Glial Cultures Differentiated from iPSCs of Patients with PARK2-Associated Parkinson’s Disease Demonstrate a Pro-Inflammatory Shift and Reduced Response to TNFα Stimulation
Abstract
1. Introduction
2. Results
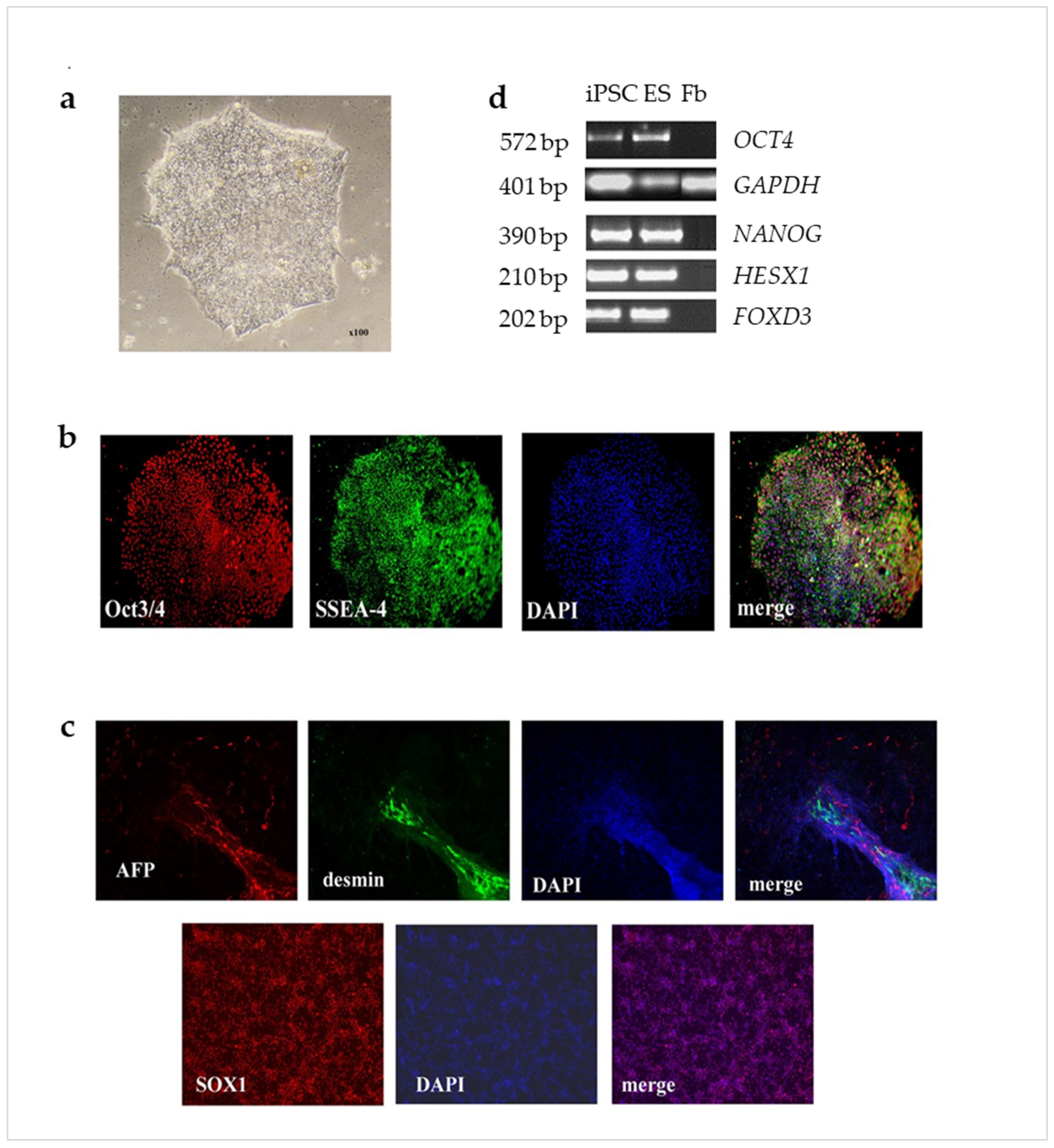
2.1. HD and PD Glial Cultures Contained High Percentage of Astrocytes
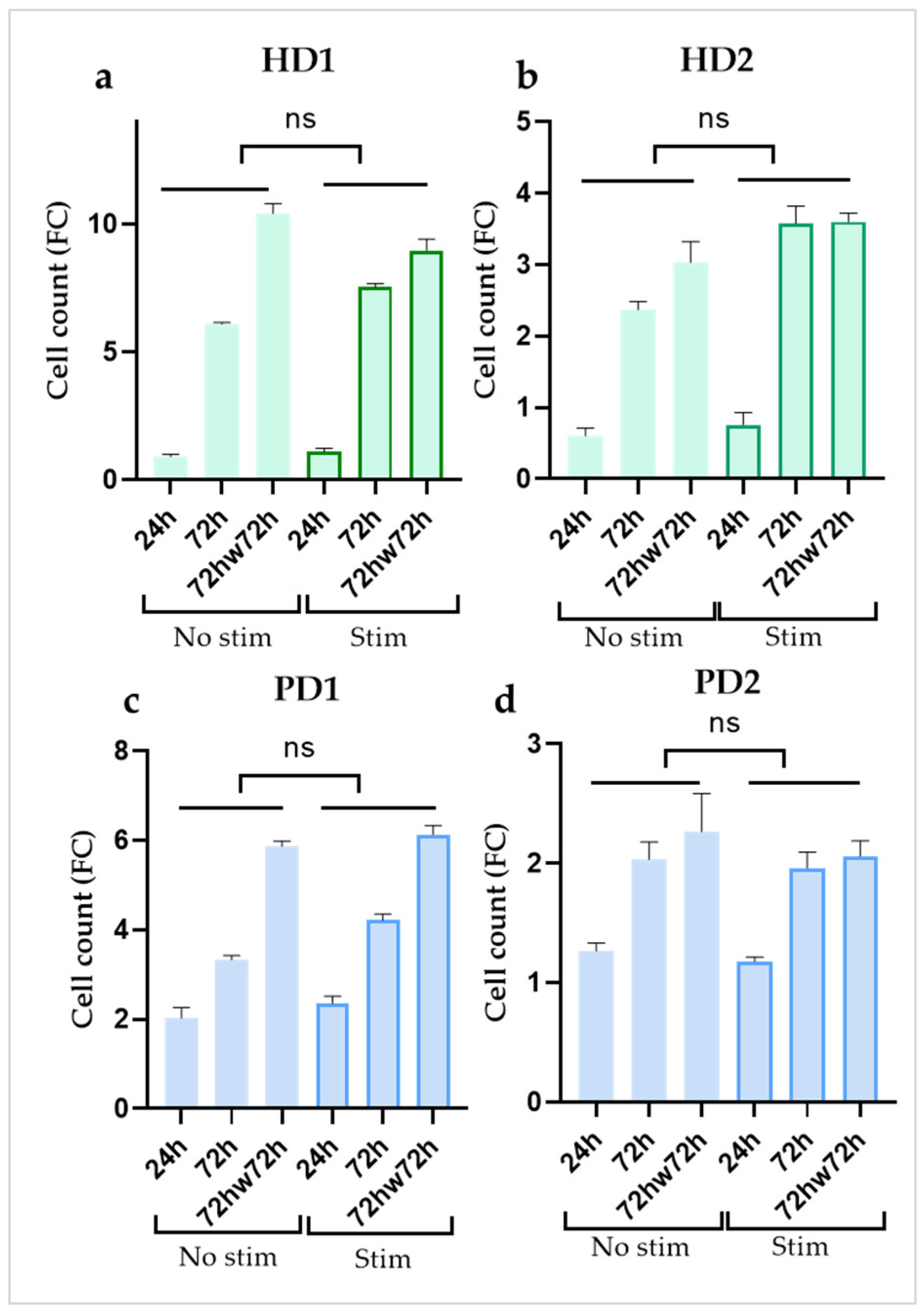
2.2. Cytokine Treatment didn’t Affect Cell Proliferation in Glial Cultures
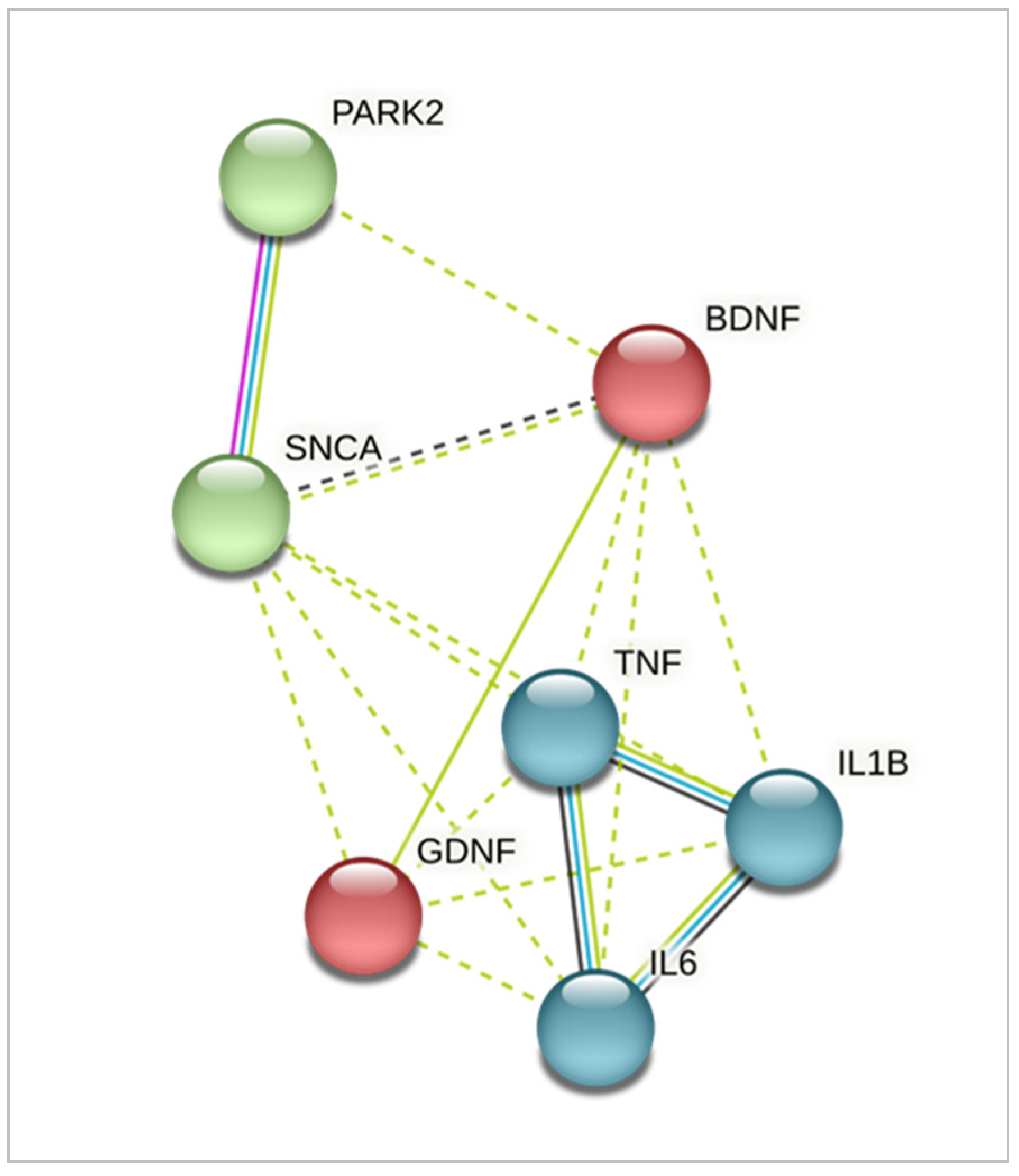
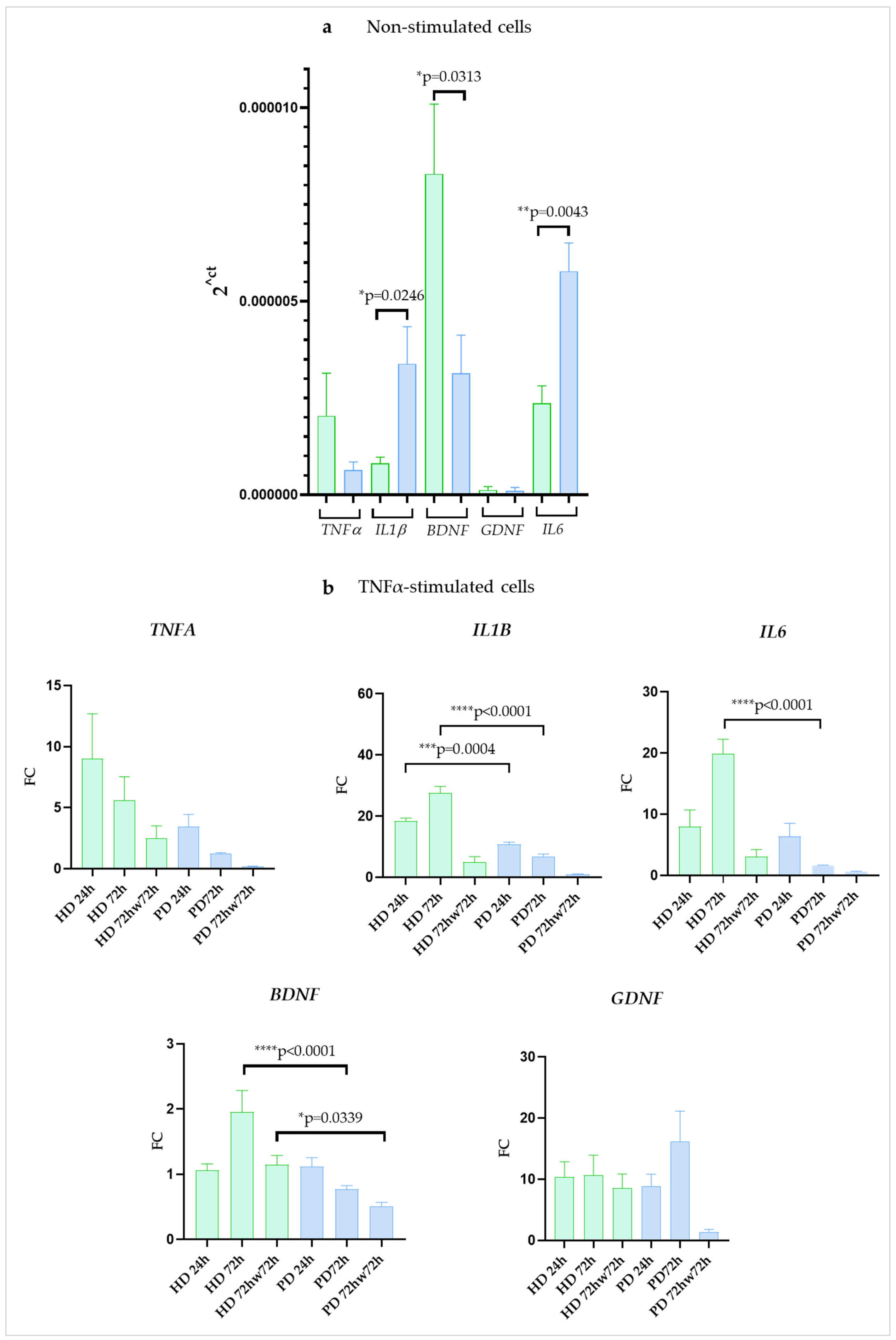
2.3. Expression of Genes Encoding Pro-Inflammatory Cytokines and Neurotrophic Factors Differed between iPSC-Derived Glia from HD and Patients with PARK2-Associated Parkinson Disease (PD) in Both Steady State and Inflammatory Conditions
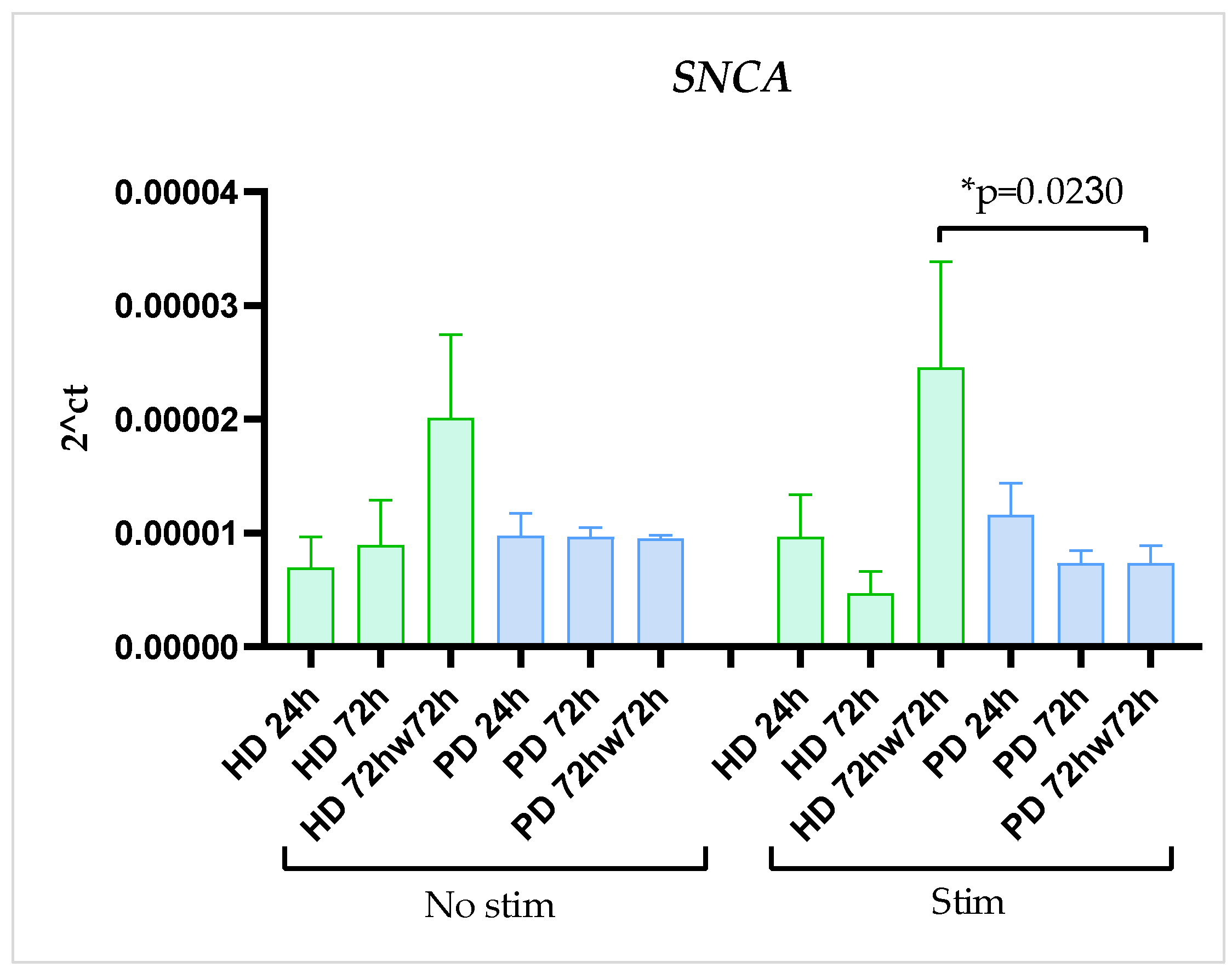
2.4. SNCA Gene Expression Was Stable
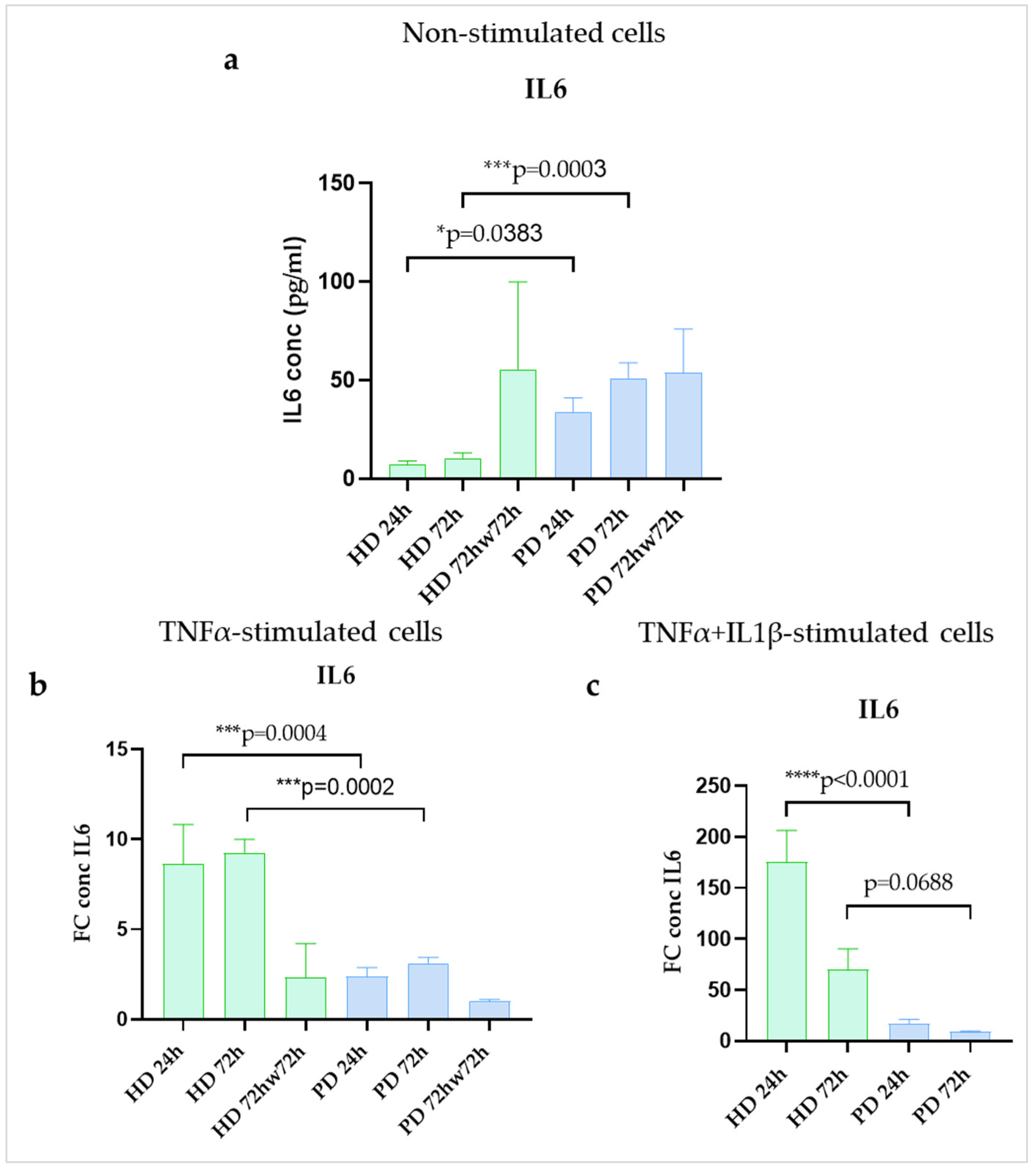
2.5. IL6 Protein Secretion Was Upregulated in Resting Glial Cultures of PD Patients with PARK2 Gene Mutations Compared to HD Cells
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Generation of iPSCs
4.3. Embryoid Bodies Formation
4.4. Obtaining of Glial Cell Lines
4.5. Immunocytochemistry
4.6. Inflammatory Stimulation of Glial Cell Cultures
4.7. MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) Assay
4.8. ELISA
4.9. Quantitative Real-Time PCR (qPCR)
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kanazawa, T.; Uchihara, T.; Takahashi, A.; Nakamura, A.; Orimo, S.; Mizusawa, H. Three-Layered Structure Shared Between Lewy Bodies and Lewy Neurites—Three-Dimensional Reconstruction of Triple-Labeled Sections. Brain Pathol. 2008, 18, 415–422. [Google Scholar] [CrossRef]
- Lee, H.-J.; Suk, J.-E.; Patrick, C.; Bae, E.-J.; Cho, J.-H.; Rho, S.; Hwang, D.; Masliah, E.; Lee, S.-J. Direct Transfer of α-Synuclein from Neuron to Astroglia Causes Inflammatory Responses in Synucleinopathies. J. Biol. Chem. 2010, 285, 9262–9272. [Google Scholar] [CrossRef]
- Ge, P.; Dawson, V.L.; Dawson, T.M. PINK1 and Parkin mitochondrial quality control: A source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener. 2020, 15, 20. [Google Scholar] [CrossRef]
- Leal, M.C.; Casabona, J.C.; Epuntel, M.; Pitossi, F.J. Interleukin-1β and tumor necrosis factor-α: Reliable targets for protective therapies in Parkinson’s Disease? Front. Cell. Neurosci. 2013, 7, 53. [Google Scholar] [CrossRef]
- Booth, H.D.; Hirst, W.D.; Wade-Martins, R. The Role of Astrocyte Dysfunction in Parkinson’s Disease Pathogenesis. Trends Neurosci. 2017, 40, 358–370. [Google Scholar] [CrossRef]
- Araújo, B.; Caridade-Silva, R.; Soares-Guedes, C.; Martins-Macedo, J.; Gomes, E.D.; Monteiro, S.; Teixeira, F.G. Neuroinflammation and Parkinson’s Disease—From Neurodegeneration to Therapeutic Opportunities. Cells 2022, 11, 2908. [Google Scholar] [CrossRef]
- Zhu, Y.; Guo, X.; Zhou, Y.; Zhang, D.; Yi, X.; Wang, H.; Liu, L.; Zhu, X. Levels of serum IL6, TNF-α and sLAG3 are changed and correlated with clinical characteristics in Parkinson’s disease patients: A case-control study. Neurol. Asia 2021, 26, 709–714. [Google Scholar] [CrossRef]
- Walker, D.G.; Lue, L.-F.; Serrano, G.; Adler, C.H.; Caviness, J.N.; Sue, L.I.; Beach, T.G. Altered Expression Patterns of Inflammation-Associated and Trophic Molecules in Substantia Nigra and Striatum Brain Samples from Parkinson’s Disease, Incidental Lewy Body Disease and Normal Control Cases. Front. Neurosci. 2016, 9, 507. [Google Scholar] [CrossRef]
- Giovannoni, F.; Quintana, F.J. The Role of Astrocytes in CNS Inflammation. Trends Immunol. 2020, 41, 805–819. [Google Scholar] [CrossRef]
- Boccazzi, M.; Van Steenwinckel, J.; Schang, A.-L.; Faivre, V.; Le Charpentier, T.; Bokobza, C.; Csaba, Z.; Verderio, C.; Fumagalli, M.; Mani, S.; et al. The immune-inflammatory response of oligodendrocytes in a murine model of preterm white matter injury: The role of TLR3 activation. Cell Death Dis. 2021, 12, 166. [Google Scholar] [CrossRef]
- Nam, J.; Park, E.S.; Won, S.-Y.; Lee, Y.A.; Kim, K.I.; Jeong, J.Y.; Baek, J.Y.; Cho, E.J.; Jin, M.; Chung, Y.C.; et al. TRPV1 on astrocytes rescues nigral dopamine neurons in Parkinson’s disease via CNTF. Brain 2015, 138, 3610–3622. [Google Scholar] [CrossRef]
- Pöyhönen, S.; Er, S.; Domanskyi, A.; Airavaara, M. Effects of Neurotrophic Factors in Glial Cells in the Central Nervous System: Expression and Properties in Neurodegeneration and Injury. Front. Physiol. 2019, 10, 486. [Google Scholar] [CrossRef]
- McBean, G.J. Astrocyte Antioxidant Systems. Antioxidants 2018, 7, 112. [Google Scholar] [CrossRef]
- Konishi, H.; Okamoto, T.; Hara, Y.; Komine, O.; Tamada, H.; Maeda, M.; Osako, F.; Kobayashi, M.; Nishiyama, A.; Kataoka, Y.; et al. Astrocytic phagocytosis is a compensatory mechanism for microglial dysfunction. EMBO J. 2020, 39, e104464. [Google Scholar] [CrossRef]
- Wang, C.; Yang, T.; Liang, M.; Xie, J.; Song, N. Astrocyte dysfunction in Parkinson’s disease: From the perspectives of transmitted α-synuclein and genetic modulation. Transl. Neurodegener. 2021, 10, 39. [Google Scholar] [CrossRef]
- Miyazaki, I.; Asanuma, M. Neuron-Astrocyte Interactions in Parkinson’s Disease. Cells 2020, 9, 2623. [Google Scholar] [CrossRef]
- Kam, T.-I.; Hinkle, J.T.; Dawson, T.M.; Dawson, V.L. Microglia and astrocyte dysfunction in parkinson’s disease. Neurobiol. Dis. 2020, 144, 105028. [Google Scholar] [CrossRef]
- Hyvärinen, T.; Hagman, S.; Ristola, M.; Sukki, L.; Veijula, K.; Kreutzer, J.; Kallio, P.; Narkilahti, S. Co-stimulation with IL-1β and TNF-α induces an inflammatory reactive astrocyte phenotype with neurosupportive characteristics in a human pluripotent stem cell model system. Sci. Rep. 2019, 9, 16944. [Google Scholar] [CrossRef]
- Yan, Y.-Q.; Ma, C.-G.; Ding, Z.-B.; Song, L.-J.; Wang, Q.; Kumar, G. Astrocytes: A double-edged sword in neurodegenerative diseases. Neural Regen. Res. 2021, 16, 1702–1710. [Google Scholar] [CrossRef]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.-S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef]
- Yun, S.P.; Kam, T.-I.; Panicker, N.; Kim, S.; Oh, Y.; Park, J.-S.; Kwon, S.-H.; Park, Y.J.; Karuppagounder, S.S.; Park, H.; et al. Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat. Med. 2018, 24, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Jeon, M.-T.; Kim, K.-S.; Kim, E.S.; Lee, S.; Kim, J.; Hoe, H.-S.; Kim, D.-G. Emerging pathogenic role of peripheral blood factors following BBB disruption in neurodegenerative disease. Ageing Res. Rev. 2021, 68, 101333. [Google Scholar] [CrossRef]
- Santos, R.; Vadodaria, K.C.; Jaeger, B.N.; Mei, A.; Lefcochilos-Fogelquist, S.; Mendes, A.P.; Erikson, G.; Shokhirev, M.; Randolph-Moore, L.; Fredlender, C.; et al. Differentiation of Inflammation-Responsive Astrocytes from Glial Progenitors Generated from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2017, 8, 1757–1769. [Google Scholar] [CrossRef]
- Trindade, P.; Loiola, E.C.; Gasparotto, J.; Ribeiro, C.T.; Cardozo, P.L.; Devalle, S.; Salerno, J.A.; Ornelas, I.M.; Ledur, P.F.; Ribeiro, F.M.; et al. Short and long TNF-alpha exposure recapitulates canonical astrogliosis events in human-induced pluripotent stem cells-derived astrocytes. Glia 2020, 68, 1396–1409. [Google Scholar] [CrossRef]
- Soubannier, V.; Maussion, G.; Chaineau, M.; Sigutova, V.; Rouleau, G.; Durcan, T.M.; Stifani, S. Characterization of human iPSC-derived astrocytes with potential for disease modeling and drug discovery. Neurosci. Lett. 2020, 731, 135028. [Google Scholar] [CrossRef]
- Novosadova, E.; Nekrasov, E.; Chestkov, I.; Surdina, A.; Vasina, E.; Bogomazova, A.; Manuilova, E.; Arsenyeva, E.; Simonova, V.; Konovalova, E.; et al. A Platform for Studying Molecular and Cellular Mechanisms of Parkinson’s Disease Based on Human Induced Pluripotent Stem Cells. Sovrem. Teh. Med. 2016, 8, 157–166. [Google Scholar] [CrossRef]
- Ishijima, T.; Nakajima, K. Inflammatory cytokines TNFα, IL-1β, and IL-6 are induced in endotoxin- stimulated microglia through different signaling cascades. Sci. Prog. 2021, 104, 368504211054985. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Lombardi, M.; Verderio, C.; Fumagalli, M. TNF Production and Release from Microglia via Extracellular Vesicles: Impact on Brain Functions. Cells 2020, 9, 2145. [Google Scholar] [CrossRef]
- Arena, G.; Sharma, K.; Agyeah, G.; Krüger, R.; Grünewald, A.; Fitzgerald, J.C. Neurodegeneration and Neuroinflammation in Parkinson’s Disease: A Self-Sustained Loop. Curr. Neurol. Neurosci. Rep. 2022, 22, 427–440. [Google Scholar] [CrossRef]
- Probert, L. TNF and its receptors in the CNS: The essential, the desirable and the deleterious effects. Neuroscience 2015, 302, 2–22. [Google Scholar] [CrossRef]
- Amin, R.; Quispe, C.; Docea, A.O.; Ydyrys, A.; Kulbayeva, M.; Daştan, S.D.; Calina, D.; Sharifi-Rad, J. The role of Tumour Necrosis Factor in neuroinflammation associated with Parkinson’s disease and targeted therapies. Neurochem. Int. 2022, 158, 105376. [Google Scholar] [CrossRef] [PubMed]
- Papazian, I.; Tsoukala, E.; Boutou, A.; Karamita, M.; Kambas, K.; Iliopoulou, L.; Fischer, R.; Kontermann, R.E.; Denis, M.C.; Kollias, G.; et al. Fundamentally different roles of neuronal TNF receptors in CNS pathology: TNFR1 and IKKβ promote microglial responses and tissue injury in demyelination while TNFR2 protects against excitotoxicity in mice. J. Neuroinflammation 2021, 18, 222. [Google Scholar] [CrossRef] [PubMed]
- Raphael, I.; Gomez-Rivera, F.; Raphael, R.A.; Robinson, R.R.; Nalawade, S.; Forsthuber, T.G. TNFR2 limits proinflammatory astrocyte functions during EAE induced by pathogenic DR2b-restricted T cells. J. Clin. Investig. 2019, 4, 132527. [Google Scholar] [CrossRef]
- Russ, K.; Teku, G.; Bousset, L.; Redeker, V.; Piel, S.; Savchenko, E.; Pomeshchik, Y.; Savistchenko, J.; Stummann, T.C.; Azevedo, C.; et al. TNF-α and α-synuclein fibrils differently regulate human astrocyte immune reactivity and impair mitochondrial respiration. Cell Rep. 2021, 34, 108895. [Google Scholar] [CrossRef] [PubMed]
- Sama, M.A.; Mathis, D.M.; Furman, J.L.; Abdul, H.M.; Artiushin, I.A.; Kraner, S.D.; Norris, C.M. Interleukin-1β-dependent Signaling between Astrocytes and Neurons Depends Critically on Astrocytic Calcineurin/NFAT Activity. J. Biol. Chem. 2008, 283, 21953–21964. [Google Scholar] [CrossRef] [PubMed]
- Erta, M.; Quintana, A.; Hidalgo, J. Interleukin-6, a Major Cytokine in the Central Nervous System. Int. J. Biol. Sci. 2012, 8, 1254–1266. [Google Scholar] [CrossRef]
- Bolin, L.; Zhaung, A.; Strychkarska-Orczyk, I.; Nelson, E.; Huang, I.; Malit, M.; Nguyen, Q. Differential inflammatory activation of IL-6 (−/−) astrocytes. Cytokine 2005, 30, 47–55. [Google Scholar] [CrossRef]
- Kummer, K.K.; Zeidler, M.; Kalpachidou, T.; Kress, M. Role of IL-6 in the regulation of neuronal development, survival and function. Cytokine 2021, 144, 155582. [Google Scholar] [CrossRef] [PubMed]
- Rothaug, M.; Becker-Pauly, C.; Rose-John, S. The role of interleukin-6 signaling in nervous tissue. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 1218–1227. [Google Scholar] [CrossRef]
- Islam, O.; Gong, X.; Rose-John, S.; Heese, K. Interleukin-6 and Neural Stem Cells: More Than Gliogenesis. Mol. Biol. Cell 2009, 20, 188–199. [Google Scholar] [CrossRef]
- Bobbo, V.C.; Engel, D.F.; Jara, C.P.; Mendes, N.F.; Haddad-Tovolli, R.; Prado, T.P.; Sidarta-Oliveira, D.; Morari, J.; Velloso, L.A.; Araujo, E.P. Interleukin-6 actions in the hypothalamus protects against obesity and is involved in the regulation of neurogenesis. J. Neuroinflam. 2021, 18, 192. [Google Scholar] [CrossRef] [PubMed]
- Barker, R.A.; Björklund, A.; Gash, D.M.; Whone, A.; Van Laar, A.; Kordower, J.H.; Bankiewicz, K.; Kieburtz, K.; Saarma, M.; Booms, S.; et al. GDNF and Parkinson’s Disease: Where Next? A Summary from a Recent Workshop. J. Park. Dis. 2020, 10, 875–891. [Google Scholar] [CrossRef]
- Azevedo, M.D.; Sander, S.; Tenenbaum, L. GDNF, A Neuron-Derived Factor Upregulated in Glial Cells during Disease. J. Clin. Med. 2020, 9, 456. [Google Scholar] [CrossRef]
- Perriot, S.; Mathias, A.; Perriard, G.; Canales, M.; Jonkmans, N.; Merienne, N.; Meunier, C.; El Kassar, L.; Perrier, A.; Laplaud, D.; et al. Human Induced Pluripotent Stem Cell-Derived Astrocytes Are Differentially Activated by Multiple Sclerosis-Associated Cytokines. Stem Cell Rep. 2018, 11, 1199–1210. [Google Scholar] [CrossRef]
- Singh, K.; Han, K.; Tilve, S.; Wu, K.; Geller, H.M.; Sack, M.N. Parkin targets NOD2 to regulate astrocyte endoplasmic reticulum stress and inflammation. Glia 2018, 66, 2427–2437. [Google Scholar] [CrossRef]
- Ledesma, M.D.; Galvan, C.; Hellias, B.; Dotti, C.; Jensen, P.H. Astrocytic but not neuronal increased expression and redistribution of parkin during unfolded protein stress. J. Neurochem. 2002, 83, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shan, B.; Liang, Y.; Wei, H.; Yuan, J. Parkin regulates NF-κB by mediating site-specific ubiquitination of RIPK1. Cell Death Dis. 2018, 9, 732. [Google Scholar] [CrossRef]
- Henn, I.H.; Bouman, L.; Schlehe, J.S.; Schlierf, A.; Schramm, J.E.; Wegener, E.; Nakaso, K.; Culmsee, C.; Berninger, B.; Krappmann, D.; et al. Parkin Mediates Neuroprotection through Activation of I B Kinase/Nuclear Factor- B Signaling. J. Neurosci. 2007, 27, 1868–1878. [Google Scholar] [CrossRef] [PubMed]
- Frank-Cannon, T.C.; Tran, T.; Ruhn, K.A.; Martinez, T.N.; Hong, J.; Marvin, M.; Hartley, M.; Treviño, I.; O’Brien, D.E.; Casey, B.; et al. Parkin Deficiency Increases Vulnerability to Inflammation-Related Nigral Degeneration. J. Neurosci. 2008, 28, 10825–10834. [Google Scholar] [CrossRef] [PubMed]
- Sonninen, T.-M.; Hämäläinen, R.H.; Koskuvi, M.; Oksanen, M.; Shakirzyanova, A.; Wojciechowski, S.; Puttonen, K.; Naumenko, N.; Goldsteins, G.; Laham-Karam, N.; et al. Metabolic alterations in Parkinson’s disease astrocytes. Sci. Rep. 2020, 10, 14474. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Shen, R.; Agnihotri, S.K.; Chen, Y.; Huang, Z.; Büeler, H. Lack of PINK1 alters glia innate immune responses and enhances inflammation-induced, nitric oxide-mediated neuron death. Sci. Rep. 2018, 8, 383. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.; DeAndrade, M.P.; Bs, H.S.N.; Lin, S.; Chang, J.; Bs, N.L.; Tomlinson, J.J.; Tansey, M.G.; LaVoie, M.J. Lysosome and Inflammatory Defects in GBA1-Mutant Astrocytes Are Normalized by LRRK2 Inhibition. Mov. Disord. 2020, 35, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Ji, X.; Liu, J. Hypoxia and Alpha-Synuclein: Inextricable Link Underlying the Pathologic Progression of Parkinson’s Disease. Front. Aging Neurosci. 2022, 14, 919343. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, M.; Okada, T. Roles of magnesium and calcium ions in cell-to-substrate adhesion. Exp. Cell Res. 1972, 74, 51–60. [Google Scholar] [CrossRef]
- Wersinger, C.; Prou, D.; Vernier, P.; Sidhu, A. Modulation of dopamine transporter function by α-synuclein is altered by impairment of cell adhesion and by induction of oxidative stress. FASEB J. 2003, 17, 1–30. [Google Scholar] [CrossRef]
- Shekoohi, S.; Rajasekaran, S.; Patel, D.; Yang, S.; Liu, W.; Huang, S.; Yu, X.; Witt, S.N. Knocking out alpha-synuclein in melanoma cells dysregulates cellular iron metabolism and suppresses tumor growth. Sci. Rep. 2021, 11, 5267. [Google Scholar] [CrossRef]
- Shuvalova, L.; Davidenko, A.; Eremeev, A.; Khomyakova, E.; Zerkalenkova, E.; Lebedeva, O.; Bogomazova, A.; Klyushnikov, S.; Illarioshkin, S.; Lagarkova, M. Generation of induced pluripotent stem cell line RCPCMi008-A derived from patient with spinocerebellar ataxia 17. Stem Cell Res. 2021, 54, 102431. [Google Scholar] [CrossRef] [PubMed]
- Novosadova, E.V.; Arsenyva, E.L.; Antonov, S.A.; Kazantseva, E.A.; Novosadova, L.V.; Kurko, O.D.; Illarioshkin, S.N.; Tarantul, V.Z.; Grivennikov, I.A. Generation and characteristics of glial cells from induced human pluripotent stem cells. Neurochem 2020, 37, 358–367. [Google Scholar] [CrossRef]
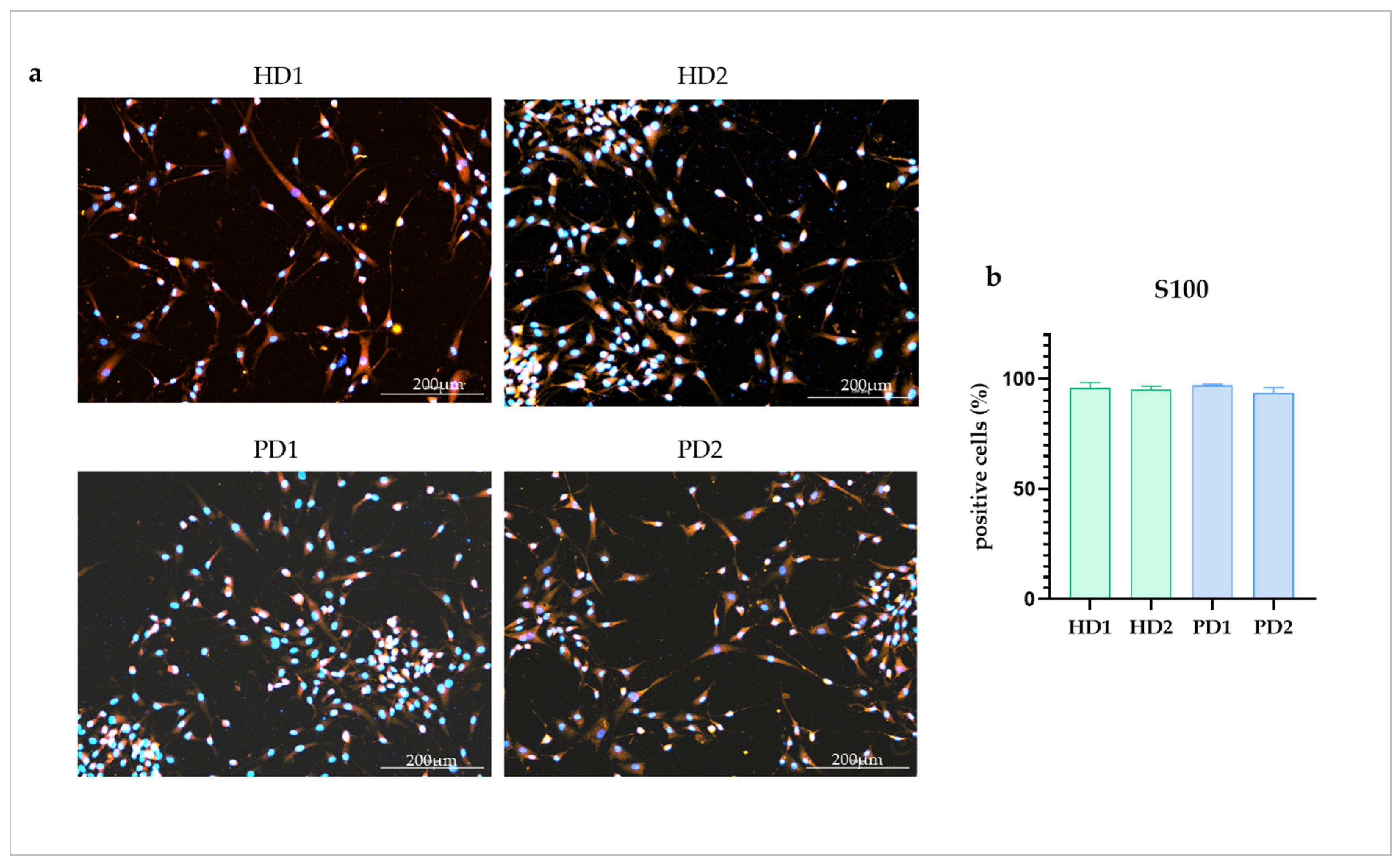
| Name of Glial Cell Lines | PD Patients and HD | Genotype | Cell Line Symbol |
|---|---|---|---|
| HD1 | Healthy male, 60 years | normal | IPSRG2L |
| HD2 | Healthy female, 18 years | normal | IPSHD1.1S |
| PD1 | Male with PD, the disease onset—40 years, biopsy—54 years | het EX2 dup PARK2 | IPSPDPS8 |
| PD2 | Male with PD, the disease onset—38 years, biopsy—40 years | het EX2 del PARK2 | IPSPDPS2d |
| Marker | Antibodies | Dilution | Company, Cat # |
|---|---|---|---|
| Pluripotency | Rabbit anti-Oct4 | 1:200 | Abcam, # ab13742 |
| Mouse anti-SSEA4 | 1:100 | Abcam, # ab16287 | |
| Differentiation | Mouse anti-AFP | 1:200 | Abcam, # ab 3980 |
| Rabbit anti-desmin | 1:200 | Abcam, # 15200 | |
| Rabbit anti-Sox1 | 1:300 | Abcam, # 87775 | |
| Rabbit anti-S100 | 1:2 | Agilent Dako, # GA50461-2 | |
| Secondary antibodies | Goat anti-Rabbit IgG (H + L), AF546 | 1:1000 | TermoFisher, # A11010 |
| Goat anti-Mouse IgG (H + L), AF488 | 1:1000 | TermoFisher, # A11008 |
| Gene | Forward | Reverse | to Annealing |
|---|---|---|---|
| OCT4 | CGACCATCTGCCGCTTTGAG | CCCCCTGTCCCCCATTCCTA | 69 °C |
| SOX2 | TCCTGATTCCAGTTTGCCTC | GCTTAGCCTCGTCGATGAAC | 69 °C |
| NANOG | CAGCCCTGATTCTTCCACCAGTCCC | TGGAAGGTTCCCAGTCGGGTTCACC | 69 °C |
| FOXD3 | CAAGCCCAAGAACAGCCTAGTGAA | TGACGAAGCAGTCGTTGAGTGAGA | 63 °C |
| HESH1 | ACCTGCAGCTCATCAGGGAAAGAT | AAAGCAGTTCTTGGTCTCGGCCT | 60 °C |
| GAPDH | GAAGGTGAAGGTCGGAGTCA | TTCACACCCATGACGAACAT | 60 °C |
| TNFA | CTCCAGGCGGTGCTTGTT | AGGCTTGTCACTCGGGGTT | 60 °C |
| IL1B | GCTCGCCAGTGAAATGATGG | GTCCTGGAAGGAGCACTTCAT | 60 °C |
| IL6 | CCTTCCAAAGATGGCTGAAA | CAGGGGTGGTTATTGCATCT | 60 °C |
| BDNF | ATTGGCTGGCGATTCATAAG | GTTTCCCTTCTGGTCATGGA | 60 °C |
| GDNF | TGGCTCTGGGCTATGAAACC | ATGCCTGCCCTACTTTGTCA | 60 °C |
| SNCA | AGTGACAAATGTTGGAGGAG | GCTTCAGGTTCGTAGTCTTG | 60 °C |
| 18S | CGGCTACCACATCCAAGGAA | GCTGGAATTACCGCGGCT | 60 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerasimova, T.; Stepanenko, E.; Novosadova, L.; Arsenyeva, E.; Shimchenko, D.; Tarantul, V.; Grivennikov, I.; Nenasheva, V.; Novosadova, E. Glial Cultures Differentiated from iPSCs of Patients with PARK2-Associated Parkinson’s Disease Demonstrate a Pro-Inflammatory Shift and Reduced Response to TNFα Stimulation. Int. J. Mol. Sci. 2023, 24, 2000. https://doi.org/10.3390/ijms24032000
Gerasimova T, Stepanenko E, Novosadova L, Arsenyeva E, Shimchenko D, Tarantul V, Grivennikov I, Nenasheva V, Novosadova E. Glial Cultures Differentiated from iPSCs of Patients with PARK2-Associated Parkinson’s Disease Demonstrate a Pro-Inflammatory Shift and Reduced Response to TNFα Stimulation. International Journal of Molecular Sciences. 2023; 24(3):2000. https://doi.org/10.3390/ijms24032000
Chicago/Turabian StyleGerasimova, Tatiana, Ekaterina Stepanenko, Lyudmila Novosadova, Elena Arsenyeva, Darya Shimchenko, Vyacheslav Tarantul, Igor Grivennikov, Valentina Nenasheva, and Ekaterina Novosadova. 2023. "Glial Cultures Differentiated from iPSCs of Patients with PARK2-Associated Parkinson’s Disease Demonstrate a Pro-Inflammatory Shift and Reduced Response to TNFα Stimulation" International Journal of Molecular Sciences 24, no. 3: 2000. https://doi.org/10.3390/ijms24032000
APA StyleGerasimova, T., Stepanenko, E., Novosadova, L., Arsenyeva, E., Shimchenko, D., Tarantul, V., Grivennikov, I., Nenasheva, V., & Novosadova, E. (2023). Glial Cultures Differentiated from iPSCs of Patients with PARK2-Associated Parkinson’s Disease Demonstrate a Pro-Inflammatory Shift and Reduced Response to TNFα Stimulation. International Journal of Molecular Sciences, 24(3), 2000. https://doi.org/10.3390/ijms24032000







