Xanthones from Gentianella acuta (Michx.) Hulten Ameliorate Colorectal Carcinoma via the PI3K/Akt/mTOR Signaling Pathway
Abstract
1. Introduction
2. Results
2.1. Isolation and Chemical Composition Analysis of GA Extracts
2.2. Intervention Effect of Different Fractions on CRC Cells
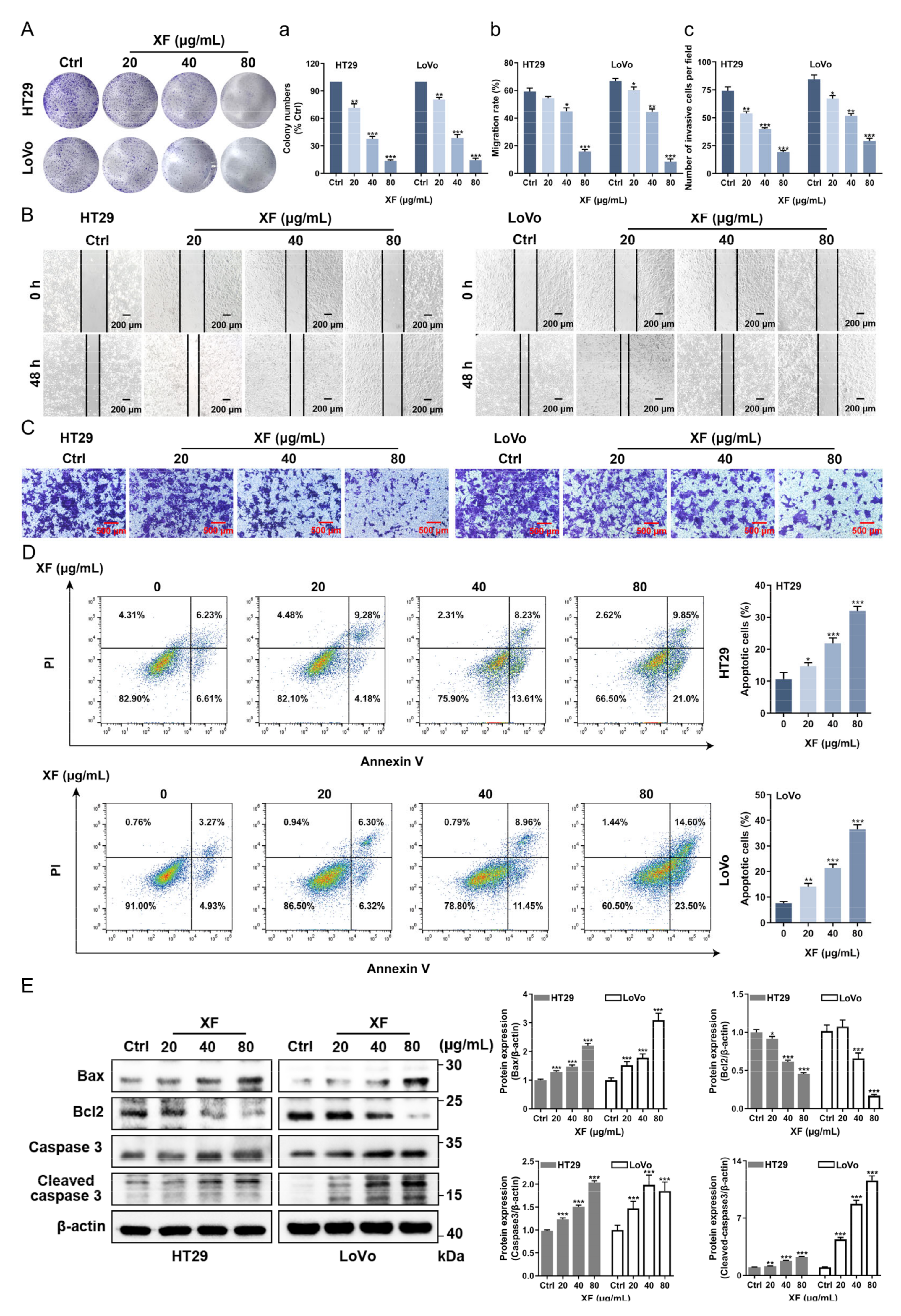
2.3. XF Inhibits the Proliferation of CRC Cells
2.4. XF Inhibits the Migration and Invasion of CRC Cell
2.5. XF Promotes the Apoptosis of CRC Cells
2.6. Xanthones in XF Inhibited Tumor Cell Proliferation In Vitro
- (1)
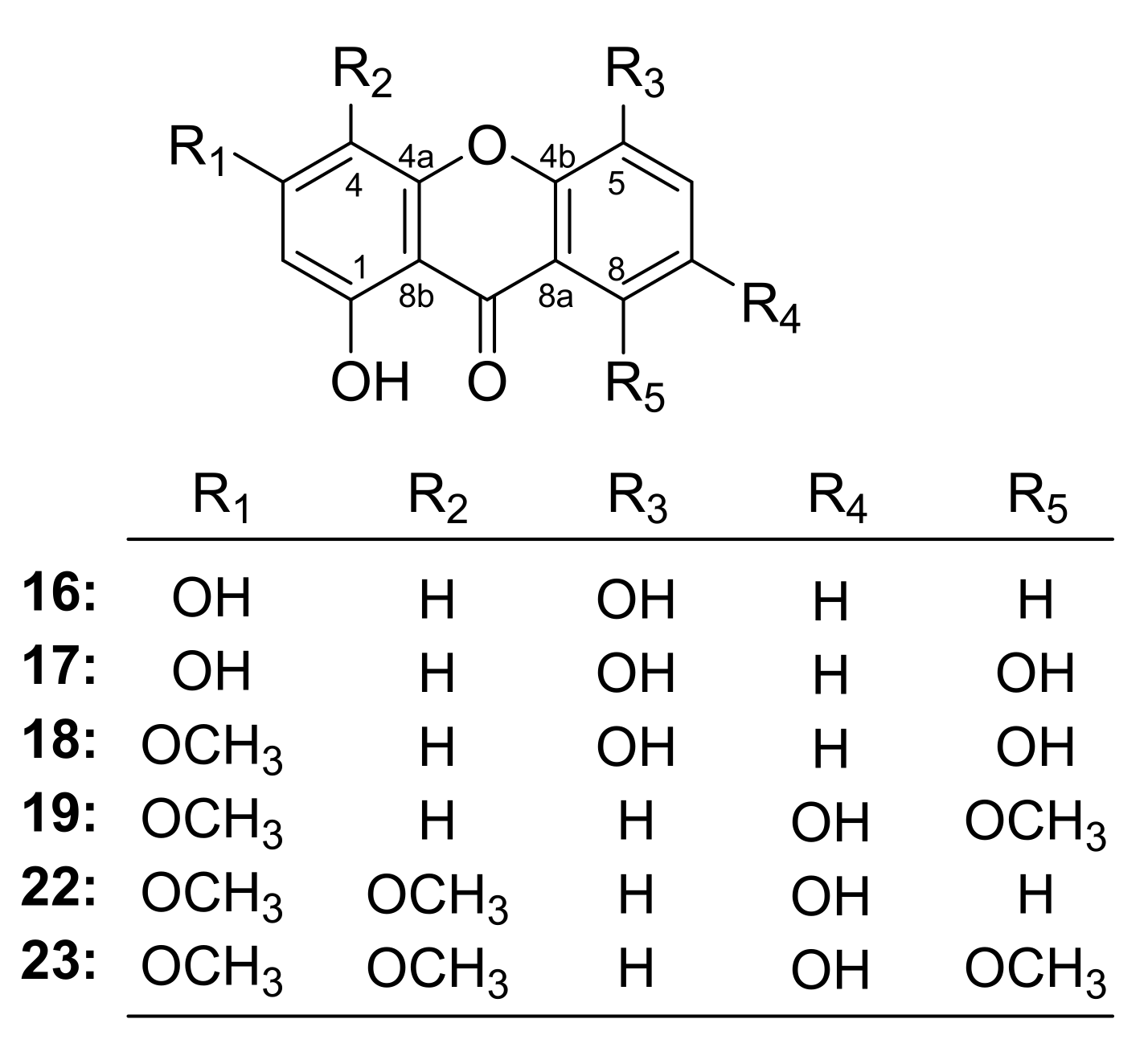
- The oxygenated substitution at C-4 in the xanthone might be the key group for its antiproliferative activity in tumor cells (22, 23 vs. 16–19).
- (2)
- The activity of xanthone substituted in C-3 by hydroxyl was superior to that substituted by methoxy (17 vs. 18).
2.7. Active Xanthones May Prevent CRC through PI3K/Akt/mTOR Pathway by Network Pharmacology Prediction
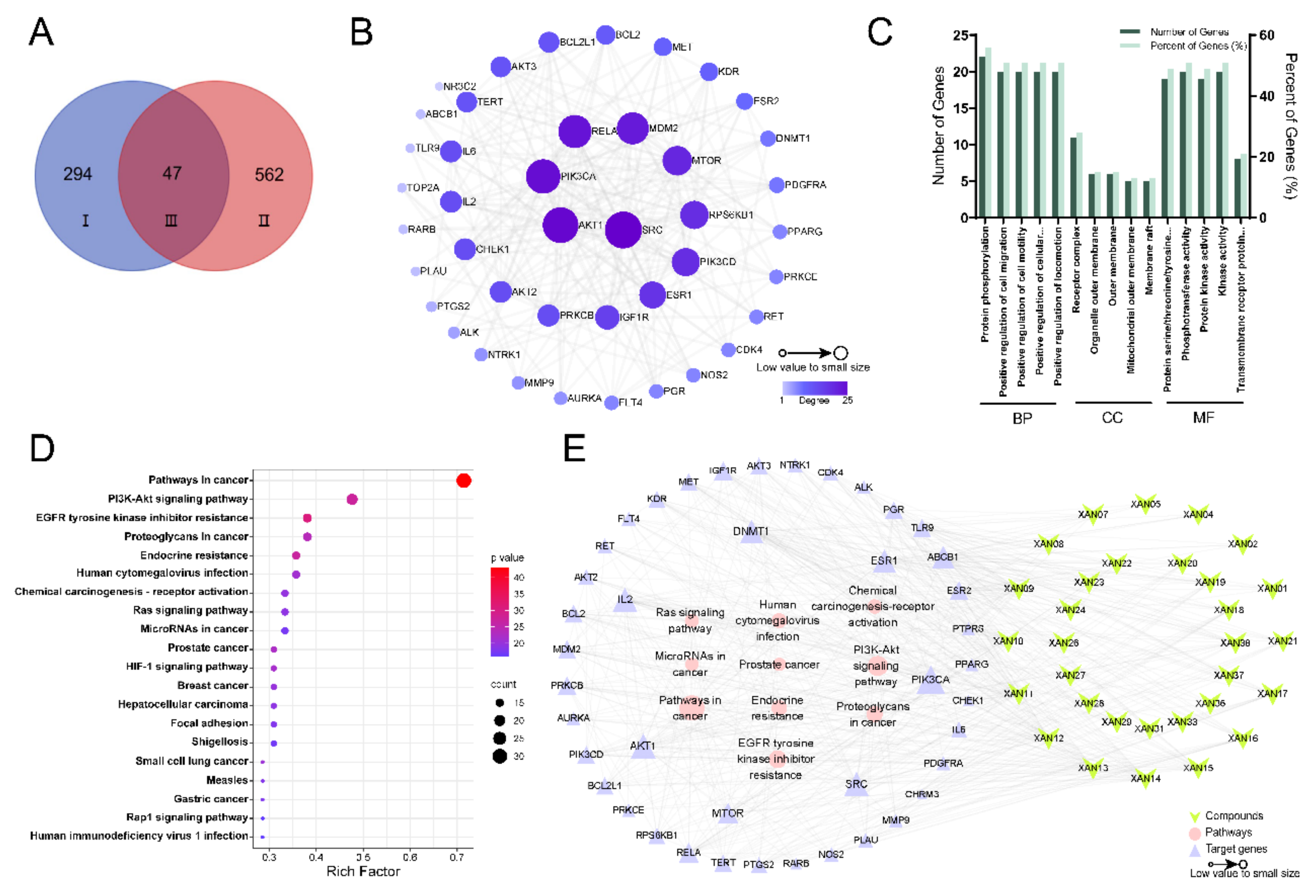
2.7.1. Components-Disease Genes Associated with CRC
2.7.2. Construction of PPI Network Diagram
2.7.3. GO Functional Annotation and KEGG Pathway Enrichment Analysis
2.7.4. Analysis of the Results of Components-Targets-Pathways Network Construction
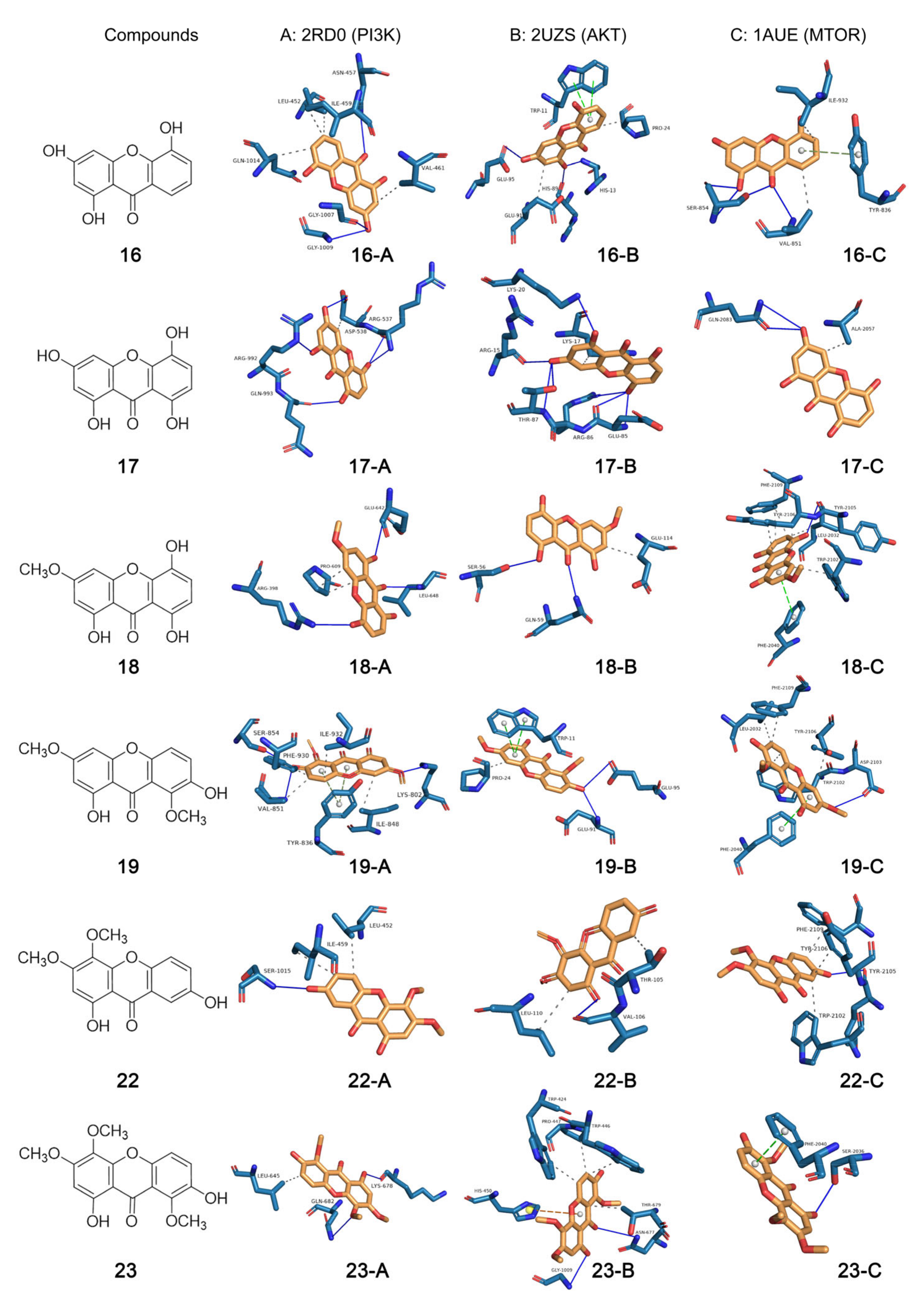
2.8. Analytical Results of Molecular Docking
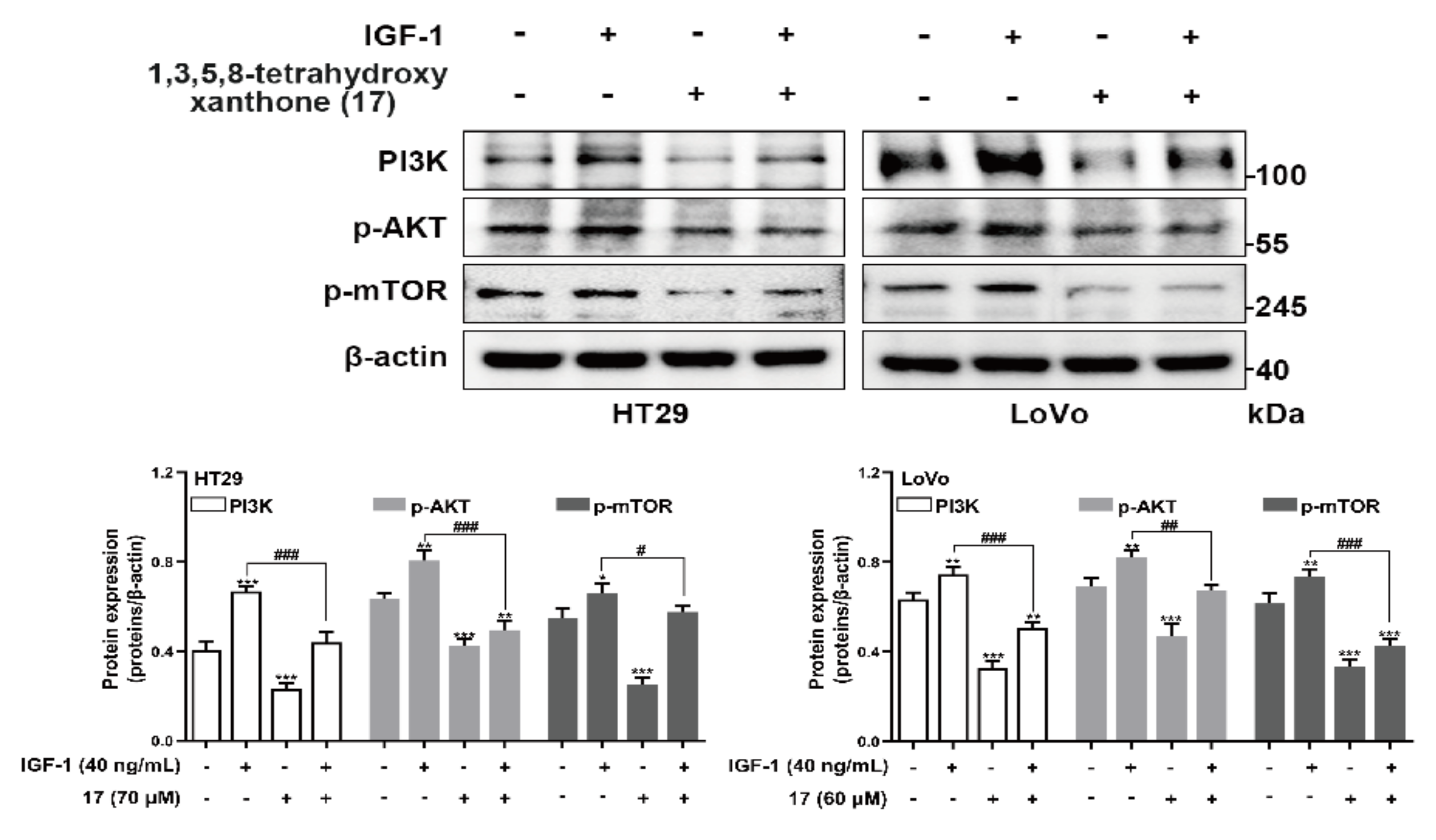
2.9. Results of Western Blotting Assay
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Sample Preparation and Chemical Composition Analysis
4.3. Cell Culture
4.4. Cell Proliferation Assay
4.5. Colony Formation Assay
4.6. Scratch Wound Assay
4.7. Cell Invasion Assay
4.8. Detection of Apoptosis by Flow Cytometry
4.9. Experiment of Network Pharmacology
4.9.1. Prediction of Target Proteins
4.9.2. Construction of Protein-Protein Interaction (PPI) Network
4.9.3. Enrichment Analysis of GO Function and KEGG Pathway
4.9.4. Construction of Component-Target-Pathway Visualization Network
4.10. Molecular Docking Simulation
4.11. Western Blot Assay
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mauri, G.; Sartore-Bianchi, A.; Russo, A.; Marsoni, S.; Bardelli, A.; Siena, S. Early-onset colorectal cancer in young individuals. Mol. Oncol. 2019, 13, 109–131. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; Nogueira, L.; Mariotto, A.; Rowland, J.; Yabroff, K.; Alfano, C.; Jemal, A.; Kramer, J.; Siegel, R. Cancer treatment and survivorship statistics. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Giacchetti, S.; Perpoint, B.; Zidani, R.; Le-Bail, N.; Faggiuolo, R.; Focan, C.; Chollet, P.; Llory, J.F.; Letourneau, Y.; Coudert, B.; et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J. Clin. Oncol. 2000, 18, 136. [Google Scholar] [CrossRef]
- Zhang, N.; Yin, Y.; Xu, S.J.; Chen, W.S. 5-Fluorouracil: Mechanisms of resistance and reversal strategies. Molecules 2008, 13, 1551–1569. [Google Scholar] [CrossRef]
- Estrada, C.C.; Maldonado, A.; Mallipattu, S.K. Therapeutic inhibition of VEGF signaling and associated nephrotoxicities. J. Am. Soc. Nephrol. 2019, 30, 187–200. [Google Scholar] [CrossRef]
- Mousa, L.; Salem, M.E.; Mikhail, S. Biomarkers of angiogenesis in colorectal cancer. Biomark. Cancer 2015, 7, 13–19. [Google Scholar] [CrossRef]
- Li, S.; So, T.; Tang, G.; Tan, H.Y.; Wang, N.; Ng, B.F.L.; Chan, C.K.W.; Yu, E.C.; Feng, Y. Chinese herbal medicine for reducing chemotherapy-associated side-effects in breast cancer patients: A aystematic review and meta-analysis. Front. Oncol. 2020, 10, 599073. [Google Scholar] [CrossRef]
- Qi, F.; Zhao, L.; Zhou, A.; Zhang, B.; Li, A.; Wang, Z.; Han, J. The advantages of using traditional Chinese medicine as an adjunctive therapy in the whole course of cancer treatment instead of only terminal stage of cancer. Biosci. Trends 2015, 9, 16–34. [Google Scholar] [CrossRef]
- Wang, Z.; Qi, F.; Cui, Y.; Zhao, L.; Sun, X.; Tang, W.; Cai, P. An update on Chinese herbal medicines as adjuvant treatment of anticancer therapeutics. Biosci. Trends 2018, 12, 220–239. [Google Scholar] [CrossRef]
- Jia, C.; Xue, J.; Gong, C.; Li, X.; Li, D.; Li, Z.; Hua, H. Chiral resolution and anticancer effect of xanthones from Garcinia paucinervis. Fitoterapia 2018, 127, 220–225. [Google Scholar] [CrossRef]
- Jia, Y.; Xiong, C. Assessing the biological activities of xanthone derivatives from Swertia macrosperma C.B. Clark. Nat. Prod. Res. 2017, 31, 704–706. [Google Scholar] [CrossRef]
- Remali, J.; Sahidin, I.; Aizat, W.M. Xanthone biosynthetic pathway in plants: A review. Front. Plant Sci. 2022, 13, 809497. [Google Scholar] [CrossRef]
- Gunter, N.V.; Teh, S.S.; Lim, Y.M.; Mah, S.H. Natural xanthones and skin inflammatory diseases: Multitargeting mechanisms of action and potential application. Front. Pharmacol. 2020, 11, 594202. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; El-Agamy, D.S.; Abdallah, H.M.; Ahmed, N.; Elkablawy, M.A.; Mohamed, G.A. Protective activity of tovophyllin A, a xanthone isolated from Garcinia mangostana pericarps, against acetaminophen-induced liver damage: Role of Nrf2 activation. Food Funct. 2018, 9, 3291–3300. [Google Scholar] [CrossRef]
- Kurniawan, Y.S.; Priyangga, K.T.A.; Jumina; Pranowo, H.D.; Sholikhah, E.N.; Zulkarnain, A.K.; Fatimi, H.A.; Julianus, J. An update on the anticancer activity of xanthone derivatives: A review. Pharmaceuticals 2021, 14, 1144. [Google Scholar] [CrossRef]
- Santos, C.M.M.; Freitas, M.; Fernandes, E. A comprehensive review on xanthone derivatives as α-glucosidase inhibitors. Eur. J. Med. Chem. 2018, 157, 1460–1479. [Google Scholar] [CrossRef]
- Zhang, K.J.; Gu, Q.L.; Yang, K.; Ming, X.J.; Wang, J.X. Anticarcinogenic effects of α-mangostin: A review. Planta Med. 2017, 83, 188–202. [Google Scholar] [CrossRef]
- Chao, A.C.; Hsu, Y.L.; Liu, C.K.; Kuo, P.L. α-Mangostin, a dietary xanthone, induces autophagic cell death by activating the AMP-activated protein kinase pathway in glioblastoma cells. J. Agric. Food Chem. 2011, 59, 2086–2096. [Google Scholar] [CrossRef]
- Kwak, H.H.; Kim, I.R.; Kim, H.J.; Park, B.S.; Yu, S.B. α-Mangostin induces apoptosis and cell cycle arrest in oral squamous cell carcinoma cell. Evid. Based Complement. Altern. Med. 2016, 2016, 5352412. [Google Scholar] [CrossRef]
- Kwok, Y.; Hurley, L.H. Topoisomerase II site-directed alkylation of DNA by psorospermin and its effect on topoisomerase II-mediated DNA cleavage. J. Biol. Chem. 1998, 273, 33020–33026. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Guan, W.; Hu, X.; Gao, Y.; Li, J. Research advance on chemical constituents and pharmacological effects of Gentianella acuta. Zhongcaoyao 2018, 49, 5468–5476. [Google Scholar]
- Liu, Y.X. Study of the Bioactive Constituents on the Isolated Mouse Intestinal Smooth Muscle Movement from the Whole Plants of Gentianella acuta. Master’s Thesis, Tianjin University of Traditional Chinese Medicine, Tianjin, China, 2017. [Google Scholar]
- Ni, Y.; Liu, M.; Yu, H.; Chen, Y.; Liu, Y.; Chen, S.; Ruan, J.; Da, A.; Zhang, Y.; Wang, T. Desmethylbellidifolin from Gentianella acuta ameliorate TNBS-Induced ulcerative colitis through antispasmodic effect and anti-inflammation. Front. Pharmacol. 2019, 10, 1104. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wu, Y.; Chen, Y.; Liu, M.; Yu, H.; Zhang, Y.; Wang, T. Desmethylbellidifolin attenuates dextran sulfate sodium-induced colitis: Impact on intestinal barrier, intestinal inflammation and gut microbiota. Planta Med. 2022, 88, 559–569. [Google Scholar] [CrossRef]
- Nadeem, M.S.; Kumar, V.; Al-Abbasi, F.A.; Kamal, M.A.; Anwar, F. Risk of colorectal cancer in inflammatory bowel diseases. Semin. Cancer Biol. 2020, 64, 51–60. [Google Scholar] [CrossRef]
- Rogler, G. Chronic ulcerative colitis and colorectal cancer. Cancer Lett. 2014, 345, 235–241. [Google Scholar] [CrossRef]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknæs, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Chen, K.; Lyu, Z.; Zhou, Z.; Li, Y. Current study and application of human colorectal cancer. Zhonghua Weichang Waike Zazhi 2015, 18, 93–97. [Google Scholar]
- Xiong, S.; Mu, T.; Wang, G.; Jiang, X. Mitochondria-mediated apoptosis in mammals. Protein Cell 2014, 5, 737–749. [Google Scholar] [CrossRef]
- Li, M.; Lin, J. The apoptotic pathways and their mechanisms. Guoji Fuchan Kexue Zazhi 2014, 41, 103–107. [Google Scholar]
- Llambi, F.; Moldoveanu, T.; Tait, S.W.G.; Bouchier-Hayes, L.; Temirov, J.; McCormick, L.L.; Dillon, C.P.; Green, D.R. A unified model of mammalian Bcl-2 protein family interactions at the mitochondria. Mol. Cell 2011, 44, 517–531. [Google Scholar] [CrossRef]
- Zhang, S.; Yu, D. Targeting Src family kinases in anti-cancer therapies: Turning promise into triumph. Trends Pharmacol. Sci. 2012, 33, 122–128. [Google Scholar] [CrossRef]
- Pal, I.; Mandal, M. PI3K and Akt as molecular targets for cancer therapy: Current clinical outcomes. Acta Pharmacol. Sin. 2012, 33, 1441–1458. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Asati, V.; Bharti, S.; Mahapatra, D.; Asati, V.; Budhwani, A. Triggering PIK3CA mutations in PI3K/Akt/mTOR axis: Exploration of newer inhibitors and rational preventive strategies. Curr. Pharm. Des. 2016, 22, 6039–6054. [Google Scholar] [CrossRef]
- Zhang, J.; Roberts, T.M.; Shivdasani, R.A. Targeting PI3K signaling as a therapeutic approach for colorectal cancer. Gastroenterology 2011, 141, 50–61. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR Signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef]
- Polivka, J.; Janku, F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol. Ther. 2014, 142, 164–175. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Nabandith, V.; Suzui, M.; Morioka, T.; Kaneshiro, T.; Kinjo, T.; Matsumoto, K.; Akao, Y.; Iinuma, M.; Yoshimi, N. Inhibitory effects of crude alpha-mangostin, a xanthone derivative, on two different categories of colon preneoplastic lesions induced by 1, 2-dimethylhydrazine in the rat. Asian Pac. J. Cancer Prev. 2004, 5, 433–438. [Google Scholar]
- Gao, G.; Bian, Y.; Qian, H.; Yang, M.; Hu, J.; Li, L.; Yu, L.; Liu, B.; Qian, X. Gambogic acid regulates the migration and invasion of colorectal cancer via microRNA-21-mediated activation of phosphatase and tensin homolog. Exp. Ther. Med. 2018, 16, 1758–1765. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Zhan, X.K.; Yu, H.; Xie, G.R.; Wang, Z.Z.; Xiao, W.; Wang, Y.G.; Xiong, F.X.; Hu, J.F.; Yang, L.; et al. An open-labeled, randomized, multicenter phase IIa study of gambogic acid injection for advanced malignant tumors. Chin. Med. J. 2013, 126, 1642–1646. [Google Scholar] [PubMed]
- Afify, S.M.; Oo, A.K.K.; Hassan, G.; Seno, A.; Seno, M. How can we turn the PI3K/AKT/mTOR pathway down? Insights into inhibition and treatment of cancer. Expert Rev. Anticancer Ther. 2021, 21, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Moafian, Z.; Maghrouni, A.; Soltani, A.; Hashemy, S.I. Cross-talk between non-coding RNAs and PI3K/AKT/mTOR pathway in colorectal cancer. Mol. Biol. Rep. 2021, 48, 4797–4811. [Google Scholar] [CrossRef]
- Tewari, D.; Patni, P.; Bishayee, A.; Sah, A.N.; Bishayee, A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Semin. Cancer Biol. 2022, 80, 1–17. [Google Scholar] [CrossRef]
- Popova, N.V.; Jücker, M. The role of mTOR signaling as a therapeutic target in cancer. Int. J. Mol. Sci. 2021, 22, 1743. [Google Scholar] [CrossRef]
- Prasad, S.; Gupta, S.C.; Tyagi, A.K. Reactive oxygen species (ROS) and cancer: Role of antioxidative nutraceuticals. Cancer Lett. 2017, 387, 95–105. [Google Scholar] [CrossRef]
- Irrazabal, T.; Thakur, B.K.; Kang, M.; Malaise, Y.; Streutker, C.; Wong, E.O.Y.; Copeland, J.; Gryfe, R.; Guttman, D.S.; Navarre, W.W.; et al. Limiting oxidative DNA damage reduces microbe–induced colitis–associated colorectal cancer. Nat. Commun. 2020, 11, e1802. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, W.; Zhang, B.; Li, C.; Zhang, X.; Wang, Q.; Wang, Y.; Zhou, Q.; Li, X.; Shen, X.L. Central role of TRAP1 in the ameliorative effect of oleanolic acid on the mitochondrial-mediated and endoplasmic reticulum stress-excitated apoptosis induced by ochratoxin A. Toxicology 2021, 450, 152681. [Google Scholar] [CrossRef]
- TargetNet. Available online: http://targetnet.scbdd.com/ (accessed on 13 December 2022).
- STITCH. Available online: http://stitch.embl.de/ (accessed on 13 December 2022).
- Swiss-Target-Prediction. Available online: http://swisstargetprediction.ch/ (accessed on 15 December 2022).
- SEA. Available online: http://sea.bkslab.org/ (accessed on 15 December 2022).
- GeneCards. Available online: https://www.genecards.org/ (accessed on 16 December 2022).
- Drugbank. Available online: https://go.drugbank.com/ (accessed on 16 December 2022).
- DisGeNET. Available online: https://www.disgenet.org/ (accessed on 16 December 2022).
- Uniprot. Available online: https://www.uniprot.org/ (accessed on 16 December 2022).
- Venny. Available online: https://bioinfogp.cnb.csic.es/tools/venny/ (accessed on 20 December 2022).
- STRING ver. 11.0. Available online: http://string-db.org/ (accessed on 20 December 2022).
- MetaScape. Available online: http://metascape.org (accessed on 21 December 2022).
- Bioinformatics. Available online: http://www.bioinformatics.com.cn/ (accessed on 13 December 2022).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 13 December 2022).
- RCSB. Available online: https://www.rcsb.org/ (accessed on 21 December 2022).
- Tanchuk, V.Y.; Tanin, V.O.; Vovk, A.I.; Poda, G. A new, improved hybrid scoring function for molecular docking and scoring based on AutoDock and AutoDock Vina. Chem. Biol. Drug Des. 2016, 87, 618–625. [Google Scholar] [CrossRef]
- Protein-Ligand Interaction Profiler. Available online: https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index (accessed on 22 December 2022).
- Zhang, Q.; Chen, W.; Zhang, B.; Zhang, Y.; Xiao, Y.; An, Y.; Han, L.; Deng, H.; Yao, S.; Wang, H.; et al. Lonp1 and Sig-1R contribute to the counteraction of ursolic acid against ochratoxin A-induced mitochondrial apoptosis. Food Chem. Toxicol. 2022, 172, e113592. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, W.; Zhang, B.; Gao, Z.; Zhang, Q.; Deng, H.; Han, L.; Shen, X.L. Perfluorooctanoic acid induces hepatocellular endoplasmic reticulum stress and mitochondrial-mediated apoptosis in vitro via endoplasmic reticulum-mitochondria communication. Chem. Biol. Interact. 2022, 354, e109844. [Google Scholar] [CrossRef]

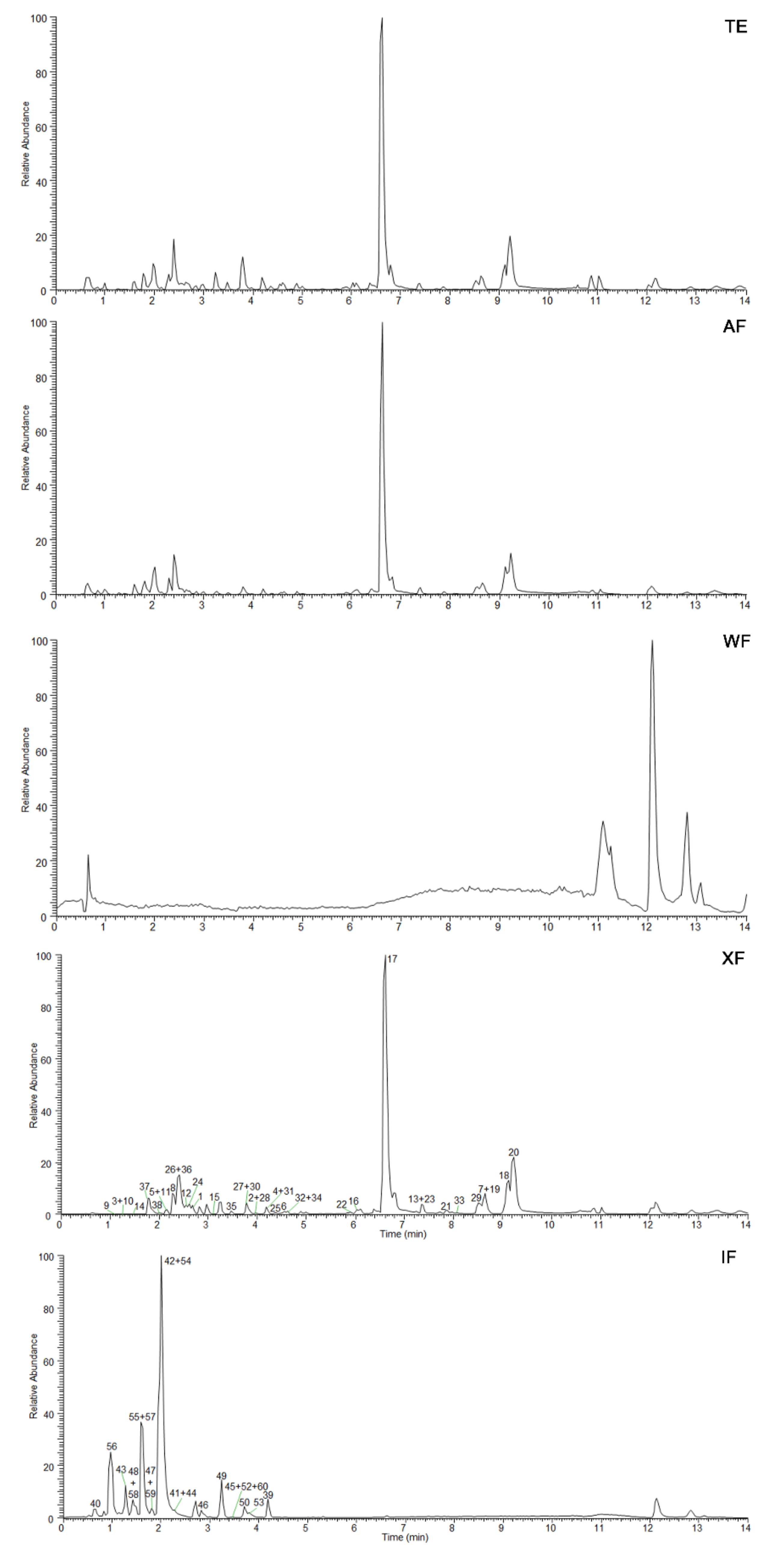

| No. | IC50 (μg/mL) | |
|---|---|---|
| HT29 | LoVo | |
| TE | 386.63 ± 4.19 | 384.20 ± 6.80 |
| PF | 366.63 ± 6.42 | 303.50 ± 6.28 |
| AF | 321.67 ± 5.76 | 256.60 ± 5.32 |
| WF | >500 | >500 |
| IF | >500 | >500 |
| XF | 85.60 ± 2.49 | 76.30 ± 2.16 |
| No. | IC50 (μM) | No. | IC50 (μM) | ||
|---|---|---|---|---|---|
| HT29 | LoVo | HT29 | LoVo | ||
| 1 | >100 | >100 | 20 | >100 | >100 |
| 2 | >100 | >100 | 21 | >100 | >100 |
| 3 | >100 | >100 | 22 | 45.86 ± 1.54 | 35.41 ± 1.45 |
| 4 | >100 | >100 | 23 | 55.79 ± 2.62 | 45.72 ± 2.14 |
| 5 | >100 | >100 | 24 | >100 | >100 |
| 6 | >100 | >100 | 25 | >100 | >100 |
| 7 | >100 | >100 | 26 | >100 | >100 |
| 8 | >100 | >100 | 27 | >100 | >100 |
| 9 | >100 | >100 | 28 | >100 | >100 |
| 10 | >100 | >100 | 29 | >100 | >100 |
| 11 | >100 | >100 | 30 | >100 | >100 |
| 12 | >100 | >100 | 31 | >100 | >100 |
| 13 | >100 | >100 | 32 | >100 | >100 |
| 14 | >100 | 37.42 ± 0.70 | 33 | >100 | 46.82 ± 2.79 |
| 15 | >100 | >100 | 34 | >100 | >100 |
| 16 | 64.31 ± 2.90 | 75.93 ± 2.73 | 35 | 88.65 ± 5.17 | >100 |
| 17 | 83.42 ± 3.88 | 69.87 ± 3.71 | 36 | >100 | >100 |
| 18 | 90.21 ± 4.57 | 85.32 ± 3.05 | 37 | >100 | >100 |
| 19 | 87.93 ± 5.03 | 88.64 ± 4.32 | 38 | >100 | >100 |
| Ligand (Compound) | Receptor PDB ID (Protein) | Affinity (kcal/mol) | Binding Site |
|---|---|---|---|
| 16 | 2RD0 (PIK3CA) | −7.8 | ASN-457, LEU-452, ILE-459, VAL-461, GLN1014, GLY-1007, GLY-1009 |
| 2UZS (Akt1) | −6.4 | TRP-11, PRO-24, GLU-95, HIS-89, HIS-13, GLU-91 | |
| 1AUE (mTOR) | −6.6 | ILE-932, TYR-836, VAL-851, SER-854 | |
| 17 | 2RD0 (PIK3CA) | −7.8 | ARG-992, GLN-993, ASP-538, ARG-537 |
| 2UZS (Akt1) | −6.2 | LYS-20, LYS-17, ARG-15, THR-87, ARG-86, GLU-85 | |
| 1AUE (mTOR) | −6.2 | GLN-2083, ALA-2057 | |
| 18 | 2RD0 (PIK3CA) | −8.2 | ARG-398, PRO-609, GLU-642, LEU-648 |
| 2UZS (Akt1) | −5.6 | SER-56, GLN-59, GLU-114, | |
| 1AUE (mTOR) | −8.2 | PHE-2019, TYR-2105, TYR-2106, LEU-2032, TRP-2102 | |
| 19 | 2RD0 (PIK3CA) | −7.9 | SER-854, PHE-930, VAL-851, ILE-932, TYR-836, ILE-848, LYS-802 |
| 2UZS (Akt1) | −6.3 | PRO-24, TRP-11, GLU-95, GLU-91 | |
| 1AUE (mTOR) | −8.0 | PHE-2019, TYR-2106, LEU-2032, TRP-2102, PHE-2040, ASP-2103 | |
| 22 | 2RD0 (PIK3CA) | −7.7 | SER-1015, ILE-459, LEU-452 |
| 2UZS (Akt1) | −5.9 | LEU-110, VAL-106, THR-105 | |
| 1AUE (mTOR) | −7.7 | PHE-2019, TYR-2106, TYR-2105, TRP-2102 | |
| 23 | 2RD0 (PIK3CA) | −7.3 | LEU-645, LYS-678, GLN-682 |
| 2UZS (Akt1) | −5.6 | TRP-424, TRP-446, PRO-447, THR-679, ASN-677, GLY-1009, HIS-450 | |
| 1AUE (mTOR) | −7.3 | PHE-2040, SER-2036 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, M.-Q.; Ruan, J.-Y.; Li, H.-M.; Yang, D.-S.; Liu, Y.-X.; Hao, M.-M.; Yu, H.-Y.; Zhang, Y.; Wang, T. Xanthones from Gentianella acuta (Michx.) Hulten Ameliorate Colorectal Carcinoma via the PI3K/Akt/mTOR Signaling Pathway. Int. J. Mol. Sci. 2023, 24, 2279. https://doi.org/10.3390/ijms24032279
Lu M-Q, Ruan J-Y, Li H-M, Yang D-S, Liu Y-X, Hao M-M, Yu H-Y, Zhang Y, Wang T. Xanthones from Gentianella acuta (Michx.) Hulten Ameliorate Colorectal Carcinoma via the PI3K/Akt/mTOR Signaling Pathway. International Journal of Molecular Sciences. 2023; 24(3):2279. https://doi.org/10.3390/ijms24032279
Chicago/Turabian StyleLu, Meng-Qi, Jing-Ya Ruan, Hui-Min Li, Ding-Shan Yang, Yan-Xia Liu, Mi-Mi Hao, Hai-Yang Yu, Yi Zhang, and Tao Wang. 2023. "Xanthones from Gentianella acuta (Michx.) Hulten Ameliorate Colorectal Carcinoma via the PI3K/Akt/mTOR Signaling Pathway" International Journal of Molecular Sciences 24, no. 3: 2279. https://doi.org/10.3390/ijms24032279
APA StyleLu, M.-Q., Ruan, J.-Y., Li, H.-M., Yang, D.-S., Liu, Y.-X., Hao, M.-M., Yu, H.-Y., Zhang, Y., & Wang, T. (2023). Xanthones from Gentianella acuta (Michx.) Hulten Ameliorate Colorectal Carcinoma via the PI3K/Akt/mTOR Signaling Pathway. International Journal of Molecular Sciences, 24(3), 2279. https://doi.org/10.3390/ijms24032279






