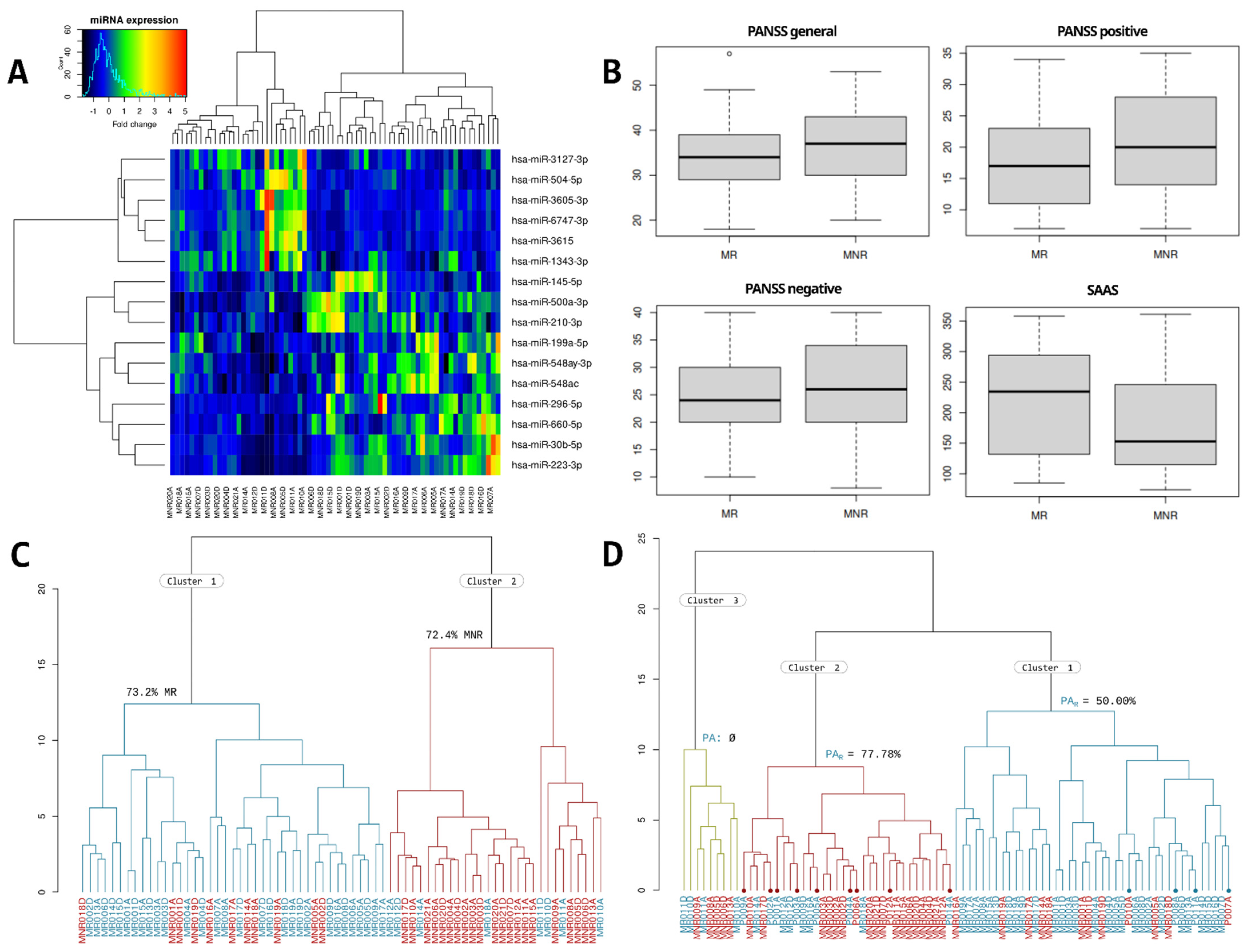
The quality of the samples, the sequencing depth, and the proportion of assigned and mapped sequences can be consulted in Supplementary Table S1. Differential expression analysis of the miRNAome of schizophrenia patients with a normal antipsychotic response (MR) vs. schizophrenia patients with antipsychotic resistance (MNR) shows a total of 16 differentially expressed miRNAs potentially related to antipsychotic resistance (TRS profile), including 6 miRNAs whose expression was upregulated and 10 miRNAs that were downregulated. As shown in Figure 1A, the miRNAs differentially expressed in the resistant schizophrenia, compared to non-resistance schizophrenia, are miR-3127-3p (↑), miR-504-5p (↑), miR-3605-3p (↑), miR-6747-3p (↑), miR-3615 (↑), miR-1343-3p (↑), miR-145-5p (↓), miR-500a-3p (↓), miR-210-3p (↓), miR-199a-5p (↓), miR-548ay-3p (↓), miR-548ac (↓), miR-296-5p (↓), miR-660-5p (↓), miR-30b-5p (↓), and miR-223-3p (↓).
Figure 1.
(A) Heatmap with the results of the hierarchical clustering of MR and MNR samples. (B) Box-plots with the MR and MNR punctuations on the PANSS general, positive, negative, and SAAS scales. The overlap between both groups can be seen. (C) Hierarchical clustering with MR samples (blue) and MNR samples (red); more than 70% of samples of both conditions were clustered homogeneously. (D) Hierarchical clustering with samples in MR, MNR, and PA groups. MR samples are colored in blue, MNR samples in red and PA samples were marked with a dot and colored in blue or red depending on the evolution of their psychotic episode.
2.2. Hierarchical Clustering Analysis Using the TRS Profile
As we described in materials and methods, hierarchical clustering was performed twice: using samples in both MR and MNR groups and using samples in the MR, MNR, and first psychotic episode at hospital arrival (PA) groups. We assessed the Positive and Negative Syndrome Scale (PANSS) and the Self-assessment Anhedonia Scale (SAAS) scores as a way to improve the discriminative power of the TRS profile (Figure 1B); however, they show a high overlap between groups and, therefore, were discarded.
In the first dendrogram of MR and MNR samples (Figure 1C), two groups were differentiated. Cluster 1 had 73.2% of the MR samples, and cluster 2 had 72.4% of the MNR samples.
In the second dendrogram of the MR, MNR, and PA samples (Figure 1D), 3 groups were differentiated: in cluster 1, 73.81% of samples were MR; in cluster 2, 59.38% of samples were MNR; and in cluster 3, 55.6% were MNR samples. Regarding the PA samples, four of them fell in cluster 1, nine in cluster 2, and none of them fell in cluster 3. Cluster 2 had 7 of the 9 PAR samples (77.78%) and 1 PANR (22.22%). Cluster 1 had the remaining 2 PAR and PANR samples (50% each). If we remove PA samples, cluster 1 has 78.9% of the MR samples, and cluster 2 has 73.9% of the MNR samples. From now on, we will refer to cluster 1 as “MR cluster” and to cluster 2 as “MNR cluster”.
2.3. Functional Analysis
The first step of the functional analysis was the target prediction. Tarbase v8 [27] predicted 13,267 targets for the 16 miRNAs of the miRNA signature. Four miRNAs, namely, hsa-miR-548ac, hsa-miR-548ay-3p, hsa-miR-660-5p, and hsa-miR-6747-3p, did not have any predicted target. Three miRNAs had overrepresented annotations, encompassing almost 75% of the total of predictions; these miRNAs were hsa-miR-210-3p, hsa-miR-1343-3p, and hsa-miR-30b-5p. Conversely, two of them had very few annotations, representing 0.86% (hsa-miR-223-3p) and 0.76% (hsa-miR-3605-3p) of the total number of annotations. More details of the results can be seen in Table 1. The ten most common targets were: ANKRD52 (shared by eight miRNAs), SPEN (seven), MIDN (seven), SRRM2 (six), SON (six), MDM2 (six), LONRF2 (six), LARP1 (six), FBXL19 (six), and DDX3X (six). More details of the results of the target prediction can be seen in Supplementary Table S2.

Table 1.
Number of targets per miRNA of the TRS profile returned by TarBase v.8 (Targets in TarBase) and found in the literature (Literature targets).
The second step of the functional analysis was a bibliographic search of relevant miRNA targets of the miRNA signature. We found 10 genes (Table 1) directly or indirectly regulated by any of the 16 miRNAs. The main functions implicated were related to the signal transducer and activator of transcription 3 (STAT3) and with several of its repressors (SOCS2, SOCS4, PTPN11). We also found some targets related to the multidrug resistance-associated proteins (ABCC1, ABCB1) and drug metabolism (CYP2C19).
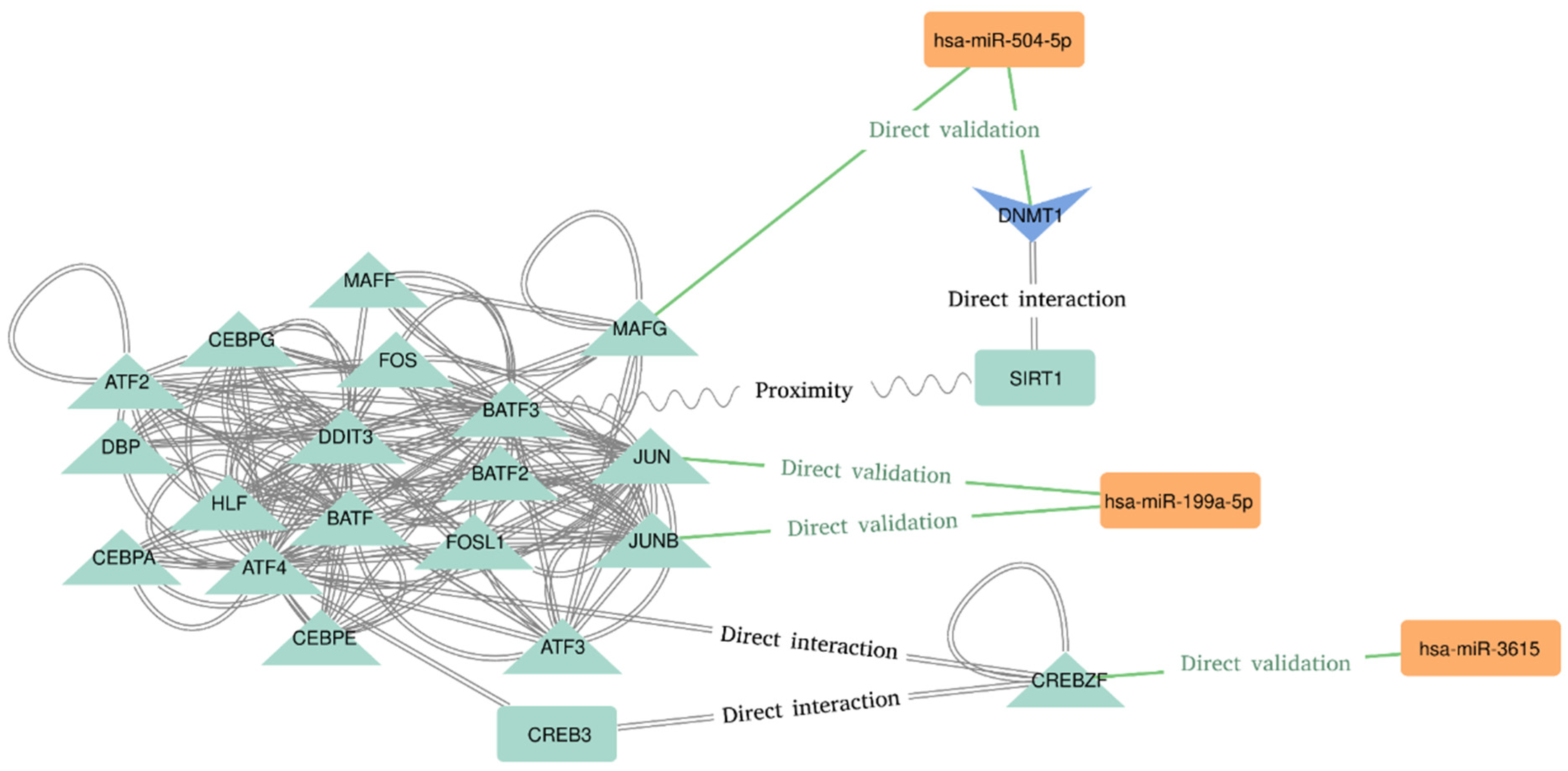
The last step was the network analysis and the functional enrichment of Pathways and GO terms. The most connected sub-network seems to be related to the AP-1 Transcription Factor Subunits (Figure 2). We found an interesting implication of hsa-miR-504-5p, hsa-miR-199a-5p, and hsa-miR-3615 in the regulation of transcription factors recognizing the cAMP response elements (CRE) sequence 5′-TGACGTCA-3′. Hsa-miR-504–5p seems to represses the transcription of “MAF BZIP Transcription Factor G” (MAFG), which directly interacts with the “Basic Leucine Zipper ATF-Like Transcription Factor 3” (BATF3) [35]. BATF3 is known to dimerize with the “Transcription Factor AP-1 Subunit Jun” (JUN) acting as transcriptional repressor of CRE elements [36,37]. On the contrary, hsa-miR-199a-5p has been proved to target JUN and JUNB transcripts, both of which dimerize with BATF3 and “Activating Transcription Factor 4” (ATF4) [38]. Moreover, hsa-miR-3615 directly represses “CREB/ATF BZIP Transcription Factor” (CREBZF) transcript, which is a known repressor of the “CAMP Responsive Element Binding Protein 3” (CREB-3), a transcription factor that binds to the CRE pattern [39]. Finally, the DNA Methyltransferase 1 (DNMT1) and the Sirtuin 1 (SIRT1) are also implied in this subnetwork. SIRT1 deacetylates DNMT1, which is a target of hsa-miR-504-5p [40]. It is worth noting the presence of more miRNA–targets interactions in this subnetwork. These interactions are hidden due to the degree filter applied to declutter the network in order to spot the most connected (and therefore, potentially relevant) nodes.
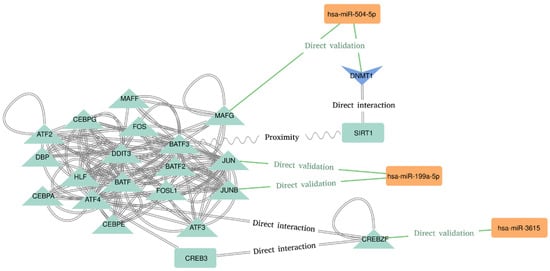
Figure 2.
Molecular interactions deduced from the most connected nodes of the network.
The top 5 pathways enriched in the Reactome database were, in order of presence in our network: 17.75% of the genes related with the metabolism of proteins (q-value = 5.03−52), 14.42% related with the transcription of genes (q-value = 1.6−51), 14.12% related to diseases (q-value = 3.89−42), 8.04% associated with RNA metabolism (q-value = 1.99−40), and 7.48% with a cellular response to stress (q-value = 1.73−44). Regarding the WikiPathway database, the top 5 enriched pathways were: 5.51% of genes related to VEGFA-VEGFR2 signaling (q-value = 4.44−29), 3.47% related to miRNA effects in Alzheimer’s disease (q-value = 3.6−19), 2.39% related to breast cancer (q-value = 1.39−16), 1.75% related to the androgen receptor signalling pathway (q-value = 1.94−15), and 1.72% related to ionising radiation damage to DNA and the subsequent cellular response via the ATR Serine/Threonine Kinase (q-value = 8.38−17). Finally, in the KEGG database, the top 5 enriched pathways were: 3.98% of genes related to amyotrophic lateral sclerosis neurodegenerative disorder (q-value = 4.07−17), 3.36% related to Huntington neurodegenerative disease (q-value = 1.41−14), 3.28% of the genes related to intestinal infection caused by Shigella (q-value = 3.57−21), 2.96% related to Parkinson neurodegenerative disorder (q-value 9.04−15), and 2.66% related to viral infection and human carcinogenesis (q-value = 6.45−17).
If we look at the genes in these pathways, we can notice that 10 of the 15 enriched pathways have TP53, PSMD4, PSMC1, and MDM2 proteins in common, and 9 pathways also have in common several 26S proteasome non-ATPase regulatory subunits, along with MTOR, EP300, CYCS, and CREBBP.
3. Discussion
Schizophrenia is a severe mental disorder that affects around 1% of the world’s population [1]. The current approach for its treatment is antipsychotic medication, but about one-fifth to one-half of patients do not respond to this pharmacological therapy [15]. Patients with resistant schizophrenia have poorer outcomes than patients with other severe mental illnesses, experiencing higher degrees of inadaptation to daily demands [41], which is an indication of a bad prognosis. Here, we present 16 miRNAs that are potentially related to treatment-resistant schizophrenia, and that will be a good starting point to biologically understand this condition and develop biological biomarkers.
3.1. miRNAs Previously Associated with Psychiatric Conditions
Seven of these 16 miRNAs have been previously related to neuropsychiatric conditions: miR-199a-5p (downregulated in MNR) has been found to target the IFNAR1 gene in a Dual-Luciferase Reporter Assay in a cell-based Parkinson’s disease model [42]. This gene encodes for type I Interferon Receptor 1, whose depletion in the organism seems to be protective against cognitive decline in mouse models of Alzheimer’s disease [43], and its presence in the human entorhinal cortex is proposed as a potential indicator of this condition [44]. Downregulation of this miRNA was also observed in dopaminergic neurons of Parkinson patients [45]. MiR-296-5p (downregulated in MNR) was found to be upregulated in schizophrenia patients compared to healthy controls [46]. MiR-504 (upregulated in MNR) has been found to target the SHANK3 gene in a luciferase assay in cultured mouse hippocampal neurons [47]. This gene encodes for scaffold proteins of the postsynaptic density, and it is also related to synapse formation and dendritic spine maturation; alterations in this gene are related to autism spectrum disorder and schizophrenia type 15 [48]. Moreover, this miRNA was found to be overexpressed in post-mortem brain tissue of patients with bipolar disorder, but not in schizophrenia patients, when compared to healthy controls [49]. MiR-660-5p (downregulated in MNR) was upregulated in the serum of patients with Alzheimer disease compared to healthy controls [50], but was found to be underexpressed in Long-Evans rats exposed to ethanol intake when compared to healthy control rats [51]. MiR-3615 (upregulated in MNR) was included in a 2020 study as one of a seven-miRNA profile for the discrimination of schizophrenia from controls [52]. A downregulation in miR-30b-5p levels (downregulated in MNR) was observed in patients with BD in comparison to healthy controls [53]. Finally, miR-6747-3p (upregulated in MNR) was found to be upregulated in the serum of Alzheimer’s disease patients compared to healthy controls [54].
3.2. miRNAs Potentially Related to Drug Resistance
A total of 3 of these 16 miRNAs have been previously related to the resistance to other medications. miR-145-5p (downregulated in MNR) has been linked to the acquired resistance to cisplatin and fluorouracil combination-based chemotherapy in gastric cancer patients [55] and also found to be underexpressed in patients with bipolar disorder type I when compared to healthy controls [56].
Downregulation of miR-210-3p (downregulated in MNR) has been related to an increase in ABCC1 expression in renal carcinoma and a subsequent reduction in chemotherapy drug sensitivity. The ABCC1 gene encodes for the multidrug resistance-associated protein 1; a member of the superfamily of ATP-binding cassette (ABC) transporters, it is related to multidrug resistance. Single nucleotide polymorphisms in this gene were associated with alterations in clozapine and norclozapine serum levels [57].
Finally, a 2020 study [29] found an upregulation of miR-1343-3p (upregulated in MNR) in the presence of the mutant allele rs4244285 of the CYP2C19 gene, which encodes a member of the cytochrome P450 enzyme superfamily. This rs4244285 allele encodes the CYP2C19*2 variant, which is known to be related to poor metabolism of compounds, such as some antidepressants, including fluvoxamine or sertraline hydrochloride [58,59]; anticonvulsants, such as valproic acid [60]; and benzodiazepines, such as phenazepam [61], among others. Moreover, CYP2C19 product is an enzyme responsible for the metabolization of clozapine [62,63].
3.3. Interpretation of the Hierarchical Clustering
In the dendrogram of MR and MNR samples (Figure 1C), a clear separation between the MR and MNR samples can be seen. Some pairs of samples (arrival and departure) are together in the MNR cluster (MR010A/D, MR011A/D, and MR012A/D) and in the MR cluster (MNR018A/D, MNR001A/D, and MNR019A/D). This probably means that the hospital environment did not significantly change their miRNA expression before and after hospitalisation, either due to similar environment variables (e.g., same hours of sleep, meals, light exposure, …) or pharmacological factors (e.g., the same medication is maintained). Interestingly, three pairs of samples (MR014A/D, MR018A/D, and MNR002A/D) change from the MNR cluster to the MR cluster after hospitalisation, whereas in two samples (MNR005A/D and MR017A/D), the opposite phenomena is observed. A change between the MNR cluster to the MR cluster could be the consequence of the hospital’s regular administration of the medication in an adequate dose, contrasting with a previous pharmacological deregularization [64,65]. This explanation is the most feasible, as two of these three samples were from patients treated with multiple antipsychotics, and the third one came from a MR patient treated with clozapine.
On the contrary, a change between the MR cluster to the MNR cluster is more difficult to explain. This implies either a MNR or MR patient was more similar to a MR patient initially, and after hospitalisation, became more similar to an MNR; in other words, a theoretical worsening of the treatment response after hospitalisation.
In the dendrogram of MR, MNR, and PA samples (Figure 1D), it can be observed that the miRNA expression of the TRS profile is more similar between PA samples and MNR samples (7 of 9 PA samples clustered in the MNR cluster, made of 73.9% MNR samples). In other words, MNR samples (treated patients) are more similar to PA (untreated) than to MR (treated). Interestingly, 4 PA samples clustered in the MR cluster, made of 78.9% MR samples. If we look at the clinical records of these four patients, we can observe that P010A and P011A had pathologies unrelated to schizophrenia—the first one, dementia related to a brain tumour and the second one, active consumption of toxic substances and aggressiveness not attributed to a psychiatric pathology. On the contrary, P008A figures as a stable non-medicated patient since the last two years, and P007A had an important mental disability previous to the psychotic episode. All this suggests that the PA samples of the MR cluster probably have conditions not intrinsically related to schizophrenia.
3.4. Interpretation of the Functional Analysis
Although at a first glance, the results of the pathway enrichment analysis could seem divergent, most of the pathways have something in common: they are related to the exposure and response of the organism to a stressor. The vulnerability–stress model has been proposed as a broad explanation for the schizophrenia aetiology since the 1980s [66,67], and it is currently used as an explanation of their aetiology and prodromes [68,69,70]. Experimental evidence, such as the association between the level of activation of the inflammatory response and schizophrenia, seems to suggest a role for stress in this condition [71,72,73,74]. Moreover, differences in this inflammatory response have also been noticed between schizophrenia and treatment-resistant schizophrenia [73,75,76,77,78].
If we closely look at the enriched pathways in Table 2, we can notice that most of them have Tumor Protein P53 (TP53) and its regulator MDM2 Proto-Oncogene (MDM2) in common, both heavily involved in the cellular stress response. MDM2 is a primary negative regulatory factor of p53 (the TP53 product) and contributes to its normal expression in the cell. This is achieved by the ubiquitination of p53 by MDM2, which leads to the p53 degradation [79]. On the other side, the protein p53 is a tumour suppressor, which responds to diverse cellular stresses to regulate expression of its target genes; it is involved in the regulation of processes such as cell cycle arrest, apoptosis, senescence, DNA repair, and changes in metabolism. The MDM2/p53 pathway has been found to play an important role in drug resistance [80,81,82,83], and its regulatory expression was found to be altered during treatments with the antipsychotic fluspirilene [84]. Interestingly, the association between the schizophrenia condition and the risk for cancer development has been debated for more than 100 years [85,86,87,88,89,90,91]. The results indicate both a decrease and an increase in the risk of cancer, which varies according to the type of tumour studied and the schizophrenia cohort [90]; still, the evidence shows that the mechanisms that regulate tumorigenesis seem to be closely related to this condition.

Table 2.
Top 5 enriched pathways per database (Reactome Pathways, WikiPathways, and KEGG Pathways). The column “Gene” is the number of genes in the network associated with each pathway term. The column “Proportion” is the ratio between the number of genes with that term divided by the total number of genes in the network (n = 3718).
3.5. A Molecular Model for Treatment Resistant Schizophrenia
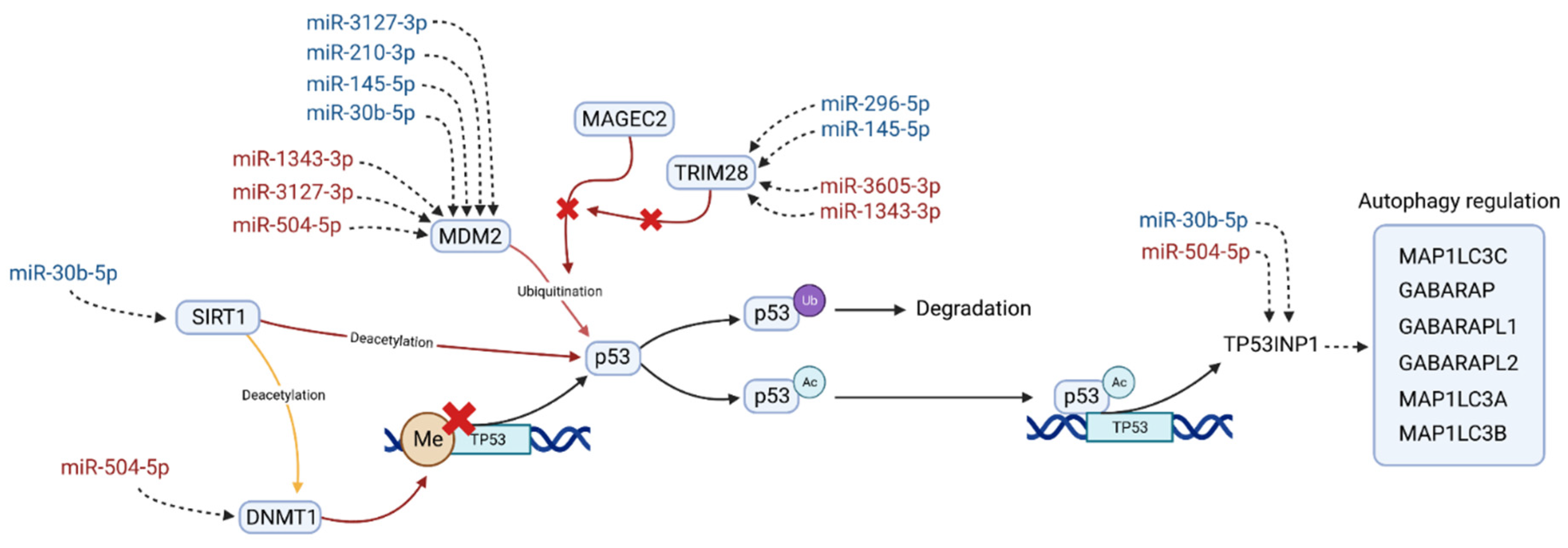
In an effort to integrate the results of the DEA, the network and pathway analysis, the miRNA target prediction, and the bibliographic search, we propose in Figure 3 a pathway in which the DE miRNAs could be potentially implied. Hopefully, it could bring a hint about the treatment-resistant schizophrenia aetiology.
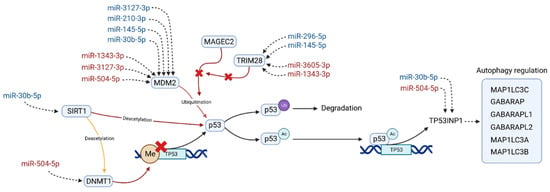
Figure 3.
Pathway in which differentially expressed miRNAs could be involved, potentially related to the antipsychotic resistance in schizophrenia. MiRNAs are colored according to their expression in the MNR group when compared to the MR group; red indicates overexpression and blue underexpression. Sirtuin 1 (SIRT1) interacts with the DNA Methyltransferase 1 (DNMT1) and the Tumor Protein P53 (p53) regulating its expression. Proteins MDM2 Proto-Oncogene (MDM2), MAGE Family Member C2 (MAGEC2), and RING Finger Protein 96 (TRIM28) are closely linked with p53 inactivation and heavily targeted by the miRNAs of our TRS profile. This inactivation of p53 is through ubiquitination (p53Ub), which leads to its degradation. Acetylated p53 (p53Ac) acts as a transcriptional factor of several genes, from which Tumor Protein P53 Inducible Nuclear Protein 1 (TP53INP1) is the one most targeted by the TRS profile. This protein is a positive regulator of the autophagy and also acts as a transcription regulator in response to cellular stress. This regulation is made through its interaction with GABA Type A Receptor-Associated Protein (GABARAP), with GABA Type A Receptor Associated Protein Like 1/2 (GABARAPL1, GABARAPL2) and with Microtubule Associated Protein 1 Light Chain 3 Alpha/Beta/Gamma (MAP1LC3A, MAP1LC3B, MAP1LC3B).
We found that p53/MDM2 interaction could possibly be the key to explain the observed differences in miRNA expression between the MR and MNR groups. First, both molecules were the most recurrent similarity in the top five enriched pathways in three different databases. Second, MDM2 and its regulators, MAGE Family Member C2 (MAGEC2) and (RING Finger Protein 96) TRIM28, were targeted by eight different miRNAs of the TRS profile. Third, SIRT1 and its deacetylation substrate, DNMT1, are both present in the most connected subnetwork and have a direct involvement in the regulation of p53 transcription. Finally, bibliographic evidence supports the proposed pathway in four aspects: (i) the implication of p53/MDM2 in the multidrug resistance process [80,81,82,83], (ii) the involvement of p53 in autophagy, a process known to be related to antipsychotic response [92,93,94,95]; (iii) the observed dysregulations of this pathway in schizophrenia patients [96,97,98], and (iv) the relationship of this pathway to negative symptoms, such as memory, learning ability impairment [99], and major depression [100].
In this pathway, SIRT1 deacetylates the methyltransferase DNMT1. This results in an activation or repression of its methyltransferase activity, which varies with the deacetylation position [40]. For its part, increased levels of DNMT1 seem to enhance the methylation in the TP53 promoter, leading to the downregulation of p53 [101,102,103]. Moreover, SIRT1 has also been found to directly deacetylate p53, repressing its transactivation [104,105,106], and play a role in the modulation of the response to stress. Two recent examples of this are the observed increment in levels of p53 and decreased levels of SIRT1 in mice brain after long-term “ultraviolet A” eye radiation [99], as well as the inverse phenomena after acute heat stress in bovine granulosa cells, where decreased levels of p53 and increased levels of SIRT1 were observed [107]. In MNR, SIRT1 is potentially targeted by the underexpressed hsa-miR-30b-5p, whereas DNMT1 is by the overexpressed hsa-miR-504-5p (Figure 3).
Other regulators of p53 are MDM2 and MAGEC2. The MDM2 transcript is heavily targeted by the miRNAs of our TRS profile, and its protein product is well-known for its role in p53 inactivation [83,108,109]. Under nonstressed conditions, MDM2 regulates p53 expression through an autoregulatory feedback loop [110,111,112]. This regulation can be made by the direct binding of MDM2 to p53 transactivation domain, by MDM2 involvement in the nuclear translocation of p53, or by promoting its degradation using its ubiquitin ligase activity [108,113]. MDM2/p53 regulation has been extensively linked to drug resistance [80,81,114], and there is evidence of its regulatory interaction being disrupted by some antipsychotics, such as fluspirilene and pimozide, or mood stabilisers, such as lithium [84,115,116]. In MNR, MDM2 is potentially targeted by seven miRNAs of the TRS profile (Figure 3), four downregulated (hsa-miR-3127-3p, hsa-miR-210-3p, hsa-miR-145-5p, and hsa-miR-30b-5p) and three overexpressed (hsa-miR-1343-3p, hsa-miR-3127-3p, and hsa-miR-504-5p).
Regarding MDM2 regulators, an important modulator of MDM2 ubiquitin ligase activity is MAGEC2, which has recently been found to inhibit the ubiquitination of p53 by directly interacting with the MHD domain of MDM2 [117]. At the same time, TRIM28 competes with MDM2 for MAGEC2 binding, thus acting as a promoter of MDM2 ubiquitin ligase activity and, consequently, triggering p53 degradation [117,118]. In MNR, TRIM28 is potentially targeted by four miRNAs of the TRS profile (Figure 3), two overexpressed (hsa-miR-3605-3p, hsa-miR-1343-3p) and two downregulated (hsa-miR-296-5p, hsa-miR-145-5p).
Finally, the acetylation of p53 is enhanced as a stress response, which results in its transcriptional activation [119,120,121]. The activated p53 acts as a transcription factor, promoting the expression of several genes, from which we selected Tumor Protein P53 Inducible Nuclear Protein 1 (TP53INP1) as the most relevant in TRS for two reasons: (i) it has been the only one targeted by two of the miRNAs of the TRS profile (Figure 3) and (ii) the known interactions of its product and several GABA Type A Receptor-Associated Proteins (GABARAP, GABARAPL1/L2) [122,123,124], which are known to mediate GABA-A receptor intracellular transport [125,126]. The availability of these GABA-A receptors has been related to the severity of symptoms in schizophrenia [127], and elevated levels of its ligand GABA had been reported in the midcingulate cortex in TRS patients [128]. In MNR, TP53INP1 is potentially targeted by two miRNAs of the TRS profile (Figure 3), one overexpressed (hsa-miR-504-5p) and one downregulated (hsa-miR-30b-5p).
Although there is not much research about the role of p53 in TRS, there is some evidence linking alterations of p53 expression, its activation, and polymorphisms in TP53 with higher schizophrenia risk and symptoms severity [97,98,129,130,131,132]. Decreased levels of SIRT1 in plasma has been linked with a higher comorbidity of depressive symptoms [133,134,135], whereas increased levels of DNMT1 had been observed in the brains of schizophrenia and bipolar patients [136,137,138,139]. Finally, MDM2 had been found to be underexpressed in the dorsolateral prefrontal cortex of schizophrenia patients [96].
All this together points to an atypical response to stress after antipsychotic administration as one key factor in the development of antipsychotic resistance. Our results indicate that this response could be potentially mediated by p53, with an important implication of its regulators MDM2, TRIM28, SIRT1, and DNMT1. Future research on TRS might focus on comparing the expression of these molecules between TRS and non-TRS patients.
3.6. Limitations and Future Perspectives
This study has the following limitations. (i) Despite the fact that we defined a TRS fingerprint based on the miRNA differential expression between MR and MNR groups, the mechanism through which those miRNAs affect the resistance condition is still unknown. (ii) Whether peripheral miRNAs represent changes in the central nervous system was not assessed. Further experiments analysing cerebral spinal fluid in human samples might be an interesting approach. (iii) Although we corrected for processing batch; sex; drug consumption; time; treatment based on the subgroup of the diazepines, oxazepines, thiazepines, and oxepins; and treatment based on other antipsychotics, it could have been of interest to also correct for treatments based on clozapine and the presence of cognitive decline. (iv) It might have been interesting to study the miRNA response to specific antipsychotic therapies. (v) Results of the hierarchical clustering suggest that the TRS profile is partially associated with the schizophrenia condition, not only with antipsychotic resistance. (vi) The sample size is modest; therefore, it is difficult to predict the scope of our conclusions. It would be necessary to assess the expression of the TRS profile in an independent sample of patients. (vii) At the time of writing this paper, four miRNAs, namely, hsa-miR-548ac, hsa-miR-548ay-3p, hsa-miR-660-5p, and hsa-miR-6747-3p, did not have any predicted target in the last version of Tarbase v.8. Therefore, our functional analysis and the conclusions drawn from it are based on 12 of the 16 miRNAs of the TRS profile. It would be interesting to repeat the functional analysis in the near future.
4. Materials and Methods
4.1. Experimental Design
We recruited schizophrenia patients with a normal antipsychotic response (MR; n = 19), schizophrenia patients with antipsychotic resistance (MNR; n = 21), patients with a first psychotic episode (P; n = 13), and 43 healthy individuals (C; n = 43). Two blood samples were collected per patient, the first one at the hospital arrival (MRA, MNRA, PA) and the second one at the hospital discharge (MRD, MNRD, PD). Scores on PANSS (positive, negative, general) and SAAS scales were recorded at the arrival [140,141]. P patients were followed up until they could be classified as responders or nonresponders to medication, resulting in eight PA with normal antipsychotic response (PAR = 9) and four PA with antipsychotic resistance (PANR = 4). As treatment resistance is a chronic condition in schizophrenia, and environmental changes during hospital admission can disturb miRNA expression, we will consider arrival and discharge samples from the same patient as two replicas of the same condition. Thus, differences between the MR and MNR groups will be more likely to be related with TRS and not with environmental variations. From now on, we will use MR to refer to MRA and MRD samples and MNR to refer to MNRA and MNRD samples.
To find a miRNA signature of the antipsychotic response, we looked for differences in miRNA expression between the MR and MNR groups. The resulting “miRNA signature” was used to perform a hierarchical clustering with the MR, MNR, and PA samples; we are interested in the clusters assigned to PA samples, as well as the overall distribution of groups between clusters. As a first approach to outline the biological context of these results, we performed a target prediction, followed by a network and a functional analysis of the miRNAs of the signature.
4.2. Human Participants Included in the Study
Patients were recruited from Álvaro Cunqueiro Hospital (Vigo, Spain) between September 2018 and November 2021. Inclusion criteria were: (i) meeting the DSM-V schizophrenia diagnostic criteria, (ii) aged 18 years or older, and (iii) signed written consent. The exclusion criteria were an additional DSM-V diagnosis, medical illnesses, and neurological conditions, as well as pregnancy or lactation. Age, sex, drug consumption (alcohol/tobacco/illegal), and current pharmacological treatment were recorded for each patient (Table 3).

Table 3.
Summary of the samples included in this study.
All patients and controls for this study were of Spanish nationality. The research was conducted in accordance with the Declaration of Helsinki, and all relevant ethical approvals were obtained. Written consent was obtained from all patients or their corresponding legal guardians.
4.3. Blood Collection
Two blood samples per patient were drawn from the cubital vein and collected into PAXgene tubes (BD Biosciences, Franklin Lakes, NJ, USA) in the morning between 8 and 10 a.m, the first one at the hospital arrival and the second one at the hospital discharge. Samples were incubated 2 h at room temperature to ensure complete lysis of blood cells. PAXgene tubes were stored in the freezer (−80 °C) following commercial specifications (Qiagen, Hilden, Germany). The tubes were first transferred to −20 °C (24–72 h) and then transferred to −80 °C freezer until further processing.
4.4. Total RNA Purification from Human Blood Samples
Total RNA, including small RNA, was purified from whole blood samples using the PAXgene® Blood microRNA Kit from Qiagen (PreAnalytiX GmbH, Hombrechtikon, Switzerland). PAXgene Blood microRNA Kit isolates total RNA >18 nucleotides (including miRNA) from human whole blood. Before isolation, samples were put at room temperature overnight. We used Automated purification of total RNA, including miRNA, on QIAcube instruments, following the manual specification (Qiagen, Hilden, Germany). Briefly, tubes were centrifuged for 10 min at 4000× g, and supernatant was removed. A total of 4 mL of RNAse-free water was added to the pellet and then vortexed until the pellet was visibly dissolved. After that, ten minutes of centrifugation at 4000× g were applied. This step was repeated once more. After that, pellet was resuspended in buffer BM1, transferred to the 2 mL processing tube, and loaded into the QIAcube Connect shaker. Proteinase K and Buffer BM2 were added prior to the 10 min period of incubation at 55 °C. Incubated tubes were transferred to PAXgene Shredder Spin Column, where 3 min of centrifugation at full speed was applied. Supernatant was transferred into new tubes, and isopropanol was added. Samples were loaded on PAXgene RNA spin columns that bind total RNA >18 nucleotides (including miRNA). Centrifugation at 10,000× g for 1 min was applied. Samples were washed once for 15 s with buffer BM3 and centrifuged at 10,000× g for 1 min. A DNA digestion was performed by incubating samples with DNase solution for 15 min at room temperature, followed by a centrifugation at 10,000× g for 1 min. Spin columns were washed, first with Buffer BM3 and then with Buffer BM4, at 10,000× g for 2 min each. Samples were eluted with 50 µL of BR5 Buffer to microcentrifuge tubes. One minute of centrifugation at 10,000× g was applied prior to the sample incubation at 65 °C for 5 min in the QIAcube Connect shaker. Finally, samples were immediately transferred to ice after incubation and then stored at −80 °C until further use.
4.5. Small RNA Sequencing
For RNA quality control, a Bioanalyzer Instrument (Agilent Technologies, Santa Clara, CA, USA) was used. For concentration determination, a NanoDrop (Thermo Scientific, Waltham, MA, USA) was used. In addition, for evaluating RNA sample quality prior to miRNA/small RNA NGS library preparation, a QIAseq miRNA Library QC Spike-ins kit was used (Qiagen, Hilden, Germany). Library preparation was done using NEBNext Multiplex Small RNA Sample Prep Set for illumina (New England biolabs, Ipswich, MA, USA) to produce high quality microRNAome data from human blood samples. Briefly, the total amount of 150 ng of RNA was taken for further cDNA preparation, fragmentation, adapter ligation, and hybridization. After pooling the libraries together, polyacrylamide gel electrophoresis was run for size selection. The insert size of 150 base pairs (bp) was chosen for quantification and purification. Quality was checked on Bioanalyzer Instrument, and libraries were quantified using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Sequencing of 8 pM concentration was performed on HiSeq 2000 sequencing system (Illumina, San Diego, CA, USA) using a 24 samples per Lane, 50 bp single read setup. As a control for Illumina sequencing runs, a PhiX Control that is a reliable adapter-ligated library was used. Sequencing data were demultiplexed using CASAVA v1.8 (Illumina, San Diego, CA, USA), and raw fastq files were generated.
4.6. Bioinformatics Analysis
The analytical methodology is detailed in Supplementary Figure S1. For quality control of the raw data, adapter removal, alignment, and quantification, we used the software myBrain-Seq v0.1.0 [23]. The biological references used were the GRCh38 human genomic build [142] for the alignment and miRBase v.22 database [143] for the miRNA annotation. We filtered miRNAs with less than 10 counts in total using a custom R script. Differential expression analyses were performed using DESeq2 v1.32.0 [144], adapting the script of myBrain-Seq to adjust for six potential confounding variables: (i) processing batch, (ii) sex, (iii) drug consumption (alcohol OR tobacco OR illegal), (iv) time (arrival, discharge), (v) treatment based in the subgroup of the diazepines, oxazepines, thiazepines, and oxepins, and (vi) treatment based in other antipsychotics. To obtain a miRNA signature for the resistance to medication, we first performed the DEA comparing MR vs. MNR, then subtracted from this profile the common miRNAs between DEA comparisons MNR vs. C and MR vs. C. This was done to filter miRNAs with potentially similar roles in MR and MNR conditions and to obtain the “miRNA profile” (Supplementary Figure S1, point 3).
4.7. Hierarchical Clustering
We performed two different hierarchical clustering analyses using the miRNAs found in the “miRNA profile”: the first one with MR and MNR samples, the second one with MR, MNR, and PA samples. Prior to the hierarchical clustering, the miRNA counts of the profiles were normalised using the Relative Log Expression method of DESeq2 1.32.0 [144]. We used the R package hclust [24] for clustering, using the Euclidean distance metric and Ward method (ward.D2).
4.8. Functional Analysis
To narrow down the potential implication of the differentially expressed miRNAs on the antipsychotic resistance, we conducted a target prediction analysis, bibliographic search, and network analysis. For the prediction of the miRNA targets, we used TarBase v8 [27] and a custom bash script. The bibliographic search was performed in order to add documented targets not present in TarBase v8; we looked for genes related to chemoresistance or the metabolism of antipsychotics, or those involved in the diffusion of molecules through the blood–brain barrier. To perform the network analysis, we used Cytoscape 3.9.1 [25]. We started by importing the miRNA–gene interactions obtained from TarBase v8 and the literature search. Then, we selected the miRNAs and ran the Cytoscape default heat diffusion algorithm [145]. Selected nodes were used as a query in the Uniprot database [146], and the resulting networks were merged with the miRNA–gene network. Finally, we used StringApp 2.0.0 [26] to annotate proteins, to filter by tissue, and to perform the functional enrichment analysis of pathways. We kept proteins expressed only in the category of “nervous system” with a tissue score of 4.50 and in the category of “blood” with a tissue score of 3.00. For the pathway enrichment analysis, we used Reactome Pathways [147], WikiPathways [148], and KEGG [62] Pathways databases. To find highly connected subnetworks, we applied a degree filter of value 10.
5. Conclusions
We presented a 16-miRNA profile related to treatment-resistant schizophrenia. With this profile, we performed a hierarchical clustering of MR and MNR samples that resulted in two clusters: the first one with 73.2% of the MR samples and the second one with 72.4% of the MNR samples. We did not use the PANSS and SAAS scores to improve the clustering accuracy, as we did not find associations between PANSS and SAAS scores and the TRS condition. By observing the dendrogram of MR, MNR, and PA samples, we deduced that similarities between the TRS profile of patients with a first psychotic episode and schizophrenia patients’ response to medication seem to be potential evidence of a first psychotic episode unrelated to schizophrenia. The network and functional analysis of the TRS profile suggests that atypical response to stress after antipsychotic administration could be one key factor in the development of antipsychotic resistance. Finally, we condensed all the results in a molecular pathway that is presumably altered in TRS patients, with a pivotal role of p53 and its regulators MDM2, TRIM28, SIRT1, DNMT1, and eight miRNAs of the TRS profile.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24031891/s1.
Author Contributions
Conceptualization, D.P.-R., H.L.-F. and R.C.A.-B.; Methodology, D.P.-R., M.A.P., T.R.-B., S.B., A.F., J.M.O., H.L.-F. and R.C.A.-B.; Software, D.P.-R., H.L.-F. and R.C.A.-B.; Formal Analysis, D.P.-R. and T.P.-C.; Investigation, D.P.-R., M.A.P., T.R.-B., J.M.O., H.L.-F. and R.C.A.-B.; Resources, J.M.O. and R.C.A.-B.; Data Curation, D.P.-R., M.A.P., T.R.-B., T.P.-C., J.M.P.-G., J.M.O., H.L.-F. and R.C.A.-B.; Writing—Original Draft Preparation, D.P.-R., H.L.-F. and R.C.A.-B.; Writing—Review & Editing, D.P.-R., M.A.P., T.R.-B., T.P.-C., S.B., A.F., J.M.P.-G., J.M.O., H.L.-F. and R.C.A.-B.; Visualization, D.P.-R.; Supervision, H.L.-F. and R.C.A.-B.; Project Administration, D.P.-R., H.L.-F. and R.C.A.-B.; Funding Acquisition, R.C.A.-B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Instituto de Salud Carlos III (ISCIII) through the project PI18/01311 (co-funded by European Regional Development Fund (FEDER), “A way to make Europe”, UE) to R.C. Agís-Balboa. D. Pérez-Rodríguez is supported by an “Investigo Program” predoctoral contract from Xunta de Galicia. This work was also partially supported by the Conselleria de Cultura, Educación e Universidade (Xunta de Galicia), under the scope of the strategic funding ED431C 2022/03-GRC Competitive Reference Group and by Ministerio de Universidades (Gobierno de España) through a “María Zambrano” contract (Hugo López-Fernández).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Galicia Sur Health Research Institute (IIS Galicia Sur) (protocol code 2016/577, 2 March 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data accessible at NCBI GEO database, accession GSE223043 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223043).
Acknowledgments
Figure 3 was created with BioRender.com. The authors would like to thank Galicia Sur Health Research Institute and the Área Sanitaria de Vigo for their support. We specially thank the Psychiatric Nursing Service and psychiatrists at the Álvaro Cunqueiro Hospital and Nicolás Peña Hospital. Finally, we acknowledge Estela Portillo Encina (ADHD Children’s Organization, ANHIDA), José Luis Fernández Sastre (Psychiatry Department, University Hospital of Vigo), and Maria Tereixa Brañas Pérez (Family medicine, SERGAS) for their intellectual and motivational help in the development of this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Saha, S.; Chant, D.; Welham, J.; McGrath, J. A Systematic Review of the Prevalence of Schizophrenia. PLoS Med. 2005, 2, e141. [Google Scholar] [CrossRef] [PubMed]
- Elkis, H. Treatment-Resistant Schizophrenia. Psychiatr. Clin. N. Am. 2007, 30, 511–533. [Google Scholar] [CrossRef] [PubMed]
- Essock, S.M.; Hargreaves, W.A.; Covell, N.H.; Goethe, J. Clozapine’s Effectiveness for Patients in State Hospitals: Results from a Randomized Trial. Psychopharmacol. Bull. 1996, 32, 683–697. [Google Scholar] [PubMed]
- Lieberman, J.A. Pathophysiologic Mechanisms in the Pathogenesis and Clinical Course of Schizophrenia. J. Clin. Psychiatr. 1999, 12, 9–12. [Google Scholar]
- Lindenmayer, J.P. Treatment Refractory Schizophrenia. Psychiatr. Q. 2000, 71, 373–384. [Google Scholar] [CrossRef]
- Correll, C.U.; Howes, O.D. Treatment-Resistant Schizophrenia: Definition, Predictors, and Therapy Options. J. Clin. Psychiatr. 2021, 82, 36608. [Google Scholar] [CrossRef]
- Kennedy, J.L.; Altar, C.A.; Taylor, D.L.; Degtiar, I.; Hornberger, J.C. The Social and Economic Burden of Treatment-Resistant Schizophrenia: A Systematic Literature Review. Int. Clin. Psychopharmacol. 2014, 29, 63–76. [Google Scholar] [CrossRef]
- Millgate, E.; Kravariti, E.; MacCabe, J.; Hide, O. S62. Cognitive deficits in treatment resistant schizophrenia. Schizophr. Bull. 2020, 46, S56–S57. [Google Scholar] [CrossRef]
- Fernández-Pereira, C.; Penedo, M.A.; Rivera-Baltanas, T.; Fernández-Martínez, R.; Ortolano, S.; Olivares, J.M.; Agís-Balboa, R.C. Insulin-like Growth Factor 2 (IGF-2) and Insulin-like Growth Factor Binding Protein 7 (IGFBP-7) Are Upregulated after Atypical Antipsychotics in Spanish Schizophrenia Patients. Int. J. Mol. Sci. 2022, 23, 9591. [Google Scholar] [CrossRef]
- Correll, C.U.; Brevig, T.; Brain, C. Patient Characteristics, Burden and Pharmacotherapy of Treatment-Resistant Schizophrenia: Results from a Survey of 204 US Psychiatrists. BMC Psychiatr. 2019, 19, 362. [Google Scholar] [CrossRef]
- Ballon, J.S.; Lieberman, J.A. Advances in the Management of Treatment- Resistant Schizophrenia. FOC 2010, 8, 475–487. [Google Scholar] [CrossRef]
- Englisch, S.; Zink, M. Treatment-Resistant Schizophrenia: Evidence-Based Strategies. Mens Sana Monogr. 2012, 10, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.J.; Zhao, S.; Qi, F.; Nyakyoma, K.; Kwong, J.S.; Adams, C.E. Electroconvulsive Therapy for Treatment-Resistant Schizophrenia. Cochrane Database Syst. Rev. 2019, 3, CD011847. [Google Scholar] [CrossRef]
- Kane, J.M.; Kishimoto, T.; Correll, C.U. Non-Adherence to Medication in Patients with Psychotic Disorders: Epidemiology, Contributing Factors and Management Strategies. World Psychiatr. 2013, 12, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Nucifora, F.C.; Woznica, E.; Lee, B.J.; Cascella, N.; Sawa, A. Treatment Resistant Schizophrenia: Clinical, Biological, and Therapeutic Perspectives. Neurobiol. Dis. 2019, 131, 104257. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, D.; López-Fernández, H.; Agís-Balboa, R.C. Application of MiRNA-Seq in Neuropsychiatry: A Methodological Perspective. Comput. Biol. Med. 2021, 135, 31–42. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, D.; López-Fernández, H.; Agís-Balboa, R.C. On the Reproducibility of MiRNA-Seq Differential Expression Analyses in Neuropsychiatric Diseases. In Proceedings of the Practical Applications of Computational Biology & Bioinformatics, 15th International Conference (PACBB 2021), Salamanca, Spain, 6–8 October 2021; Rocha, M., Fdez-Riverola, F., Mohamad, M.S., Casado-Vara, R., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 41–51. [Google Scholar]
- Pandey, A.; Kalita, K.N. Treatment-Resistant Schizophrenia: How Far Have We Traveled? Front. Psychiatr. 2022, 13, 994425. [Google Scholar] [CrossRef]
- Potkin, S.G.; Kane, J.M.; Correll, C.U.; Lindenmayer, J.-P.; Agid, O.; Marder, S.R.; Olfson, M.; Howes, O.D. The Neurobiology of Treatment-Resistant Schizophrenia: Paths to Antipsychotic Resistance and a Roadmap for Future Research. NPJ Schizophr. 2020, 6, 1. [Google Scholar] [CrossRef]
- Delgado-Morales, R.; Agís-Balboa, R.C.; Esteller, M.; Berdasco, M. Epigenetic Mechanisms during Ageing and Neurogenesis as Novel Therapeutic Avenues in Human Brain Disorders. Clin. Epigenet. 2017, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, D.; Pérez-Rodríguez, M.; Agís-Balboa, R.C.; López-Fernández, H. Towards a Flexible and Portable Workflow for Analyzing MiRNA-Seq Neuropsychiatric Data: An Initial Replicability Assessment. In Proceedings of the Practical Applications of Computational Biology and Bioinformatics, 16th International Conference (PACBB 2022), L’Aquila, Italy, 13–15 July 2022; Fdez-Riverola, F., Rocha, M., Mohamad, M.S., Caraiman, S., Gil-González, A.B., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 31–42. [Google Scholar]
- Pérez-Rodríguez, D.; López-Fernández, H.; Agís-Balboa, R.C. MyBrain-Seq. 2022. Available online: https://github.com/sing-group/my-brain-seq (accessed on 10 June 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A Decade-Long Collection of Experimentally Supported MiRNA–Gene Interactions. Nucl. Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef]
- Li, S.; Yang, J.; Wang, J.; Gao, W.; Ding, Y.; Ding, Y.; Jia, Z. Down-Regulation of MiR-210-3p Encourages Chemotherapy Resistance of Renal Cell Carcinoma via Modulating ABCC1. Cell Biosci. 2018, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.R.; Vohra, M.; Shukla, V.; Guddattu, V.; Razak Uk, A.; Shetty, R.; Umakanth, S.; Satyamoorthy, K.; Rai, P.S. Coding SNPs in Hsa-MiR-1343-3p and Hsa-MiR-6783-3p Target Sites of CYP2C19 Modulates Clopidogrel Response in Individuals with Cardiovascular Diseases. Life Sci. 2020, 245, 117364. [Google Scholar] [CrossRef]
- Lin, X.; Yu, S.; Ren, P.; Sun, X.; Jin, M. Human MicroRNA-30 Inhibits Influenza Virus Infection by Suppressing the Expression of SOCS1, SOCS3, and NEDD4. Cell. Microbiol. 2020, 22, e13150. [Google Scholar] [CrossRef]
- Duan, Z.; Choy, E.; Harmon, D.; Liu, X.; Susa, M.; Mankin, H.; Hornicek, F. MicroRNA-199a-3p Is Downregulated in Human Osteosarcoma and Regulates Cell Proliferation and Migration. Mol. Cancer Ther. 2011, 10, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Long, J.; Liu, B.; Xu, M.; Wang, W.; Xie, X.; Wang, X.; Kuang, M. MiR-500a-3p Promotes Cancer Stem Cells Properties via STAT3 Pathway in Human Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 99. [Google Scholar] [CrossRef]
- Wang, X.; Meng, R.; Hu, Q.-M. LINC00319-Mediated MiR-3127 Repression Enhances Bladder Cancer Progression Through Upregulation of RAP2A. Front. Genet. 2020, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Zavala, M.L.; Lopez-Castillejos, E.S.; Requenez-Contreras, J.L.; Granados-Riveron, J.T.; Aquino-Jarquin, G. A Single MiRNA and MiRNA Sponge Expression System for Efficient Modulation of MiR-223 Availability in Mammalian Cells. J. Gene Med. 2019, 21, e3100. [Google Scholar] [CrossRef]
- Reinke, A.W.; Baek, J.; Ashenberg, O.; Keating, A.E. Networks of BZIP Protein-Protein Interactions Diversified over a Billion Years of Evolution. Science 2013, 340, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Iacobelli, M.; Wachsman, W.; McGuire, K.L. Repression of IL-2 Promoter Activity by the Novel Basic Leucine Zipper P21SNFT Protein1 2. J. Immunol. 2000, 165, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Bower, K.E.; Zeller, R.W.; Wachsman, W.; Martinez, T.; McGuire, K.L. Correlation of Transcriptional Repression by P21SNFTwith Changes in DNA·NF-AT Complex Interactions. J. Biol. Chem. 2002, 277, 34967–34977. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, J.A.; Reinke, A.W.; Bhimsaria, D.; Keating, A.E.; Ansari, A.Z. Combinatorial BZIP Dimers Display Complex DNA-Binding Specificity Landscapes. eLife 2017, 6, e19272. [Google Scholar] [CrossRef]
- Misra, V.; Rapin, N.; Akhova, O.; Bainbridge, M.; Korchinski, P. Zhangfei Is a Potent and Specific Inhibitor of the Host Cell Factor-Binding Transcription Factor Luman. J. Biol. Chem. 2005, 280, 15257–15266. [Google Scholar] [CrossRef]
- Peng, L.; Yuan, Z.; Ling, H.; Fukasawa, K.; Robertson, K.; Olashaw, N.; Koomen, J.; Chen, J.; Lane, W.S.; Seto, E. SIRT1 Deacetylates the DNA Methyltransferase 1 (DNMT1) Protein and Alters Its Activities. Mol. Cell. Biol. 2011, 31, 4720–4734. [Google Scholar] [CrossRef]
- Iasevoli, F.; Giordano, S.; Balletta, R.; Latte, G.; Formato, M.V.; Prinzivalli, E.; De Berardis, D.; Tomasetti, C.; de Bartolomeis, A. Treatment Resistant Schizophrenia Is Associated with the Worst Community Functioning among Severely-Ill Highly-Disabling Psychiatric Conditions and Is the Most Relevant Predictor of Poorer Achievements in Functional Milestones. Prog. Neuro-Psychopharmacol. Biol. Psychiatr. 2016, 65, 34–48. [Google Scholar] [CrossRef]
- Xu, F.; Wang, H.; Tian, J.; Xu, H. Down-Regulation of ID2-AS1 Alleviates the Neuronal Injury Induced by 1-Methy1-4-Phenylpyridinium in Human Neuroblastoma Cell Line SH-SY5Y Cells Through Regulating MiR-199a-5p/IFNAR1/JAK2/STAT1 Axis. Neurochem. Res. 2021, 46, 2192–2203. [Google Scholar] [CrossRef]
- Minter, M.R.; Moore, Z.; Zhang, M.; Brody, K.M.; Jones, N.C.; Shultz, S.R.; Taylor, J.M.; Crack, P.J. Deletion of the Type-1 Interferon Receptor in APPSWE/PS1ΔE9 Mice Preserves Cognitive Function and Alters Glial Phenotype. Acta Neuropathol. Commun. 2016, 4, 72. [Google Scholar] [CrossRef]
- Madar, I.H.; Sultan, G.; Tayubi, I.A.; Hasan, A.N.; Pahi, B.; Rai, A.; Sivanandan, P.K.; Loganathan, T.; Begum, M.; Rai, S. Identification of Marker Genes in Alzheimer’s Disease Using a Machine-Learning Model. Bioinformation 2021, 17, 348–355. [Google Scholar] [CrossRef]
- Tolosa, E.; Botta-Orfila, T.; Morató, X.; Calatayud, C.; Ferrer-Lorente, R.; Martí, M.-J.; Fernández, M.; Gaig, C.; Raya, Á.; Consiglio, A.; et al. MicroRNA Alterations in IPSC-Derived Dopaminergic Neurons from Parkinson Disease Patients. Neurobiol. Aging 2018, 69, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Q.; Qiu, Y.; Meng, Y.; Wei, L.; Wang, H.; Mo, R.; Zou, D.; Liu, C. A Potential Autophagy-Related Competing Endogenous RNA Network and Corresponding Diagnostic Efficacy in Schizophrenia. Front. Psychiatr. 2021, 12, 628361. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-Y.; Pang, K.; Kim, J.Y.; Ryu, J.R.; Kang, H.; Liu, Z.; Kim, W.-K.; Sun, W.; Kim, H.; Han, K. Post-Transcriptional Regulation of SHANK3 Expression by MicroRNAs Related to Multiple Neuropsychiatric Disorders. Mol. Brain 2015, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- MalaCards—Human Disease Database. Available online: https://www.malacards.org/ (accessed on 10 November 2022).
- Kim, A.H.; Reimers, M.; Maher, B.; Williamson, V.; McMichael, O.; McClay, J.L.; van den Oord, E.J.C.G.; Riley, B.P.; Kendler, K.S.; Vladimirov, V.I. MicroRNA Expression Profiling in the Prefrontal Cortex of Individuals Affected with Schizophrenia and Bipolar Disorders. Schizophr. Res. 2010, 124, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Fan, G.; Zhang, J.; Wu, C.; Du, Y.; Ye, H.; Li, Z.; Wang, L.; Zhang, Z.; Zhang, L.; et al. A 9-MicroRNA Signature in Serum Serves as a Noninvasive Biomarker in Early Diagnosis of Alzheimer’s Disease. J. Alzheimers Dis. 2017, 60, 1365–1377. [Google Scholar] [CrossRef]
- Ignacio, C.; Hicks, S.D.; Burke, P.; Lewis, L.; Szombathyne-Meszaros, Z.; Middleton, F.A. Alterations in Serum MicroRNA in Humans with Alcohol Use Disorders Impact Cell Proliferation and Cell Death Pathways and Predict Structural and Functional Changes in Brain. BMC Neurosci. 2015, 16, 55. [Google Scholar] [CrossRef]
- Pala, E.; Denkçeken, T. Evaluation of MiRNA Expression Profiles in Schizophrenia Using Principal-Component Analysis-Based Unsupervised Feature Extraction Method. J. Comput. Biol. 2020, 27, 1253–1263. [Google Scholar] [CrossRef]
- Ceylan, D.; Tufekci, K.U.; Keskinoglu, P.; Genc, S.; Özerdem, A. Circulating Exosomal MicroRNAs in Bipolar Disorder. J. Affect. Disord. 2020, 262, 99–107. [Google Scholar] [CrossRef]
- Lu, L.; Dai, W.-Z.; Zhu, X.-C.; Ma, T. Analysis of Serum MiRNAs in Alzheimer’s Disease. Am. J. Alzheimers Dis. Other Demen. 2021, 36, 15333175211021712. [Google Scholar] [CrossRef]
- Sun, J.; Zhao, J.; Yang, Z.; Zhou, Z.; Lu, P. Identification of Gene Signatures and Potential Therapeutic Targets for Acquired Chemotherapy Resistance in Gastric Cancer Patients. J. Gastrointest Oncol. 2021, 12, 407–422. [Google Scholar] [CrossRef]
- Tekin, S.S.; Erdal, M.E.; Asoğlu, M.; Ay, Ö.İ.; Ay, M.E.; Yılmaz, Ş.G. Biomarker Potential of Hsa-MiR-145-5p in Peripheral Whole Blood of Manic Bipolar I Patients. Braz. J. Psychiatr. 2022, 44, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Piatkov, I.; Caetano, D.; Assur, Y.; Lau, S.L.; Jones, T.; Boyages, S.C.; McLean, M. ABCB1 and ABCC1 Single-Nucleotide Polymorphisms in Patients Treated with Clozapine. Pharm. Pers. Med. 2017, 10, 235–242. [Google Scholar] [CrossRef]
- Sakatis, M.Z.; Reese, M.J.; Harrell, A.W.; Taylor, M.A.; Baines, I.A.; Chen, L.; Bloomer, J.C.; Yang, E.Y.; Ellens, H.M.; Ambroso, J.L.; et al. Preclinical Strategy to Reduce Clinical Hepatotoxicity Using in Vitro Bioactivation Data for >200 Compounds. Chem. Res. Toxicol. 2012, 25, 2067–2082. [Google Scholar] [CrossRef] [PubMed]
- Milosavljević, F.; Bukvić, N.; Pavlović, Z.; Miljević, Č.; Pešić, V.; Molden, E.; Ingelman-Sundberg, M.; Leucht, S.; Jukić, M.M. Association of CYP2C19 and CYP2D6 Poor and Intermediate Metabolizer Status with Antidepressant and Antipsychotic Exposure. JAMA Psychiatr. 2021, 78, 270–280. [Google Scholar] [CrossRef]
- Wang, S.; Li, J.; Song, M.; Yan, P.; Ju, X.; Liu, J.; Wang, C.; Shi, J. Effect of CYP2C19 Polymorphisms on Serum Valproic Level Acid in Chinese Han Patients with Schizophrenia. Sci. Rep. 2021, 11, 23150. [Google Scholar] [CrossRef]
- Zastrozhin, M.S.; Skryabin, V.Y.; Torrado, M.; Petrovna, A.; Sorokin, A.S.; Grishina, E.A.; Ryzhikova, K.A.; Bedina, I.A.; Buzik, O.Z.; Chumakov, E.M.; et al. Effects of CYP2C19*2 Polymorphisms on the Efficacy and Safety of Phenazepam in Patients with Anxiety Disorder and Comorbid Alcohol Use Disorder. Pharmacogenomics 2020, 21, 111–123. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl. Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Pirmohamed, M.; Williams, D.; Madden, S.; Templeton, E.; Park, B.K. Metabolism and Bioactivation of Clozapine by Human Liver in Vitro. J. Pharmacol. Exp. Ther. 1995, 272, 984–990. [Google Scholar]
- Haddad, P.M.; Brain, C.; Scott, J. Nonadherence with Antipsychotic Medication in Schizophrenia: Challenges and Management Strategies. Patient Relat. Outcome Meas. 2014, 5, 43–62. [Google Scholar] [CrossRef]
- Valenstein, M.; Blow, F.C.; Copeland, L.A.; McCarthy, J.F.; Zeber, J.E.; Gillon, L.; Bingham, C.R.; Stavenger, T. Poor Antipsychotic Adherence Among Patients With Schizophrenia: Medication and Patient Factors. Schizophr. Bull. 2004, 30, 255–264. [Google Scholar] [CrossRef]
- Lukoff, D.; Snyder, K.; Ventura, J.; Nuechterlein, K.H. Life Events, Familial Stress, and Coping in the Developmental Course of Schizophrenia. Schizophr. Bull. 1984, 10, 258–292. [Google Scholar] [CrossRef] [PubMed]
- Nuechterlein, K.H.; Dawson, M.E. A Heuristic Vulnerability/Stress Model of Schizophrenic Episodes. Schizophr. Bull. 1984, 10, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.C.; Walsh, E.; Schepp, K.G. Vulnerability, Stress, and Support in the Disease Trajectory from Prodrome to Diagnosed Schizophrenia: Diathesis–Stress–Support Model. Arch. Psychiatr. Nurs. 2016, 30, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Walder, D.J.; Faraone, S.V.; Glatt, S.J.; Tsuang, M.T.; Seidman, L.J. Genetic Liability, Prenatal Health, Stress and Family Environment: Risk Factors in the Harvard Adolescent Family High Risk for Schizophrenia Study. Schizophr. Res. 2014, 157, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Hahlweg, K.; Baucom, D.H. Family Therapy for Persons with Schizophrenia: Neglected yet Important. Eur. Arch. Psychiatr. Clin. Neurosci. 2022. [Google Scholar] [CrossRef]
- Müller, N. Inflammation in Schizophrenia: Pathogenetic Aspects and Therapeutic Considerations. Schizophr. Bull. 2018, 44, 973–982. [Google Scholar] [CrossRef]
- Black, P.H. Stress and the Inflammatory Response: A Review of Neurogenic Inflammation. Brain Behav. Immun. 2002, 16, 622–653. [Google Scholar] [CrossRef]
- Rodrigues-Amorim, D.; Rivera-Baltanás, T.; Spuch, C.; Caruncho, H.J.; González-Fernandez, Á.; Olivares, J.M.; Agís-Balboa, R.C. Cytokines Dysregulation in Schizophrenia: A Systematic Review of Psychoneuroimmune Relationship. Schizophr. Res. 2018, 197, 19–33. [Google Scholar] [CrossRef]
- Rodrigues-Amorim, D.; Rivera-Baltanás, T.; Bessa, J.; Sousa, N.; de Carmen Vallejo-Curto, M.; Rodríguez-Jamardo, C.; de Las Heras, M.E.; Díaz, R.; Agís-Balboa, R.C.; Olivares, J.M.; et al. The Neurobiological Hypothesis of Neurotrophins in the Pathophysiology of Schizophrenia: A Meta-Analysis. J. Psychiatr. Res. 2018, 106, 43–53. [Google Scholar] [CrossRef]
- Labonté, C.; Zhand, N.; Park, A.; Harvey, P.D. Complete Blood Count Inflammatory Markers in Treatment-Resistant Schizophrenia: Evidence of Association between Treatment Responsiveness and Levels of Inflammation. Psychiatr. Res. 2022, 308, 114382. [Google Scholar] [CrossRef]
- Lin, A.; Kenis, G.; Bignotti, S.; Tura, G.-J.-B.; De Jong, R.; Bosmans, E.; Pioli, R.; Altamura, C.; Scharpé, S.; Maes, M. The Inflammatory Response System in Treatment-Resistant Schizophrenia: Increased Serum Interleukin-6. Schizophr. Res. 1998, 32, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Leboyer, M.; Godin, O.; Terro, E.; Boukouaci, W.; Lu, C.; Andre, M.; Aouizerate, B.; Berna, F.; Barau, C.; Capdevielle, D.; et al. Immune Signatures of Treatment-Resistant Schizophrenia: A FondaMental Academic Centers of Expertise for Schizophrenia (FACE-SZ) Study. Schizophr. Bull. Open 2021, 2, sgab012. [Google Scholar] [CrossRef] [PubMed]
- Penedo, M.A.; Rivera-Baltanas, T.; Perez-Rodriguez, D.; Allen, J.; Borrajom, A.; Alonso-Crespo, D.; Fernández-Pereira, C.; Nieto-Araujo, M.; Ramos-García, S.; Barreiro-Villar, C.; et al. The Role of Dopamine Receptors in Lymphocytes and Their Changes in Schizophrenia. Brain Behav. Immun.–Health 2021, 12, 100199. [Google Scholar] [CrossRef] [PubMed]
- Bose, I.; Ghosh, B. The P53-MDM2 Network: From Oscillations to Apoptosis. J. Biosci. 2007, 32, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Zhao, K.; Zhang, J.; Zhu, J.; Xiao, J. YB-1 Modulates the Drug Resistance of Glioma Cells by Activation of MDM2/P53 Pathway. Drug Des. Devel. 2019, 13, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Sun, D.; Zhang, X. The Role of MDM2 Amplification and Overexpression in Therapeutic Resistance of Malignant Tumors. Cancer Cell Int. 2019, 19, 216. [Google Scholar] [CrossRef] [PubMed]
- Tweddle, D.A.; Pearson, A.D.J.; Haber, M.; Norris, M.D.; Xue, C.; Flemming, C.; Lunec, J. The P53 Pathway and Its Inactivation in Neuroblastoma. Cancer Lett. 2003, 197, 93–98. [Google Scholar] [CrossRef]
- Zafar, A.; Wang, W.; Liu, G.; Xian, W.; McKeon, F.; Zhou, J.; Zhang, R. Targeting the P53-MDM2 Pathway for Neuroblastoma Therapy: Rays of Hope. Cancer Lett. 2021, 496, 16–29. [Google Scholar] [CrossRef]
- Patil, S.P.; Pacitti, M.F.; Gilroy, K.S.; Ruggiero, J.C.; Griffin, J.D.; Butera, J.J.; Notarfrancesco, J.M.; Tran, S.; Stoddart, J.W. Identification of Antipsychotic Drug Fluspirilene as a Potential P53-MDM2 Inhibitor: A Combined Computational and Experimental Study. J. Comput. Aided Mol. Des. 2015, 29, 155–163. [Google Scholar] [CrossRef]
- Annual Report of the Commissioners in Lunacy to the Lord Chancellor. Available online: https://wellcomecollection.org/works/sgjdfxwv/items (accessed on 10 November 2022).
- Mortensen, P.B. Neuroleptic Treatment and Other Factors Modifying Cancer Risk in Schizophrenic Patients. Acta Psychiatr. Scand. 1987, 75, 585–590. [Google Scholar] [CrossRef]
- Dalton, S.O.; Johansen, C.; Poulsen, A.H.; Nørgaard, M.; Sørensen, H.T.; McLaughlin, J.K.; Mortensen, P.B.; Friis, S. Cancer Risk among Users of Neuroleptic Medication: A Population-Based Cohort Study. Br. J. Cancer 2006, 95, 934–939. [Google Scholar] [CrossRef] [PubMed]
- Grinshpoon, A.; Barchana, M.; Ponizovsky, A.; Lipshitz, I.; Nahon, D.; Tal, O.; Weizman, A.; Levav, I. Cancer in Schizophrenia: Is the Risk Higher or Lower? Schizophr. Res. 2005, 73, 333–341. [Google Scholar] [CrossRef] [PubMed]
- McGinty, E.E.; Zhang, Y.; Guallar, E.; Ford, D.E.; Steinwachs, D.; Dixon, L.B.; Keating, N.L.; Daumit, G.L. Cancer Incidence in a Sample of Maryland Residents With Serious Mental Illness. Psychiatr. Serv. 2012, 63, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Mi, Y.; Guo, N.; Xu, H.; Jiang, P.; Zhang, R.; Xu, L.; Gou, X. Glioma in Schizophrenia: Is the Risk Higher or Lower? Front. Cell. Neurosci. 2018, 12, 289. [Google Scholar] [CrossRef]
- Ge, F.; Huo, Z.; Liu, Y.; Du, X.; Wang, R.; Lin, W.; Wang, R.; Chen, J.; Lu, Y.; Wen, Y.; et al. Association between Schizophrenia and Prostate Cancer Risk: Results from a Pool of Cohort Studies and Mendelian Randomization Analysis. Compr. Psychiatr. 2022, 115, 152308. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Gu, S.; Gu, Y.; Yin, J.; Fang, C.; Liu, L. Alteration in the MRNA Expression Profile of the Autophagy-Related MTOR Pathway in Schizophrenia Patients Treated with Olanzapine. BMC Psychiatr. 2021, 21, 388. [Google Scholar] [CrossRef] [PubMed]
- Vucicevic, L.; Misirkic-Marjanovic, M.; Paunovic, V.; Kravic-Stevovic, T.; Martinovic, T.; Ciric, D.; Maric, N.; Petricevic, S.; Harhaji-Trajkovic, L.; Bumbasirevic, V.; et al. Autophagy Inhibition Uncovers the Neurotoxic Action of the Antipsychotic Drug Olanzapine. Autophagy 2014, 10, 2362–2378. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, C.; Xu, Y.; Hou, W.; Chen, J.; Li, Q.; Liu, Z.; Dou, G.; Sun, Y.; Li, R.; Ma, X.; et al. Mechanistic/Mammalian Target of Rapamycin and Side Effects of Antipsychotics: Insights into Mechanisms and Implications for Therapy. Transl. Psychiatr. 2022, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, M.; Riscal, R.; Arena, G.; Linares, L.K.; Le Cam, L. Metabolic Functions of the Tumor Suppressor P53: Implications in Normal Physiology, Metabolic Disorders, and Cancer. Mol. Metab. 2020, 33, 2–22. [Google Scholar] [CrossRef]
- Andrews, J.L.; Goodfellow, F.J.; Matosin, N.; Snelling, M.K.; Newell, K.A.; Huang, X.-F.; Fernandez-Enright, F. Alterations of Ubiquitin Related Proteins in the Pathology and Development of Schizophrenia: Evidence from Human and Animal Studies. J. Psychiatr. Res. 2017, 90, 31–39. [Google Scholar] [CrossRef]
- Maqsood, Q.; Sumrin, A.; Mahnoor, M.; Waseem, M.; Tabassum, N.; Bhattacharya, R.; Saraf, D.; Bose, D. Tumor Suppressor Protein P53 and Association of Its Gene TP53 with Schizophrenia Patients. Gene Rep. 2021, 25, 101402. [Google Scholar] [CrossRef]
- Ni, X.; Trakalo, J.; Valente, J.; Azevedo, M.H.; Pato, M.T.; Pato, C.N.; Kennedy, J.L. Human P53 Tumor Suppressor Gene (TP53) and Schizophrenia: Case–Control and Family Studies. Neurosci. Lett. 2005, 388, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Hiramoto, K.; Yamate, Y.; Sato, E.F. P53 and Clock Genes Play an Important Role in Memory and Learning Ability Depression Due to Long-Term Ultraviolet A Eye Irradiation. Photochem. Photobiol. Sci. 2021, 20, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, M.; Asghar, R.; Firdoos, S.; Ahmad, N.; Nazir, A.; Ullah, K.M.; Li, N.; Zhuang, F.; Chen, Z.; Deng, Y. A Systematic Review of Circulatory MicroRNAs in Major Depressive Disorder: Potential Biomarkers for Disease Prognosis. Int. J. Mol. Sci. 2022, 23, 1294. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Young, M.-J.; Chang, H.-P.; Liu, C.-Y.; Lee, C.-C.; Tseng, Y.-L.; Wang, Y.-C.; Chang, W.-C.; Hung, J.-J. Estradiol-Mediated Inhibition of DNMT1 Decreases P53 Expression to Induce M2-Macrophage Polarization in Lung Cancer Progression. Oncogenesis 2022, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Li, X.; Awan, F.M.; Yang, Z.; Fang, L.; Lyu, J.; Li, F.; Peng, C.; Krylov, S.N.; et al. A Circular RNA Circ-DNMT1 Enhances Breast Cancer Progression by Activating Autophagy. Oncogene 2018, 37, 5829–5842. [Google Scholar] [CrossRef]
- Li, J.; Bian, E.-B.; He, X.-J.; Ma, C.-C.; Zong, G.; Wang, H.-L.; Zhao, B. Epigenetic Repression of Long Non-Coding RNA MEG3 Mediated by DNMT1 Represses the P53 Pathway in Gliomas. Int. J. Oncol. 2016, 48, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Langley, E.; Pearson, M.; Faretta, M.; Bauer, U.-M.; Frye, R.A.; Minucci, S.; Pelicci, P.G.; Kouzarides, T. Human SIR2 Deacetylates P53 and Antagonizes PML/P53-Induced Cellular Senescence. EMBO J. 2002, 21, 2383–2396. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Gu, W. SIRT1: Regulator of P53 Deacetylation. Genes Cancer 2013, 4, 112–117. [Google Scholar] [CrossRef]
- Vaziri, H.; Dessain, S.K.; Eaton, E.N.; Imai, S.-I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. HSIR2SIRT1 Functions as an NAD-Dependent P53 Deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef]
- Sammad, A.; Luo, H.; Hu, L.; Zhu, H.; Wang, Y. Transcriptome Reveals Granulosa Cells Coping through Redox, Inflammatory and Metabolic Mechanisms under Acute Heat Stress. Cells 2022, 11, 1443. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In Vivo Activation of the P53 Pathway by Small-Molecule Antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Lievens, S.; Gerlo, S.; Lemmens, I.; De Clercq, D.J.H.; Risseeuw, M.D.P.; Vanderroost, N.; De Smet, A.-S.; Ruyssinck, E.; Chevet, E.; Van Calenbergh, S.; et al. Kinase Substrate Sensor (KISS), a Mammalian In Situ Protein Interaction Sensor. Mol. Cell. Proteom. 2014, 13, 3332–3342. [Google Scholar] [CrossRef] [PubMed]
- Oliner, J.D.; Kinzler, K.W.; Meltzer, P.S.; George, D.L.; Vogelstein, B. Amplification of a Gene Encoding a P53-Associated Protein in Human Sarcomas. Nature 1992, 358, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Bayle, J.H.; Olson, D.; Levine, A.J. The P53-Mdm-2 Autoregulatory Feedback Loop. Genes Dev. 1993, 7, 1126–1132. [Google Scholar] [CrossRef]
- Ashcroft, M.; Taya, Y.; Vousden, K.H. Stress Signals Utilize Multiple Pathways To Stabilize P53. Mol. Cell. Biol. 2000, 20, 3224–3233. [Google Scholar] [CrossRef]
- Michael, D.; Oren, M. The P53-Mdm2 Module and the Ubiquitin System. Semin. Cancer Biol. 2003, 13, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Tang, L. MDM2 Increases Drug Resistance in Cancer Cells by Inducing EMT Independent of P53. Curr. Med. Chem. 2016, 23, 4529–4539. [Google Scholar] [CrossRef]
- Li, J.; Qu, P.; Zhou, X.-Z.; Ji, Y.-X.; Yuan, S.; Liu, S.-P.; Zhang, Q.-G. Pimozide Inhibits the Growth of Breast Cancer Cells by Alleviating the Warburg Effect through the P53 Signaling Pathway. Biomed Pharm. 2022, 150, 113063. [Google Scholar] [CrossRef]
- Chen, R.W.; Chuang, D.M. Long Term Lithium Treatment Suppresses P53 and Bax Expression but Increases Bcl-2 Expression. A Prominent Role in Neuroprotection against Excitotoxicity. J. Biol. Chem. 1999, 274, 6039–6042. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, B.; Hu, L.; Ye, J.; Tian, W.; He, X. The Dual Roles of MAGE-C2 in P53 Ubiquitination and Cell Proliferation through E3 Ligases MDM2 and TRIM28. Front. Cell Dev. Biol. 2022, 10, 922675. [Google Scholar] [CrossRef]
- Jin, J.-O.; Lee, G.D.; Nam, S.H.; Lee, T.H.; Kang, D.H.; Yun, J.K.; Lee, P.C.-W. Sequential Ubiquitination of P53 by TRIM28, RLIM, and MDM2 in Lung Tumorigenesis. Cell Death Differ. 2021, 28, 1790–1803. [Google Scholar] [CrossRef]
- Luo, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative Control of P53 by Sir2α Promotes Cell Survival under Stress. Cell 2001, 107, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, K.; Herrera, J.E.; Saito, S.; Miki, T.; Bustin, M.; Vassilev, A.; Anderson, C.W.; Appella, E. DNA Damage Activates P53 through a Phosphorylation-Acetylation Cascade. Genes Dev. 1998, 12, 2831–2841. [Google Scholar] [CrossRef]
- Brooks, C.L.; Gu, W. Ubiquitination, Phosphorylation and Acetylation: The Molecular Basis for P53 Regulation. Curr. Opin. Cell Biol. 2003, 15, 164–171. [Google Scholar] [CrossRef]
- Francis, V.A.; Zorzano, A.; Teleman, A.A. DDOR Is an EcR Coactivator That Forms a Feed-Forward Loop Connecting Insulin and Ecdysone Signaling. Curr. Biol. 2010, 20, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A Reference Map of the Human Binary Protein Interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Seillier, M.; Peuget, S.; Gayet, O.; Gauthier, C.; N’Guessan, P.; Monte, M.; Carrier, A.; Iovanna, J.L.; Dusetti, N.J. TP53INP1, a Tumor Suppressor, Interacts with LC3 and ATG8-Family Proteins through the LC3-Interacting Region (LIR) and Promotes Autophagy-Dependent Cell Death. Cell Death Differ. 2012, 19, 1525–1535. [Google Scholar] [CrossRef]
- Kittler, J.T.; Moss, S.J. Modulation of GABAA Receptor Activity by Phosphorylation and Receptor Trafficking: Implications for the Efficacy of Synaptic Inhibition. Curr. Opin. Neurobiol. 2003, 13, 341–347. [Google Scholar] [CrossRef]
- Ye, J.; Zou, G.; Zhu, R.; Kong, C.; Miao, C.; Zhang, M.; Li, J.; Xiong, W.; Wang, C. Structural Basis of GABARAP-Mediated GABAA Receptor Trafficking and Functions on GABAergic Synaptic Transmission. Nat. Commun. 2021, 12, 297. [Google Scholar] [CrossRef]
- Marques, T.R.; Ashok, A.H.; Angelescu, I.; Borgan, F.; Myers, J.; Lingford-Hughes, A.; Nutt, D.J.; Veronese, M.; Turkheimer, F.E.; Howes, O.D. GABA-A Receptor Differences in Schizophrenia: A Positron Emission Tomography Study Using [11C]Ro154513. Mol. Psychiatr. 2021, 26, 2616–2625. [Google Scholar] [CrossRef]
- Ueno, F.; Nakajima, S.; Iwata, Y.; Honda, S.; Torres-Carmona, E.; Mar, W.; Tsugawa, S.; Truong, P.; Plitman, E.; Noda, Y.; et al. Gamma-Aminobutyric Acid (GABA) Levels in the Midcingulate Cortex and Clozapine Response in Patients with Treatment-Resistant Schizophrenia: A Proton Magnetic Resonance Spectroscopy (1H-MRS) Study. Psychiatr. Clin. Neurosci. 2022, 76, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Lee, H.J.; Kim, J.W.; Park, Y.H.; Lee, S.S.; Chang, H.I.; Song, J.Y.; Yoon, D.J.; Bahn, G.H.; Shin, Y.H.; et al. Differences in P53 Gene Polymorphisms between Korean Schizophrenia and Lung Cancer Patients. Schizophr. Res. 2004, 67, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Catts, V.S.; Catts, S.V. Apoptosis and Schizophrenia: Is the Tumour Suppressor Gene, P53, a Candidate Susceptibility Gene? Schizophr. Res. 2000, 41, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xiao, Z.; Chen, W.; Sang, H.; Guan, Y.; Peng, Y.; Zhang, D.; Gu, Z.; Qian, M.; He, G.; et al. Tumor Suppressor Gene TP53 Is Genetically Associated with Schizophrenia in the Chinese Population. Neurosci. Lett. 2004, 369, 126–131. [Google Scholar] [CrossRef]
- Molina, V.; Papiol, S.; Sanz, J.; Rosa, A.; Arias, B.; Fatjó-Vilas, M.; Calama, J.; Hernández, A.I.; Bécker, J.; Fañanás, L. Convergent Evidence of the Contribution of TP53 Genetic Variation (Pro72Arg) to Metabolic Activity and White Matter Volume in the Frontal Lobe in Schizophrenia Patients. NeuroImage 2011, 56, 45–51. [Google Scholar] [CrossRef]
- Fang, X.; Chen, Y.; Wang, Y.; Ren, J.; Zhang, C. Depressive Symptoms in Schizophrenia Patients: A Possible Relationship between SIRT1 and BDNF. Prog. Neuro-Psychopharmacol. Biol. Psychiatr. 2019, 95, 109673. [Google Scholar] [CrossRef]
- Wang, D.; Tang, W.; Zhao, J.; Fan, W.; Zhang, Y.; Zhang, C. A Comprehensive Analysis of the Effect of SIRT1 Variation on the Risk of Schizophrenia and Depressive Symptoms. Front. Genet. 2020, 11, 832. [Google Scholar] [CrossRef]
- Niu, J.; Cao, Y.; Ji, Y. Resveratrol, a SIRT1 Activator, Ameliorates MK-801-Induced Cognitive and Motor Impairments in a Neonatal Rat Model of Schizophrenia. Front. Psychiatr. 2020, 11, 716. [Google Scholar] [CrossRef]
- Dong, E.; Agis-Balboa, R.C.; Simonini, M.V.; Grayson, D.R.; Costa, E.; Guidotti, A. Reelin and Glutamic Acid Decarboxylase 67 Promoter Remodeling in an Epigenetic Methionine-Induced Mouse Model of Schizophrenia. Proc. Natl. Acad. Sci. USA 2005, 102, 12578–12583. [Google Scholar] [CrossRef]
- Saxena, S.; Choudhury, S.; Maroju, P.A.; Anne, A.; Kumar, L.; Mohan, K.N. Dysregulation of Schizophrenia-Associated Genes and Genome-Wide Hypomethylation in Neurons Overexpressing DNMT1. Epigenomics 2021, 13, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Dong, E.; Ruzicka, W.B.; Grayson, D.R.; Guidotti, A. DNA-Methyltransferase1 (DNMT1) Binding to CpG Rich GABAergic and BDNF Promoters Is Increased in the Brain of Schizophrenia and Bipolar Disorder Patients. Schizophr. Res. 2015, 167, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Veldic, M.; Kadriu, B.; Maloku, E.; Agis-Balboa, R.C.; Guidotti, A.; Davis, J.M.; Costa, E. Epigenetic Mechanisms Expressed in Basal Ganglia GABAergic Neurons Differentiate Schizophrenia from Bipolar Disorder. Schizophr. Res. 2007, 91, 51–61. [Google Scholar] [CrossRef]
- Kay, S.R.; Fiszbein, A.; Opler, L.A. The Positive and Negative Syndrome Scale (PANSS) for Schizophrenia. Schizophr. Bull. 1987, 13, 261–276. [Google Scholar] [CrossRef]
- Olivares, J.M.; Berrios, G.E.; Bousoño, M. The Self-Assessment Anhedonia Scale (SAAS). Neurol. Psychiatr. Brain Res. 2005, 12, 121–134. [Google Scholar]
- GRCh38—Hg38—Genome—Assembly—NCBI. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/ (accessed on 14 November 2022).
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucl. Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Carlin, D.E.; Demchak, B.; Pratt, D.; Sage, E.; Ideker, T. Network Propagation in the Cytoscape Cyberinfrastructure. PLoS Comput. Biol. 2017, 13, e1005598. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2021. Nucl. Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The Reactome Pathway Knowledgebase 2022. Nucl. Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; Miller, R.A.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: Connecting Communities. Nucl. Acids Res. 2021, 49, D613–D621. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).