Coordinated Loss and Acquisition of NK Cell Surface Markers Accompanied by Generalized Cytokine Dysregulation in COVID-19
Abstract
1. Introduction
2. Results
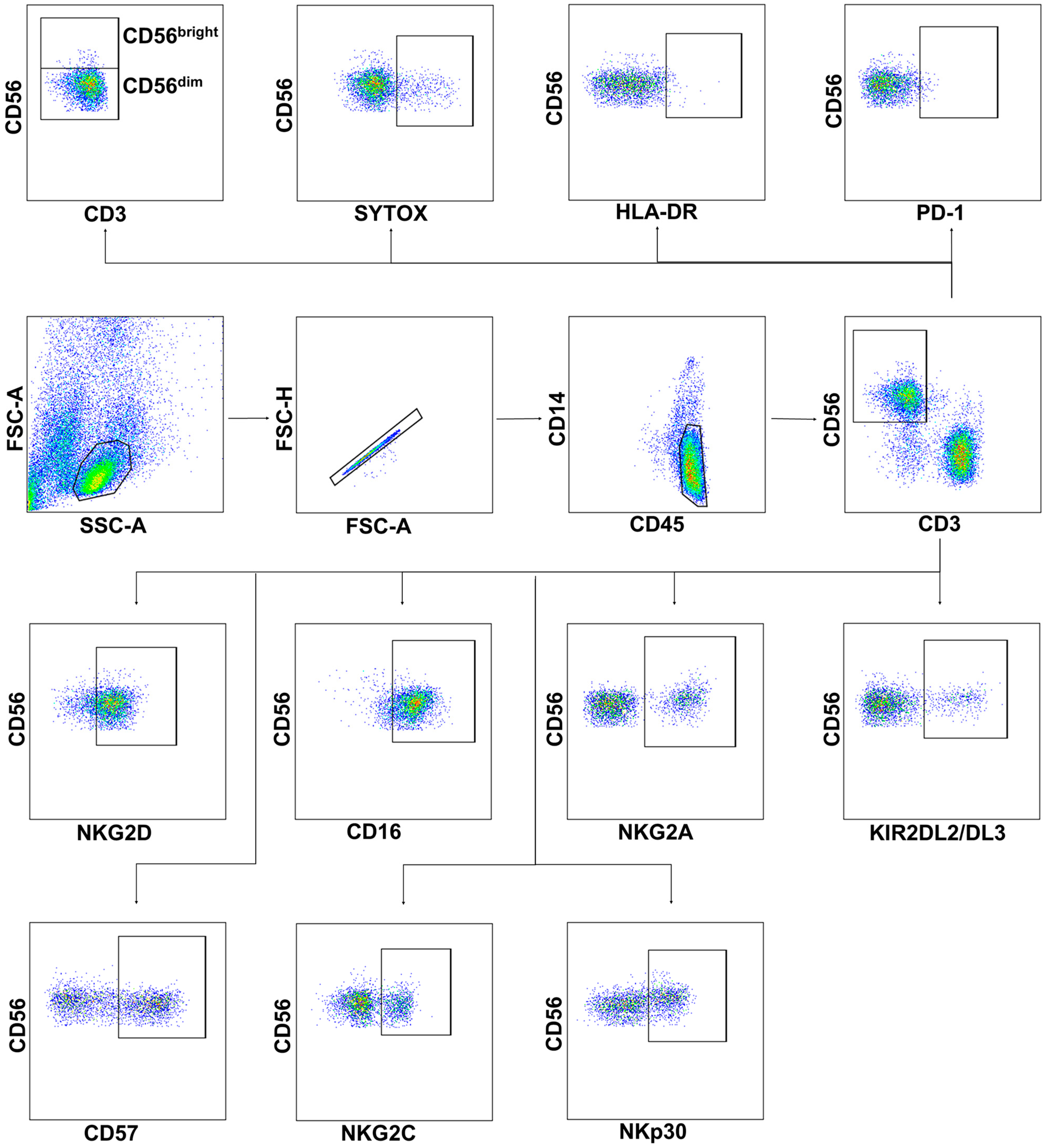
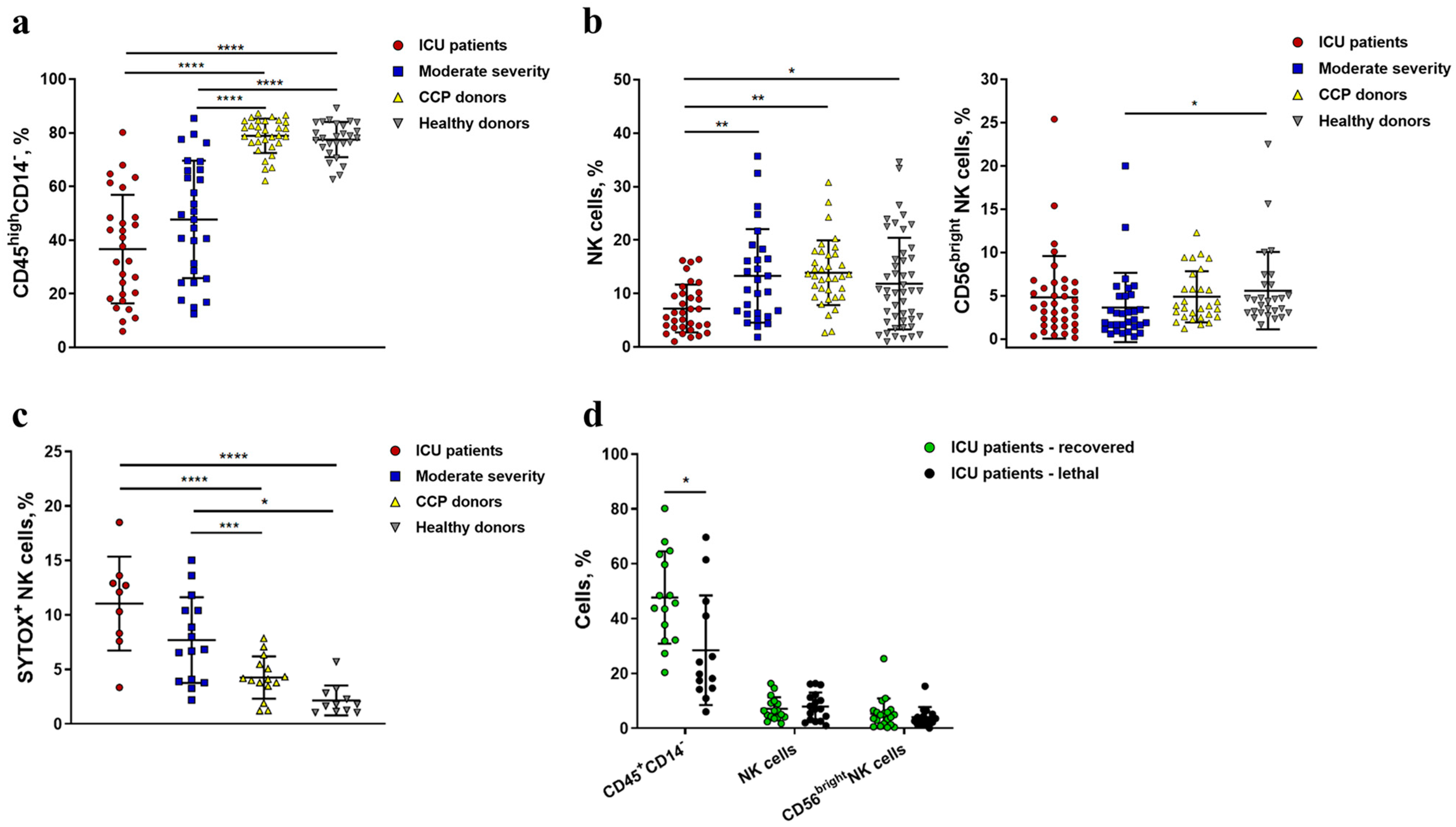
2.1. Severe COVID-19 Is Accompanied by the Depletion of Circulating Lymphocytes and a Decrease in the Content and Viability of NK Cells
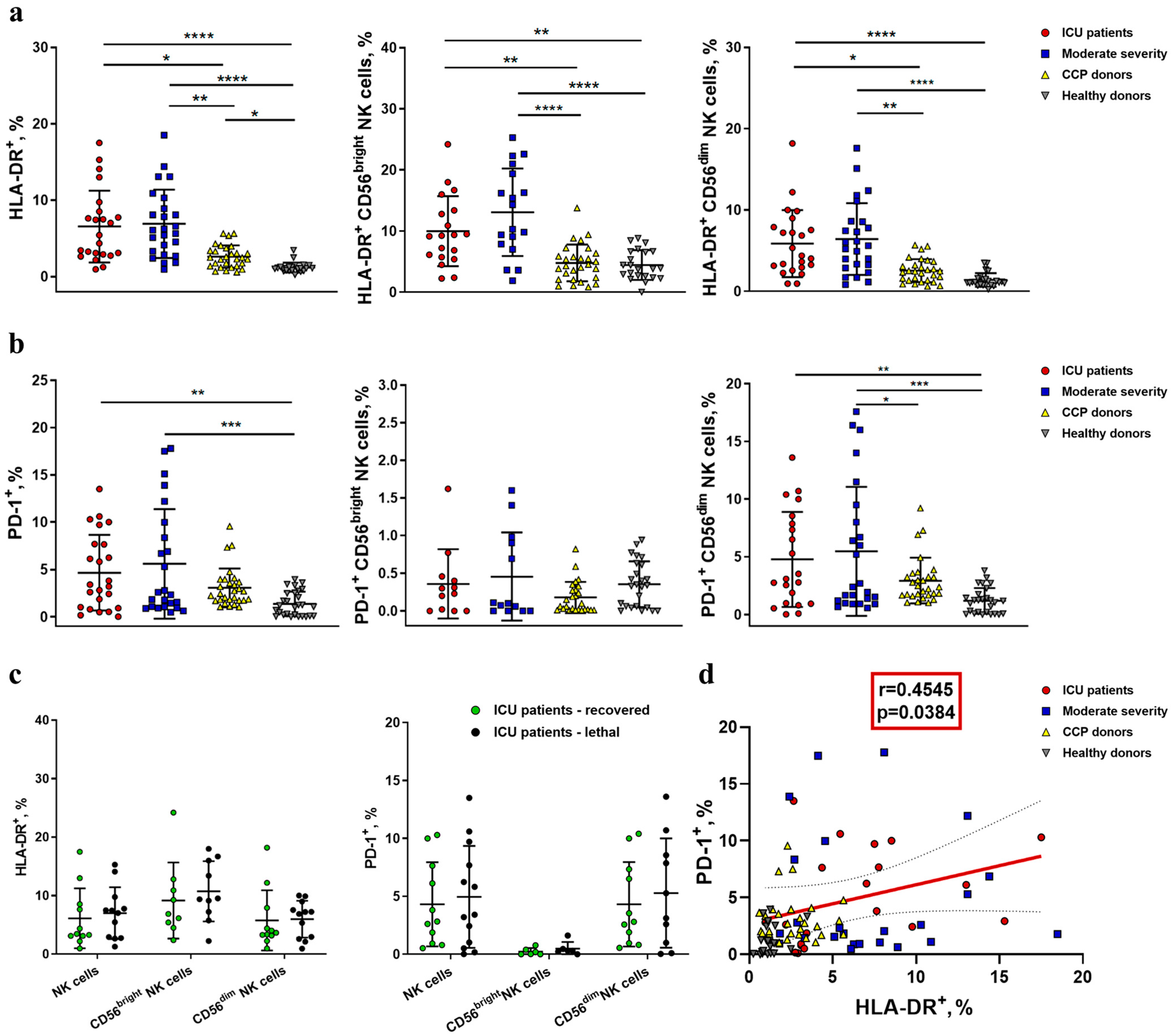
2.2. NK Cells in COVID-19 Show an Activated Profile with Signs of Exhaustion
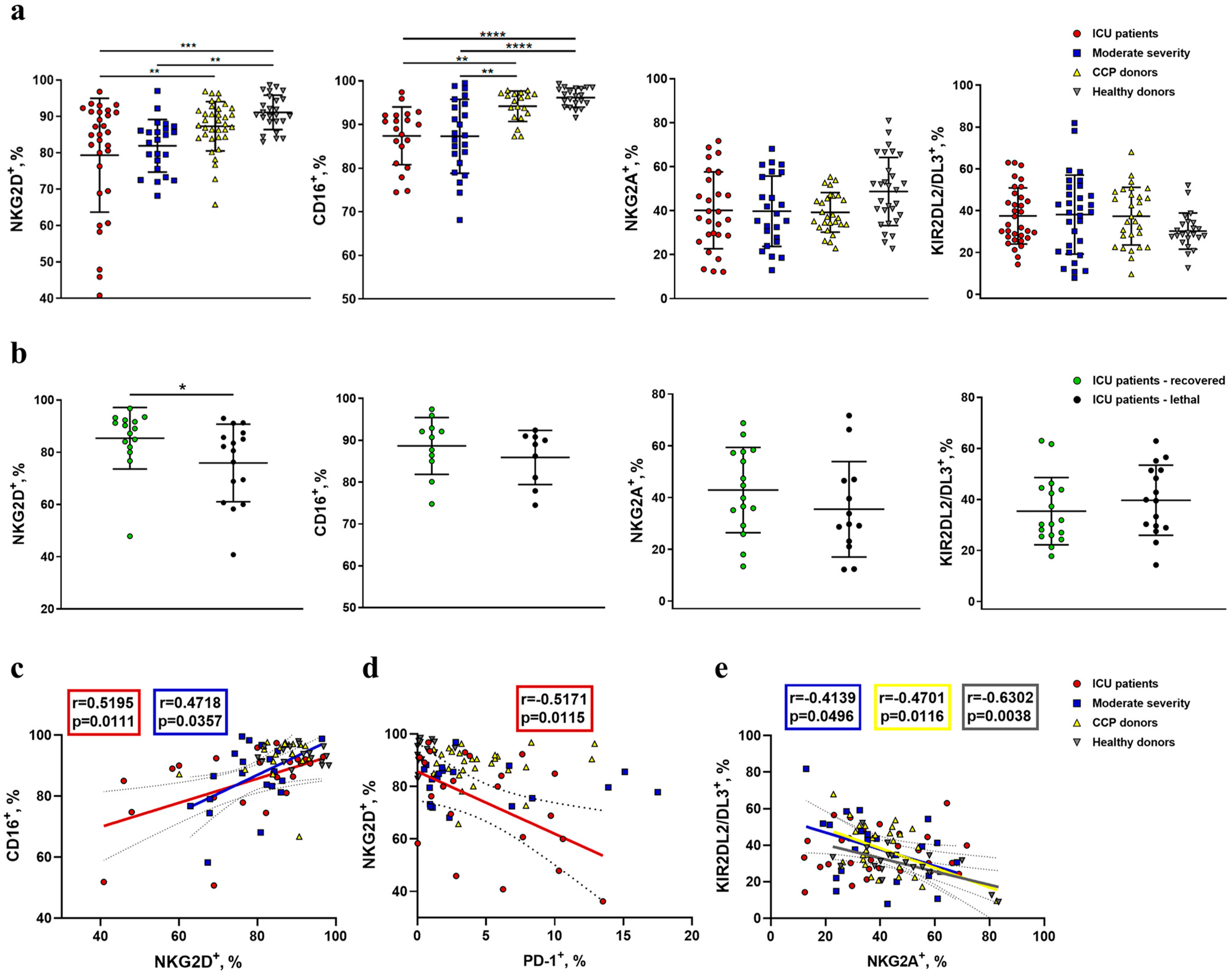
2.3. Reduced Expression of NKG2D and CD16 Receptors Is Observed on the Surface of NK Cells during COVID-19
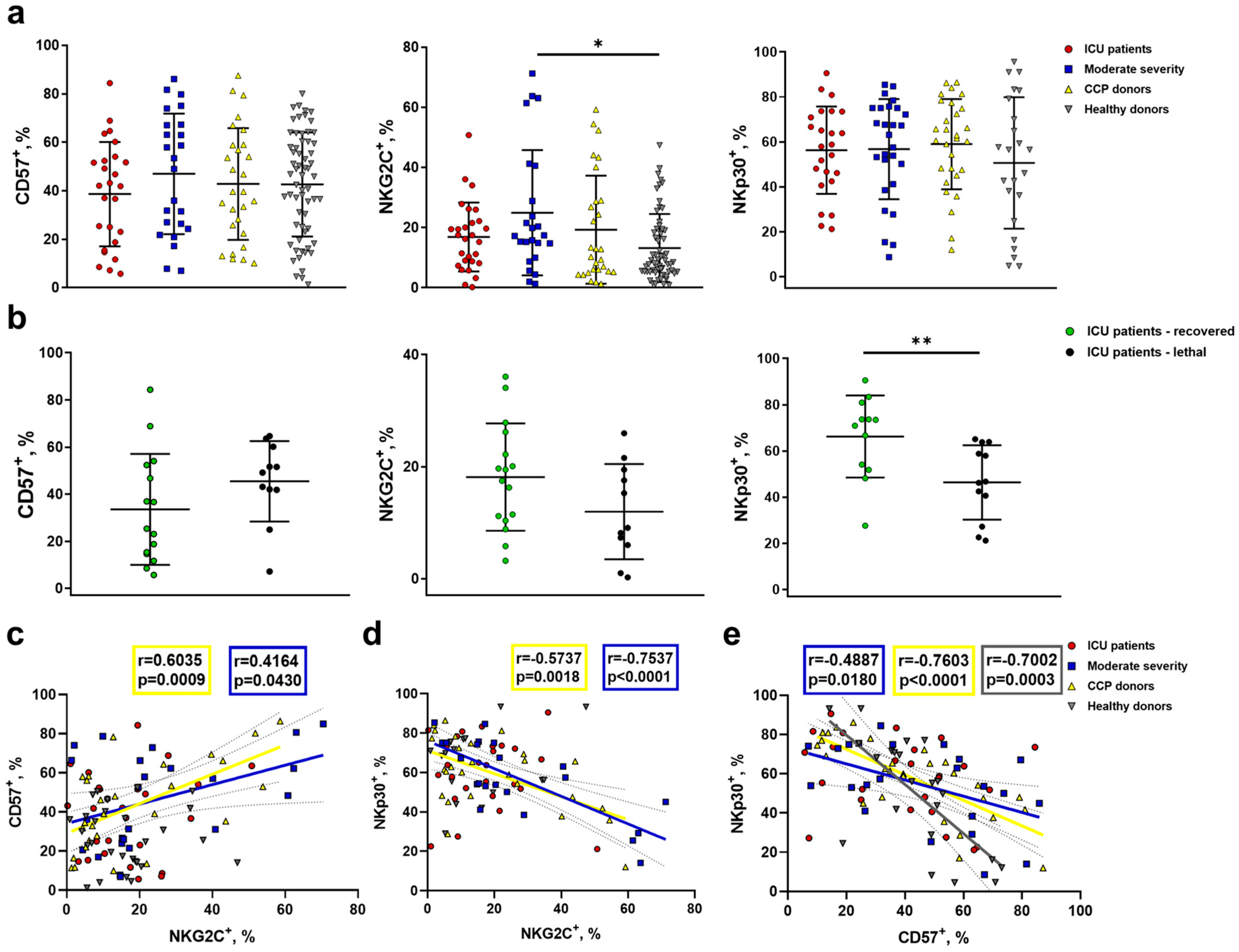
2.4. Increased NKG2C Expression Accompanies the Formation of Adaptive-Like NK Cells in Moderate COVID-19, While in Severe COVID-19, Lethal Outcome Is Associated with a Decrease in the NKp30+ NK Cell Proportion
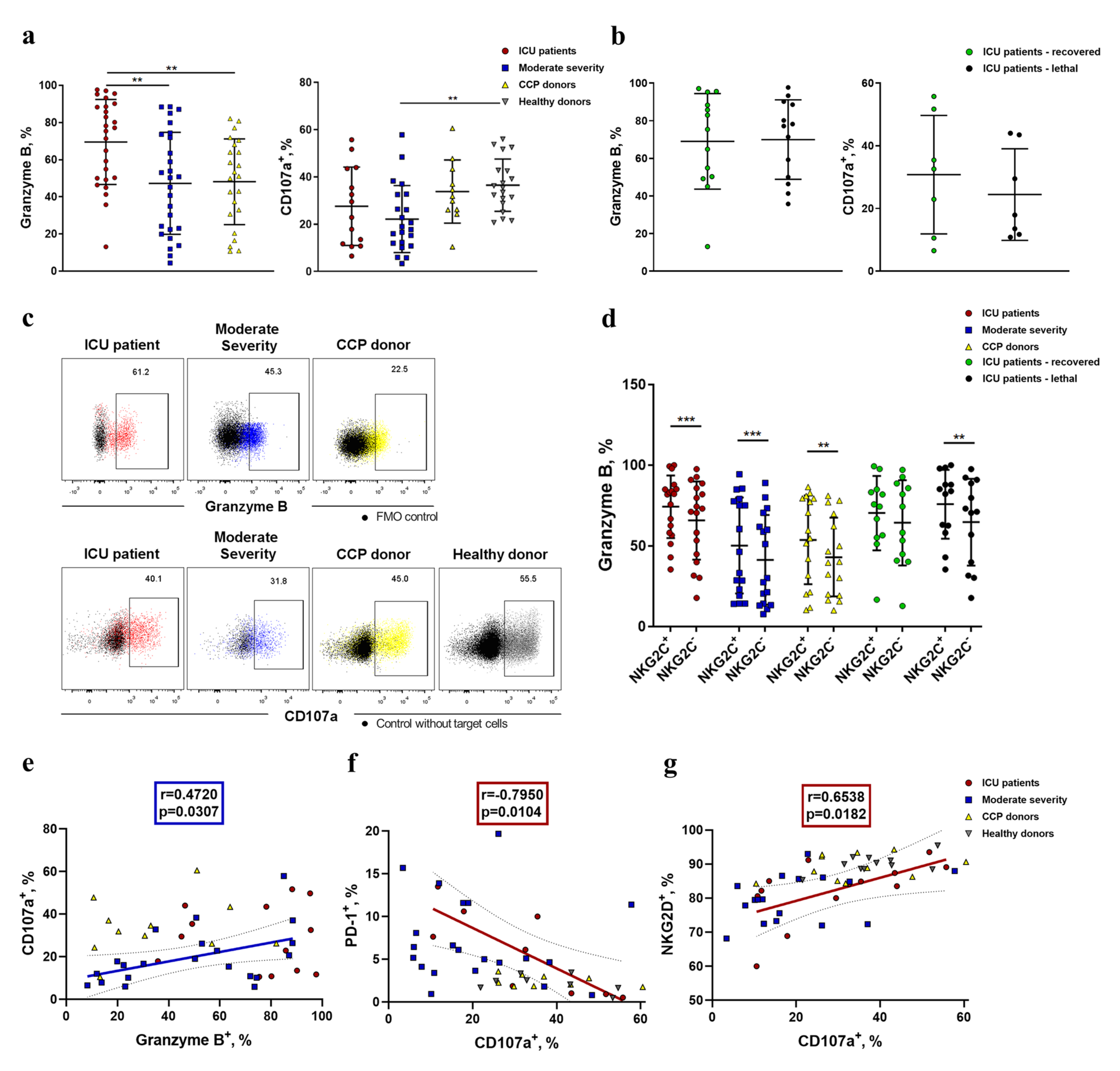
2.5. NK Cells Demonstrate an Increased Level of Granzyme B and Decreased K562-Cell-Induced Degranulation in COVID-19
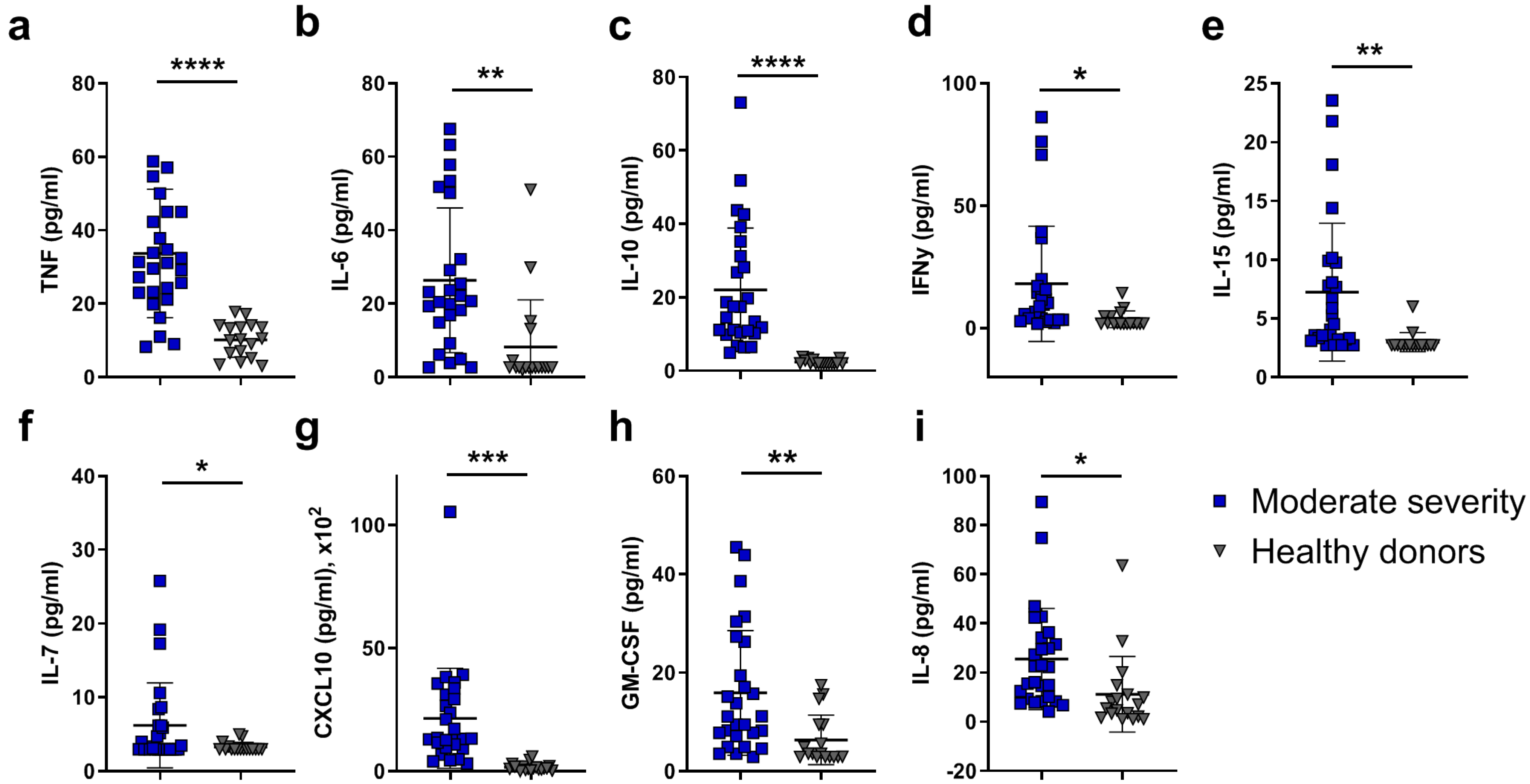
3. Discussion
4. Materials and Methods
4.1. Patient and Donor Characteristics
4.2. Sample Collection
4.3. Phenotype and Viability Analysis
4.4. Granzyme B Staining
4.5. Functional Test
4.6. Cytokine Production
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- COVID Live-Coronavirus Statistics–Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 22 April 2022).
- Platto, S.; Xue, T.; Carafoli, E. COVID19: An Announced Pandemic. Cell Death Dis. 2020, 11, 799. [Google Scholar] [CrossRef]
- Camporota, L.; Chiumello, D.; Busana, M.; Gattinoni, L.; Marini, J.J. Pathophysiology of COVID-19-Associated Acute Respiratory Distress Syndrome. Lancet Respir. Med. 2021, 9, e1. [Google Scholar] [CrossRef]
- Björkström, N.K.; Strunz, B.; Ljunggren, H.G. Natural Killer Cells in Antiviral Immunity. Nat. Rev. Immunol. 2022, 22, 112. [Google Scholar] [CrossRef] [PubMed]
- di Vito, C.; Calcaterra, F.; Coianiz, N.; Terzoli, S.; Voza, A.; Mikulak, J.; della Bella, S.; Mavilio, D. Natural Killer Cells in SARS-CoV-2 Infection: Pathophysiology and Therapeutic Implications. Front. Immunol. 2022, 13, 888248. [Google Scholar] [CrossRef] [PubMed]
- Bi, J. NK Cell Dysfunction in Patients with COVID-19. Cell Mol. Immunol. 2022, 19, 127–129. [Google Scholar] [CrossRef]
- Witkowski, M.; Tizian, C.; Ferreira-Gomes, M.; Niemeyer, D.; Jones, T.C.; Heinrich, F.; Frischbutter, S.; Angermair, S.; Hohnstein, T.; Mattiola, I.; et al. Untimely TGFβ Responses in COVID-19 Limit Antiviral Functions of NK Cells. Nature 2021, 600, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional Exhaustion of Antiviral Lymphocytes in COVID-19 Patients. Cell Mol. Immunol. 2020, 17, 533. [Google Scholar] [CrossRef] [PubMed]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host. Microbe. 2020, 27, 992. [Google Scholar] [CrossRef]
- Maucourant, C.; Filipovic, I.; Ponzetta, A.; Aleman, S.; Cornillet, M.; Hertwig, L.; Strunz, B.; Lentini, A.; Reinius, B.; Brownlie, D.; et al. Natural Killer Cell Immunotypes Related to COVID-19 Disease Severity. Sci. Immunol. 2020, 5, 6832. [Google Scholar] [CrossRef] [PubMed]
- Rendeiro, A.F.; Casano, J.; Vorkas, C.K.; Singh, H.; Morales, A.; DeSimone, R.A.; Ellsworth, G.B.; Soave, R.; Kapadia, S.N.; Saito, K.; et al. Profiling of Immune Dysfunction in COVID-19 Patients Allows Early Prediction of Disease Progression. Life Sci. Alliance 2021, 4, e202000955. [Google Scholar] [CrossRef]
- Jiang, Y.; Wei, X.; Guan, J.; Qin, S.; Wang, Z.; Lu, H.; Qian, J.; Wu, L.; Chen, Y.; Chen, Y.; et al. COVID-19 Pneumonia: CD8+ T and NK Cells Are Decreased in Number but Compensatory Increased in Cytotoxic Potential. Clin. Immunol. 2020, 218, 108516. [Google Scholar] [CrossRef] [PubMed]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A Single-Cell Atlas of the Peripheral Immune Response in Patients with Severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Carsetti, R.; Zaffina, S.; Piano Mortari, E.; Terreri, S.; Corrente, F.; Capponi, C.; Palomba, P.; Mirabella, M.; Cascioli, S.; Palange, P.; et al. Different Innate and Adaptive Immune Responses to SARS-CoV-2 Infection of Asymptomatic, Mild, and Severe Cases. Front. Immunol. 2020, 11, 610300. [Google Scholar] [CrossRef]
- Krämer, B.; Knoll, R.; Bonaguro, L.; ToVinh, M.; Raabe, J.; Astaburuaga-García, R.; Schulte-Schrepping, J.; Kaiser, K.M.; Rieke, G.J.; Bischoff, J.; et al. Early IFN-α Signatures and Persistent Dysfunction Are Distinguishing Features of NK Cells in Severe COVID-19. Immunity 2021, 54, 2650. [Google Scholar] [CrossRef] [PubMed]
- Rieke, G.J.; van Bremen, K.; Bischoff, J.; Vinh, M.T.; Monin, M.B.; Schlabe, S.; Raabe, J.; Kaiser, K.M.; Finnemann, C.; Odainic, A.; et al. Induction of NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity (ADCC) against SARS-CoV-2 after Natural Infection Is More Potent than after Vaccination. J. Infect. Dis. 2022, 225, 1688–1693. [Google Scholar] [CrossRef]
- Li, M.; Guo, W.; Dong, Y.; Wang, X.; Dai, D.; Liu, X.; Wu, Y.; Li, M.; Zhang, W.; Zhou, H.; et al. Elevated Exhaustion Levels of NK and CD8+ T Cells as Indicators for Progression and Prognosis of COVID-19 Disease. Front. Immunol. 2020, 11, 580237. [Google Scholar] [CrossRef]
- Niedźwiedzka-Rystwej, P.; Majchrzak, A.; Aksak-Wąs, B.; Serwin, K.; Czajkowski, Z.; Grywalska, E.; Korona-Głowniak, I.; Roliński, J.; Parczewski, M. Programmed Cell Death-1/Programmed Cell Death-1 Ligand as Prognostic Markers of Coronavirus Disease 2019 Severity. Cells 2022, 11, 1978. [Google Scholar] [CrossRef]
- Niu, C.; Li, M.; Zhu, S.; Chen, Y.; Zhou, L.; Xu, D.; Xu, J.; Li, Z.; Li, W.; Cui, J. PD-1-Positive Natural Killer Cells Have a Weaker Antitumor Function than That of PD-1-Negative Natural Killer Cells in Lung Cancer. Int. J. Med. Sci. 2020, 17, 1964. [Google Scholar] [CrossRef]
- Yunis, E.J.; Romero, V.; Diaz-Giffero, F.; Zuñiga, J.; Koka, P. Natural Killer Cell Receptor NKG2A/HLA-E Interaction Dependent Differential Thymopoiesis of Hematopoietic Progenitor Cells Influences the Outcome of HIV Infection. J. Stem. Cells 2007, 2, 237. [Google Scholar]
- Ljunggren, H.G.; Heggernes Ask, E.; Cornillet, M.; Strunz, B.; Chen, P.; Rao Muvva, J.; Akber, M.; Buggert, M.; Chambers, B.J.; Cuapio Gomez, A.; et al. The Karolinska KI/K COVID-19 Immune Atlas: An Open Resource for Immunological Research and Educational Purposes. Scand. J. Immunol. 2022, 96, 13195. [Google Scholar] [CrossRef]
- Bergantini, L.; D’alessandro, M.; Cameli, P.; Cavallaro, D.; Gangi, S.; Cekorja, B.; Sestini, P.; Bargagli, E. Nk and t Cell Immunological Signatures in Hospitalized Patients with COVID-19. Cells 2021, 10, 3182. [Google Scholar] [CrossRef] [PubMed]
- Vollmers, S.; Lobermeyer, A.; Körner, C. The New Kid on the Block: HLA-C, a Key Regulator of Natural Killer Cells in Viral Immunity. Cells 2021, 10, 3108. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Shao, Z.; Ni, W.; Sun, P.; Qiao, J.; Wan, H.; Huang, Y.; Liu, X.; Zhai, H.; Xiao, M.; et al. The KIR2DL2/HLA-C1C1 Gene Pairing Is Associated with an Increased Risk of SARS-CoV-2 Infection. Front. Immunol. 2022, 13, 3557. [Google Scholar] [CrossRef] [PubMed]
- Frazao, A.; Rethacker, L.; Messaoudene, M.; Avril, M.F.; Toubert, A.; Dulphy, N.; Caignard, A. NKG2D/NKG2-Ligand Pathway Offers New Opportunities in Cancer Treatment. Front. Immunol. 2019, 10, 661. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol. Res. 2015, 3, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Varchetta, S.; Mele, D.; Oliviero, B.; Mantovani, S.; Ludovisi, S.; Cerino, A.; Bruno, R.; Castelli, A.; Mosconi, M.; Vecchia, M.; et al. Unique Immunological Profile in Patients with COVID-19. Cell Mol. Immunol. 2021, 18, 604–612. [Google Scholar] [CrossRef]
- Wang, W.; Erbe, A.K.; Hank, J.A.; Morris, Z.S.; Sondel, P.M. NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity in Cancer Immunotherapy. Front. Immunol. 2015, 6, 386. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, X.; Sun, J.; Xie, T.; Lei, Y.; Muhammad, J.; Li, X.; Zeng, X.; Zhou, F.; Qin, H.; et al. Clinical Characteristics and Immune Injury Mechanisms in 71 Patients with COVID-19. Msphere 2020, 5, e00362-20. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, M.; Zhang, X.; Li, S.; Lu, Q.; Zeng, H.; Hou, H.; Li, H.; Zhang, M.; Jiang, F.; et al. Antibody-Dependent Cellular Cytotoxicity Response to SARS-CoV-2 in COVID-19 Patients. Signal Transduct. Target. Ther. 2021, 6, 346. [Google Scholar] [CrossRef]
- Lopez-Vergès, S.; Milush, J.M.; Schwartz, B.S.; Pando, M.J.; Jarjoura, J.; York, V.A.; Houchins, J.P.; Miller, S.; Kang, S.M.; Norris, P.J.; et al. Expansion of a Unique CD57 +NKG2C Hi Natural Killer Cell Subset during Acute Human Cytomegalovirus Infection. Proc. Natl. Acad. Sci. USA 2011, 108, 14725–14732. [Google Scholar] [CrossRef]
- Schlums, H.; Cichocki, F.; Tesi, B.; Theorell, J.; Beziat, V.; Holmes, T.D.; Han, H.; Chiang, S.C.C.; Foley, B.; Mattsson, K.; et al. Cytomegalovirus Infection Drives Adaptive Epigenetic Diversification of NK Cells with Altered Signaling and Effector Function. Immunity 2015, 42, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Gumá, M.; Budt, M.; Sáez, A.; Brckalo, T.; Hengel, H.; Angulo, A.; López-Botet, M. Expansion of CD94/NKG2C+ NK Cells in Response to Human Cytomegalovirus-Infected Fibroblasts. Blood 2006, 107, 3624–3631. [Google Scholar] [CrossRef] [PubMed]
- Geary, C.D.; Sun, J.C. Memory Responses of Natural Killer Cells. Semin. Immunol. 2017, 31, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, A.; Su, Y.; Calabrese, D.R.; Chen, D.; Arakawa-Hoyt, J.; Roybal, K.T.; Heath, J.R.; Greenland, J.R.; Lanier, L.L. Diminished Cell Proliferation Promotes Natural Killer Cell Adaptive-like Phenotype by Limiting FcεRIγ Expression. J. Exp. Med. 2022, 219, e20220551. [Google Scholar] [CrossRef]
- Vietzen, H.; Zoufaly, A.; Traugott, M.; Aberle, J.; Aberle, S.W.; Puchhammer-Stöckl, E. Deletion of the NKG2C Receptor Encoding KLRC2 Gene and HLA-E Variants Are Risk Factors for Severe COVID-19. Genet. Med. 2021, 23, 963. [Google Scholar] [CrossRef]
- Song, J.W.; Zhang, C.; Fan, X.; Meng, F.P.; Xu, Z.; Xia, P.; Cao, W.J.; Yang, T.; Dai, X.P.; Wang, S.Y.; et al. Immunological and Inflammatory Profiles in Mild and Severe Cases of COVID-19. Nat. Commun. 2020, 11, 3410. [Google Scholar] [CrossRef]
- del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An Inflammatory Cytokine Signature Predicts COVID-19 Severity and Survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling Serum Cytokines in COVID-19 Patients Reveals IL-6 and IL-10 Are Disease Severity Predictors. Emerg. Microbes. Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef]
- Wilson, J.G.; Simpson, L.J.; Ferreira, A.M.; Rustagi, A.; Roque, J.; Asuni, A.; Ranganath, T.; Grant, P.M.; Subramanian, A.; Rosenberg-Hasson, Y.; et al. Cytokine Profile in Plasma of Severe COVID-19 Does Not Differ from ARDS and Sepsis. JCI Insight 2020, 5, 140289. [Google Scholar] [CrossRef]
- Zhang, F.; Mears, J.R.; Shakib, L.; Beynor, J.I.; Shanaj, S.; Korsunsky, I.; Nathan, A.; Donlin, L.T.; Raychaudhuri, S. IFN-γ and TNF-α Drive a CXCL10+ CCL2+ Macrophage Phenotype Expanded in Severe COVID-19 Lungs and Inflammatory Diseases with Tissue Inflammation. Genome Med. 2021, 13, 64. [Google Scholar] [CrossRef]
- Kaiser, R.; Leunig, A.; Pekayvaz, K.; Popp, O.; Joppich, M.; Polewka, V.; Escaig, R.; Anjum, A.; Hoffknecht, M.L.; Gold, C.; et al. Self-Sustaining IL-8 Loops Drive a Prothrombotic Neutrophil Phenotype in Severe COVID-19. JCI Insight 2021, 6, e150862. [Google Scholar] [CrossRef] [PubMed]
- Demaria, O.; Carvelli, J.; Batista, L.; Thibult, M.-L.; Morel, A.; André, P.; Morel, Y.; Vély, F.; Vivier, E. Identification of Druggable Inhibitory Immune Checkpoints on Natural Killer Cells in COVID-19. Cell. Mol. Immunol. 2020, 17, 995–997. [Google Scholar] [CrossRef] [PubMed]
- Vavilova, J.D.; Boyko, A.A.; Ponomareva, N.V.; Fokin, V.F.; Fedotova, E.Y.; Streltsova, M.A.; Kust, S.A.; Grechikhina, M.V.; Bril, E.V.; Zimnyakova, O.S.; et al. Reduced Immunosenescence of Peripheral Blood T Cells in Parkinson’s Disease with CMV Infection Background. Int. J. Mol. Sci. 2021, 22, 13119. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, E.I.; Streltsova, M.A.; Kanevskiy, L.M.; Erokhina, S.A.; Telford, W.G. Identification of Human Memory-like NK Cells. Curr. Protoc. Cytom. 2017, 79, 9.50.1–9.50.11. [Google Scholar] [CrossRef]
- Nielsen, C.M.; White, M.J.; Goodier, M.R.; Riley, E.M. Functional Significance of CD57 Expression on Human NK Cells and Relevance to Disease. Front. Immunol. 2013, 4, 422. [Google Scholar] [CrossRef]
- Bayard, C.; Lepetitcorps, H.; Roux, A.; Larsen, M.; Fastenackels, S.; Salle, V.; Vieillard, V.; Marchant, A.; Stern, M.; Boddaert, J.; et al. Coordinated Expansion of Both Memory T Cells and NK Cells in Response to CMV Infection in Humans. Eur. J. Immunol. 2016, 46, 1168–1179. [Google Scholar] [CrossRef]
- Sintsov, A.V.; Kovalenko, E.I.; Khanin, M.A. Apoptosis Induced by Granzyme B. Bioorg. Khim. 2008, 34, 647–654. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, M.; Chen, X.; Montaner, L.J. Cytokine Storm and Leukocyte Changes in Mild versus Severe SARS-CoV-2 Infection: Review of 3939 COVID-19 Patients in China and Emerging Pathogenesis and Therapy Concepts. J. Leukoc. Biol. 2020, 108, 17–41. [Google Scholar] [CrossRef]
- Malengier-Devlies, B.; Filtjens, J.; Ahmadzadeh, K.; Boeckx, B.; Vandenhaute, J.; de Visscher, A.; Bernaerts, E.; Mitera, T.; Jacobs, C.; Vanderbeke, L.; et al. Severe COVID-19 Patients Display Hyper-Activated NK Cells and NK Cell-Platelet Aggregates. Front. Immunol. 2022, 13, 861251. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Erokhina, S.A.; Streltsova, M.A.; Kanevskiy, L.M.; Grechikhina, M.V.; Sapozhnikov, A.M.; Kovalenko, E.I. HLA-DR-Expressing NK Cells: Effective Killers Suspected for Antigen Presentation. J. Leukoc. Biol. 2021, 109, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S.; Qiu, J.; Rupert, A.W.; Hu, Z.; Higgins, J.; Dewar, R.L.; Stevens, R.; Rehm, C.A.; Metcalf, J.A.; Sherman, B.T.; et al. Interleukin-15 (IL-15) Strongly Correlates with Increasing HIV-1 Viremia and Markers of Inflammation. PLoS ONE 2016, 11, 167091. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-de Prada, L.; Gorgojo-Galindo, Ó.; Fierro, I.; Martínez-García, A.M.; de Quintana, G.S.L.; Gutiérrez-Bustillo, R.; Pelaez-Jareño, M.T.; Álvarez-Fuente, E.; Gómez-Sánchez, E.; Tamayo, E.; et al. Time Evolution of Cytokine Profiles Associated with Mortality in COVID-19 Hospitalized Patients. Front. Immunol. 2022, 13, 946730. [Google Scholar] [CrossRef] [PubMed]
- Schönrich, G.; Raftery, M.J. The PD-1/PD-L1 Axis and Virus Infections: A Delicate Balance. Front. Cell Infect. Microbiol. 2019, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Judge, S.J.; Dunai, C.; Aguilar, E.G.; Vick, S.C.; Sturgill, I.R.; Khuat, L.T.; Stoffel, K.M.; van Dyke, J.; Longo, D.L.; Darrow, M.A.; et al. Minimal PD-1 Expression in Mouse and Human NK Cells under Diverse Conditions. J. Clin. Invest. 2020, 130, 3051–3068. [Google Scholar] [CrossRef]
- Kandeel, E.Z.; Refaat, L.; Bayoumi, A.; Nooh, H.A.; Hammad, R.; Khafagy, M.; Abdellateif, M.S. The Role of Lymphocyte Subsets, PD-1, and FAS (CD95) in COVID-19 Cancer Patients. Viral. Immunol. 2022, 35, 491–502. [Google Scholar] [CrossRef]
- Osman, M.S.; van Eeden, C.; Cohen Tervaert, J.W. Fatal COVID-19 Infections: Is NK Cell Dysfunction a Link with Autoimmune HLH? Autoimmun. Rev. 2020, 19, 102561. [Google Scholar] [CrossRef]
- Fielding, C.A.; Sabberwal, P.; Williamson, J.C.; Greenwood, E.J.D.; Crozier, T.W.M.; Zelek, W.; Seow, J.; Graham, C.; Huettner, I.; Edgeworth, J.D.; et al. SARS-CoV-2 Host-Shutoff Impacts Innate NK Cell Functions, but Antibody-Dependent NK Activity Is Strongly Activated through Non-Spike Antibodies. Elife 2022, 11, 74489. [Google Scholar] [CrossRef]
- de Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; lo Tartaro, D.; Mattioli, M.; et al. Marked T Cell Activation, Senescence, Exhaustion and Skewing towards TH17 in Patients with COVID-19 Pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef]
- Zwirner, N.W.; Fuertes, M.B.; Girart, M.V.; Domaica, C.I.; Rossi, L.E. Cytokine-Driven Regulation of NK Cell Functions in Tumor Immunity: Role of the MICA-NKG2D System. Cytokine Growth Factor Rev. 2007, 18, 159–170. [Google Scholar] [CrossRef]
- Kim, H.; Byun, J.E.; Yoon, S.R.; Koohy, H.; Jung, H.; Choi, I. SARS-CoV-2 Peptides Bind to NKG2D and Increase NK Cell. Cell Immunol. 2022, 371, 104454. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, K.; Riecken, K.; Jung, J.M.; Hildebrandt, H.; Menzel, S.; Bunders, M.J.; Fehse, B.; Koch-Nolte, F.; Heinrich, F.; Peine, S.; et al. Natural Killer Cell-mediated ADCC in SARS-CoV-2-infected Individuals and Vaccine Recipients. Eur. J. Immunol. 2022, 52, 1297. [Google Scholar] [CrossRef] [PubMed]
- Herrera, L.; Martin-Inaraja, M.; Santos, S.; Inglés-Ferrándiz, M.; Azkarate, A.; Perez-Vaquero, M.A.; Vesga, M.A.; Vicario, J.L.; Soria, B.; Solano, C.; et al. Identifying SARS-CoV-2 “memory” NK Cells from COVID-19 Convalescent Donors for Adoptive Cell Therapy. Immunology 2022, 165, 234–249. [Google Scholar] [CrossRef] [PubMed]
- Vietzen, H.; Danklmaier, V.; Zoufaly, A.; Puchhammer-Stöckl, E. High-Affinity FcγRIIIa Genetic Variants and Potent NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity (ADCC) Responses Contributing to Severe COVID-19. Genet. Med. 2022, 24, 1449. [Google Scholar] [CrossRef]
- Antonioli, L.; Fornai, M.; Pellegrini, C.; Blandizzi, C. NKG2A and COVID-19: Another Brick in the Wall. Cell Mol. Immunol. 2020, 17, 672–674. [Google Scholar] [CrossRef]
- Hammer, Q.; Dunst, J.; Christ, W.; Picarazzi, F.; Wendorff, M.; Momayyezi, P.; Huhn, O.; Netskar, H.K.; Maleki, K.T.; García, M.; et al. SARS-CoV-2 Nsp13 Encodes for an HLA-E-Stabilizing Peptide That Abrogates Inhibition of NKG2A-Expressing NK Cells. Cell Rep. 2022, 38, 110503. [Google Scholar] [CrossRef]
- Kobyzeva, P.A.; Streltsova, M.A.; Erokhina, S.A.; Kanevskiy, L.M.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko, E.I. CD56dim CD57- NKG2C+ NK Cells Retaining Proliferative Potential Are Possible Precursors of CD57+ NKG2C+ Memory-like NK Cells. J. Leukoc. Biol. 2020, 108, 1379–1395. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495.e20. [Google Scholar] [CrossRef]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Rotola, A.; Rizzo, R. SARS-CoV-2 Spike 1 Protein Controls Natural Killer Cell Activation via the HLA-E/NKG2A Pathway. Cells 2020, 9, 1975. [Google Scholar] [CrossRef]
- Zenarruzabeitia, O.; Astarloa-Pando, G.; Terrén, I.; Orrantia, A.; Pérez-Garay, R.; Seijas-Betolaza, I.; Nieto-Arana, J.; Imaz-Ayo, N.; Pérez-Fernández, S.; Arana-Arri, E.; et al. T Cell Activation, Highly Armed Cytotoxic Cells and a Shift in Monocytes CD300 Receptors Expression Is Characteristic of Patients with Severe COVID-19. Front. Immunol. 2021, 12, 655934. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ustiuzhanina, M.O.; Vavilova, J.D.; Boyko, A.A.; Streltsova, M.A.; Kust, S.A.; Kanevskiy, L.M.; Sapozhnikov, A.M.; Iskhakov, R.N.; Gubernatorova, E.O.; Drutskaya, M.S.; et al. Coordinated Loss and Acquisition of NK Cell Surface Markers Accompanied by Generalized Cytokine Dysregulation in COVID-19. Int. J. Mol. Sci. 2023, 24, 1996. https://doi.org/10.3390/ijms24031996
Ustiuzhanina MO, Vavilova JD, Boyko AA, Streltsova MA, Kust SA, Kanevskiy LM, Sapozhnikov AM, Iskhakov RN, Gubernatorova EO, Drutskaya MS, et al. Coordinated Loss and Acquisition of NK Cell Surface Markers Accompanied by Generalized Cytokine Dysregulation in COVID-19. International Journal of Molecular Sciences. 2023; 24(3):1996. https://doi.org/10.3390/ijms24031996
Chicago/Turabian StyleUstiuzhanina, Maria O., Julia D. Vavilova, Anna A. Boyko, Maria A. Streltsova, Sofya A. Kust, Leonid M. Kanevskiy, Alexander M. Sapozhnikov, Rustam N. Iskhakov, Ekaterina O. Gubernatorova, Marina S. Drutskaya, and et al. 2023. "Coordinated Loss and Acquisition of NK Cell Surface Markers Accompanied by Generalized Cytokine Dysregulation in COVID-19" International Journal of Molecular Sciences 24, no. 3: 1996. https://doi.org/10.3390/ijms24031996
APA StyleUstiuzhanina, M. O., Vavilova, J. D., Boyko, A. A., Streltsova, M. A., Kust, S. A., Kanevskiy, L. M., Sapozhnikov, A. M., Iskhakov, R. N., Gubernatorova, E. O., Drutskaya, M. S., Bychinin, M. V., Zhukova, O. A., Novikova, O. N., Sotnikova, A. G., Yusubalieva, G. M., Baklaushev, V. P., & Kovalenko, E. I. (2023). Coordinated Loss and Acquisition of NK Cell Surface Markers Accompanied by Generalized Cytokine Dysregulation in COVID-19. International Journal of Molecular Sciences, 24(3), 1996. https://doi.org/10.3390/ijms24031996






