Impact of Rhamnolipids (RLs), Natural Defense Elicitors, on Shoot and Root Proteomes of Brassica napus by a Tandem Mass Tags (TMTs) Labeling Approach
Abstract
:1. Introduction
2. Results
2.1. RLs Significantly Modify Protein Abundance in Rapeseed Shoots and Roots upon Elicitation
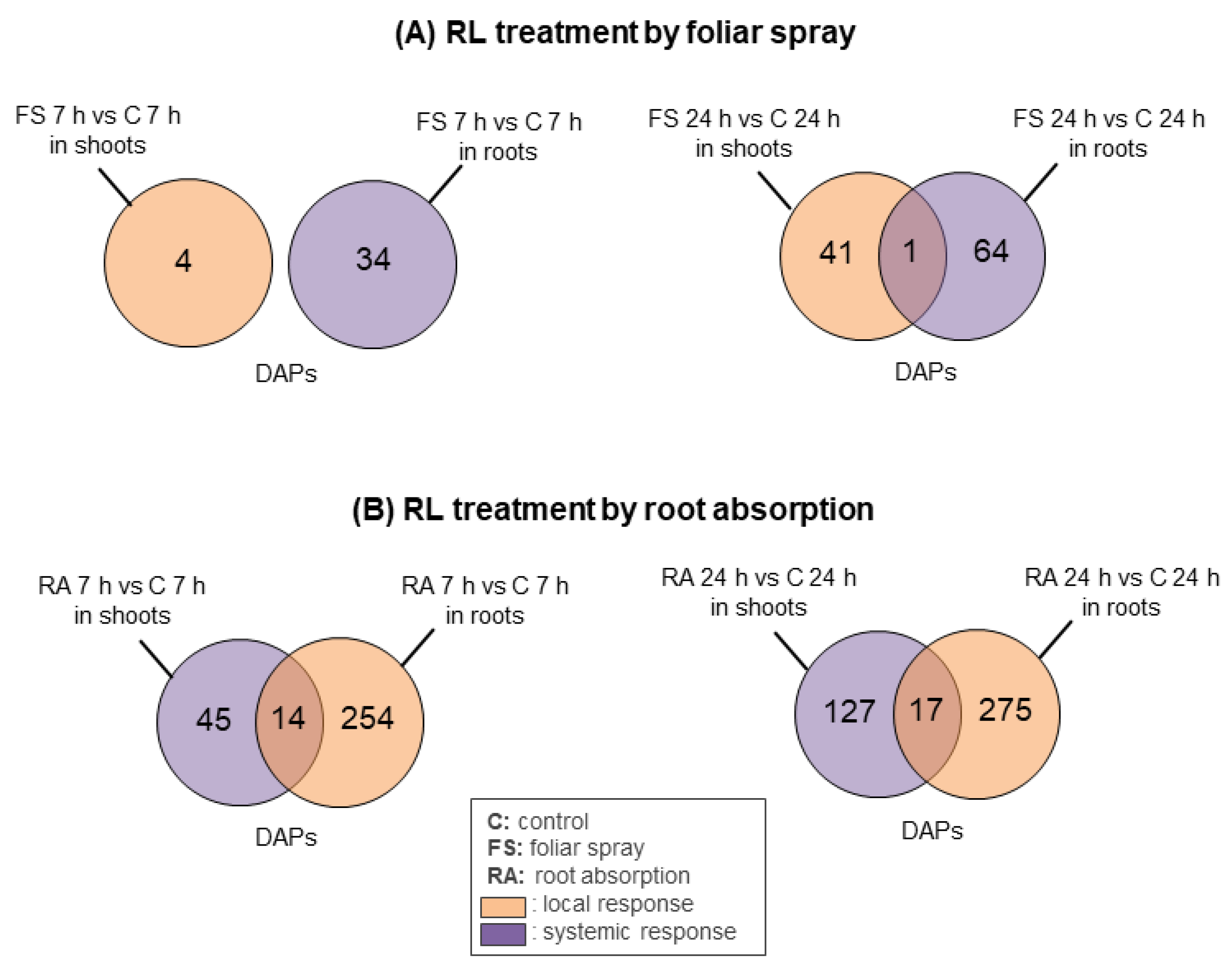
2.2. Mode of RL Application Differently Influences Protein Accumulation at the Local and Systemic Level in Rapeseed
2.3. Plant Defense/Stress, Transport and Secondary Metabolism Proteins Are DAPs Most Widely Represented in Shoots and Roots of Rapeseed upon RL Treatment at T 7 h
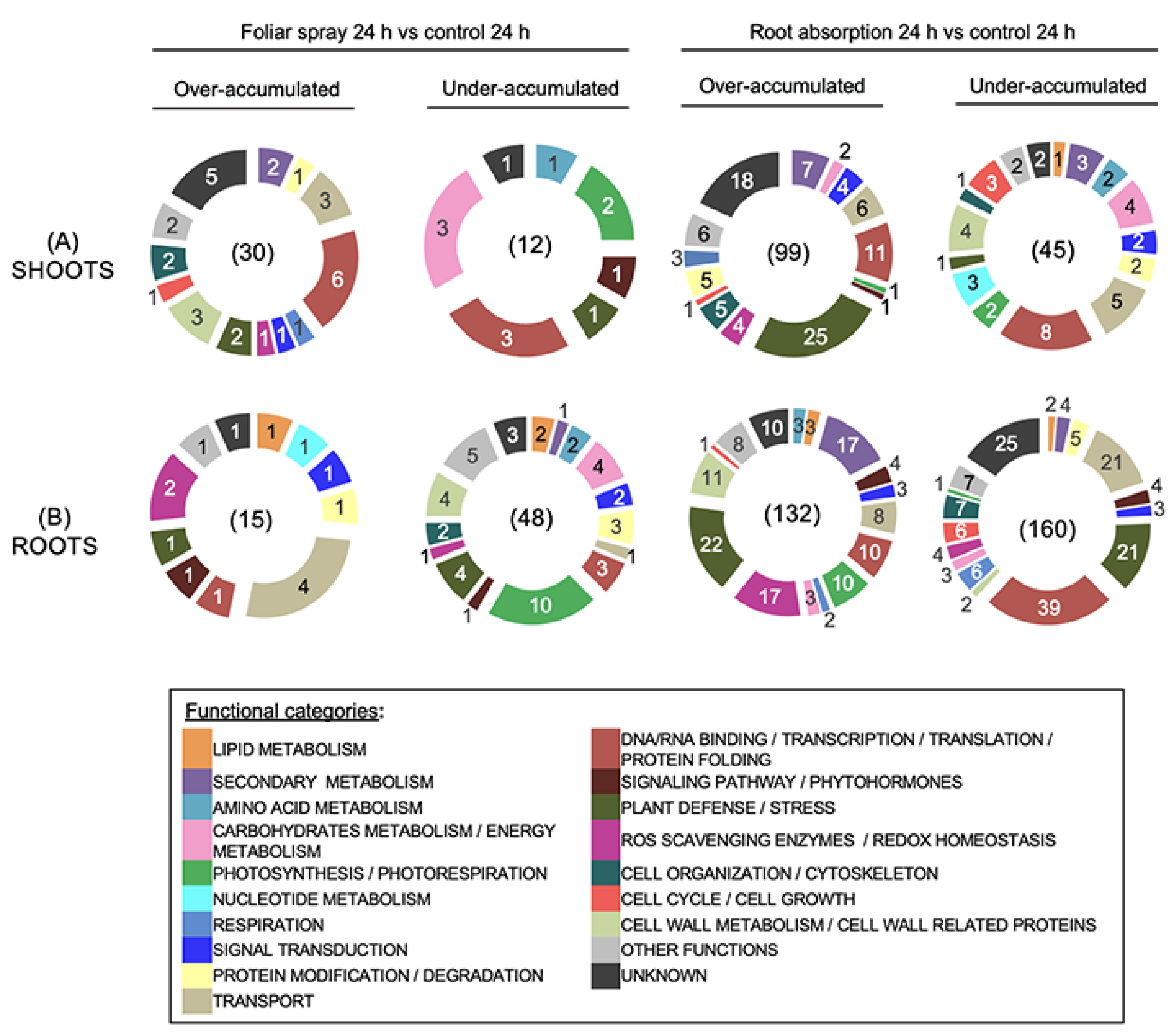
2.4. RLs Modify More Diverse Functional Protein Categories in Rapeseed upon Elicitation at 24 h
3. Discussion
4. Materials and Methods
4.1. Biological Materials and Culture Conditions
4.2. Preparation of RL Solutions and Applications
4.3. Proteomic Analysis
4.4. Functional Classification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Paul, E.; McRoberts, N.; Nelson, A. The Global Burden of Pathogens and Pests on Major Food Crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Burketova, L.; Trda, L.; Ott, P.G.; Valentova, O. Bio-Based Resistance Inducers for Sustainable Plant Protection against Pathogens. Biotechnol. Adv. 2015, 33, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (Microbe-Associated Molecular Pattern) Triggered Immunity in Plants. Front. Plant Sci. 2013, 4, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.E.; Mesarich, C.H.; Thomma, B.P. Understanding Plant Immunity as a Surveillance System to Detect Invasion. Phytopathol. 2015, 53, 536–541. [Google Scholar] [CrossRef]
- Monnier, N.; Sarazin, C.; Rippa, S. Transcriptomic Dataset from Arabidopsis Thaliana Seedlings in Response to Pseudomonas Aeruginosa Mono-Rhamnolipids. Data Br. 2021, 38, 107397. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–407. [Google Scholar] [CrossRef]
- Fischer, T.; Byerlee, D.; Edmeades, G. Crop Yields and Global Food Security: Will Copyright Act 1968 Yield Increase Continue to Feed the World? Aust. Cent. Int. Agric. Res. 2014, 634. [Google Scholar]
- Singh, A.K.; Cameotra, S.S. Rhamnolipids Production by Multi-Metal-Resistant and Plant-Growth-Promoting Rhizobacteria. Appl. Biochem. Biotechnol. 2013, 170, 1038–1056. [Google Scholar] [CrossRef]
- Verma, S.S.; Yajima, W.R.; Rahman, M.H.; Shah, S.; Liu, J.J.; Ekramoddoullah, A.K.M.; Kav, N.N.V. A Cysteine-Rich Antimicrobial Peptide from Pinus Monticola (PmAMP1) Confers Resistance to Multiple Fungal Pathogens in Canola (Brassica Napus). Plant Mol. Biol. 2012, 79, 61–74. [Google Scholar] [CrossRef]
- Chopra, A.; Bobate, S.; Rahi, P.; Banpurkar, A.; Mazumder, P.B.; Satpute, S. Pseudomonas Aeruginosa RTE4: A Tea Rhizobacterium With Potential for Plant Growth Promotion and Biosurfactant Production. Front. Bioeng. Biotechnol. 2020, 8, 1–14. [Google Scholar] [CrossRef]
- Adesemoye, A.O.; Kloepper, J.W. Plant-Microbes Interactions in Enhanced Fertilizer-Use Efficiency. Appl. Microbiol. Biotechnol. 2009, 85, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Benhamou, N.; Rey, P. Stimulateurs Des Défenses Naturelles Des Plantes: Une Nouvelle Stratégie Phytosanitaire Dans Un Contexte d ’ Écoproduction Durable. I. Principes de La Résistance Induite Elicitors of Natural Plant Defense Mechanisms: A New Management Strategy in the C. Phytoprotection 2012, 92, 1. [Google Scholar] [CrossRef] [Green Version]
- Crouzet, J.; Arguelles-Arias, A.; Dhondt-Cordelier, S.; Cordelier, S.; Pršić, J.; Hoff, G.; Mazeyrat-Gourbeyre, F.; Baillieul, F.; Clément, C.; Ongena, M.; et al. Biosurfactants in Plant Protection Against Diseases: Rhamnolipids and Lipopeptides Case Study. Front. Bioeng. Biotechnol. 2020, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Toribio, J.; Escalante, A.E.; Soberón-Chávez, G. Rhamnolipids: Production in Bacteria Other than Pseudomonas Aeruginosa. Eur. J. Lipid Sci. Technol. 2010, 112, 1082–1087. [Google Scholar] [CrossRef]
- Robineau, M.; Le Guenic, S.; Sanchez, L.; Chaveriat, L.; Lequart, V.; Joly, N.; Calonne, M.; Jacquard, C.; Declerck, S.; Martin, P.; et al. Synthetic Mono-Rhamnolipids Display Direct Antifungal Effects and Trigger an Innate Immune Response in Tomato against Botrytis Cinerea. Molecules 2020, 25, 3108. [Google Scholar] [CrossRef] [PubMed]
- Hogan, D.E.; Tian, F.; Malm, S.W.; Olivares, C.; Palos Pacheco, R.; Simonich, M.T.; Hunjan, A.S.; Tanguay, R.L.; Klimecki, W.T.; Polt, R.; et al. Biodegradability and Toxicity of Monorhamnolipid Biosurfactant Diastereomers. J. Hazard. Mater. 2019, 364, 600–607. [Google Scholar] [CrossRef]
- Sekhon Randhawa, K.K.; Rahman, P.K.S.M.; Neubauer, P. Rhamnolipid Biosurfactants-Past, Present, and Future Scenario of Global Market. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Rikalović, M.G.; Vrvić, M.M.; Karadžić, I.M. Rhamnolipid Biosurfactant from Pseudomonas Aeruginosa - From Discovery to Application in Contemporary Technology. J. Serbian Chem. Soc. 2015, 80, 279–304. [Google Scholar] [CrossRef]
- Vatsa, P.; Sanchez, L.; Clement, C.; Baillieul, F.; Dorey, S. Rhamnolipid Biosurfactants as New Players in Animal and Plant Defense against Microbes. Int. J. Mol. Sci. 2010, 11, 5095–5108. [Google Scholar] [CrossRef] [Green Version]
- Varnier, A.L.; Sanchez, L.; Vatsa, P.; Boudesocque, L.; Garcia-Brugger, A.; Rabenoelina, F.; Sorokin, A.; Renault, J.H.; Kauffmann, S.; Pugin, A.; et al. Bacterial Rhamnolipids Are Novel MAMPs Conferring Resistance to Botrytis Cinerea in Grapevine. Plant Cell Environ. 2009, 32, 178–193. [Google Scholar] [CrossRef]
- Sanchez, L.; Courteaux, B.; Hubert, J.; Kauffmann, S.; Renault, J.H.; Clément, C.; Baillieul, F.; Dorey, S. Rhamnolipids Elicit Defense Responses and Induce Disease Resistance against Biotrophic, Hemibiotrophic, and Necrotrophic Pathogens That Require Different Signaling Pathways in Arabidopsis and Highlight a Central Role for Salicylic Acid. Plant Physiol. 2012, 160, 1630–1641. [Google Scholar] [CrossRef] [PubMed]
- Monnier, N.; Furlan, A.; Botcazon, C.; Dahi, A.; Mongelard, G.; Cordelier, S.; Clément, C.; Dorey, S.; Sarazin, C.; Rippa, S. Rhamnolipids from Pseudomonas Aeruginosa Are Elicitors Triggering Brassica Napus Protection against Botrytis Cinerea without Physiological Disorders. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monnier, N.; Cordier, M.; Dahi, A.; Santoni, V.; Guénin, S.; Clément, C.; Sarazin, C.; Penaud, A.; Dorey, S.; Cordelier, S.; et al. Semipurified Rhamnolipid Mixes Protect Brassica Napus Against Leptosphaeria Maculans Early Infections. Phytopathology 2020, 110, 834–842. [Google Scholar] [CrossRef]
- Monnier, N.; Furlan, A.L.; Buchoux, S.; Deleu, M.; Dauchez, M.; Rippa, S.; Sarazin, C. Exploring the Dual Interaction of Natural Rhamnolipids with Plant and Fungal Biomimetic Plasma Membranes through Biophysical Studies. Int. J. Mol. Sci. Artic. 2019, 20, 1009. [Google Scholar] [CrossRef] [Green Version]
- Schellenberger, R.; Touchard, M.; Clément, C.; Baillieul, F.; Cordelier, S.; Crouzet, J.; Dorey, S. Apoplastic Invasion Patterns Triggering Plant Immunity: Plasma Membrane Sensing at the Frontline. Mol. Plant Pathol. 2019, 20, 1602–1616. [Google Scholar] [CrossRef]
- Botcazon, C.; Bergia, T.; Lecouturier, D.; Dupuis, C.; Rochex, A.; Acket, S.; Nicot, P.; Leclère, V.; Sarazin, C.; Rippa, S. Rhamnolipids and Fengycins, Very Promising Amphiphilic Antifungal Compounds from Bacteria Secretomes, Act on Sclerotiniaceae Fungi through Different Mechanisms. Front. Microbiol. 2022, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.; Aranda, F.J.; Teruel, J.A.; Espuny, M.J.; Marqués, A.; Manresa, Á.; Ortiz, A. Permeabilization of Biological and Artificial Membranes by a Bacterial Dirhamnolipid Produced by Pseudomonas Aeruginosa. J. Colloid Interface Sci. 2010, 341, 240–247. [Google Scholar] [CrossRef]
- Yan, F.; Xu, S.; Guo, J.; Chen, Q.; Meng, Q.; Zheng, X. Biocontrol of Post-Harvest Alternaria Alternata Decay of Cherry Tomatoes with Rhamnolipids and Possible Mechanisms of Action. J. Sci. Food Agric. 2015, 95, 1469–1474. [Google Scholar] [CrossRef]
- Yan, F.; Hu, H.; Lu, L.; Zheng, X. Rhamnolipids Induce Oxidative Stress Responses in Cherry Tomato Fruit to Alternaria Alternata. Pest Manag. Sci. 2016, 72, 1500–1507. [Google Scholar] [CrossRef]
- Sharma, A.; Jansen, R.; Nimtz, M.; Johri, B.N.; Wray, V. Rhamnolipids from the Rhizosphere Bacterium Pseudomonas Sp. GRP3 That Reduces Damping-off Disease in Chilli and Tomato Nurseries. J. Nat. Prod. 2007, 70, 941–947. [Google Scholar] [CrossRef]
- Takemoto, J.Y.; Bensaci, M.; de Lucca, A.J.; Cleveland, T.E.; Gandhi, N.R.; Skebba, V.P. Inhibition of Fungi from Diseased Grape by Syringomycin E-Rhamnolipid Mixture. Am. J. Enol. Vitic. 2010, 61, 120–124. [Google Scholar] [CrossRef]
- Chen, Y.; Ren, X.; Zheng, Y.; Zhou, X.; Huang, L.; Yan, L.; Jiao, Y.; Chen, W.; Huang, S.; Wan, L.; et al. Genetic Mapping of Yield Traits Using RIL Population Derived from Fuchuan Dahuasheng and ICG6375 of Peanut (Arachis hypogaea L.). Mol. Breed. 2017, 37. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Shao, B.; Long, X.; Yao, Y.; Meng, Q. Foliar Penetration Enhanced by Biosurfactant Rhamnolipid. Colloids Surf. B Biointerfaces 2016, 145, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Chivasa, S.; Hamilton, J.M.; Pringle, R.S.; Ndimba, B.K.; Simon, W.J.; Lindsey, K.; Slabas, A.R. Proteomic Analysis of Differentially Expressed Proteins in Fungal Elicitor-Treated Arabidopsis Cell Cultures. J. Exp. Bot. 2006, 57, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Lemaître-Guillier, C.; Hovasse, A.; Schaeffer-Reiss, C.; Recorbet, G.; Poinssot, B.; Trouvelot, S.; Daire, X.; Adrian, M.; Héloir, M.C. Proteomics towards the Understanding of Elicitor Induced Resistance of Grapevine against Downy Mildew. J. Proteomics 2017, 156, 113–125. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, S.; Liu, K.; Wang, S.; Huang, L.; Guo, L. Proteomics: A Powerful Tool to Study Plant Responses to Biotic Stress. Plant Methods 2019, 15, 1–20. [Google Scholar] [CrossRef]
- Bassal, M.; Abukhalaf, M.; Majovsky, P.; Thieme, D.; Herr, T.; Ayash, M.; Tabassum, N.; Al Shweiki, M.R.; Proksch, C.; Hmedat, A.; et al. Reshaping of the Arabidopsis Thaliana Proteome Landscape and Co-Regulation of Proteins in Development and Immunity. Mol. Plant 2020, 13, 1709–1732. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van Der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [Green Version]
- Czékus, Z.; Kukri, A.; Hamow, K.Á.; Szalai, G.; Tari, I.; Ördög, A.; Poór, P. Activation of Local and Systemic Defence Responses by Flg22 Is Dependent on Daytime and Ethylene in Intact Tomato Plants. Int. J. Mol. Sci. 2021, 22, 8354. [Google Scholar] [CrossRef]
- Ceron-Garcia, A.; Vargas-Arispuro, I.; Aispuro-Hernandez, E.; Martinez-Tellez, M.A. Oligoglucan Elicitor Effects During Plant Oxidative Stress. In Cell Metabolism-Cell Homeostasis and Stress Response; Bululya, P., Ed.; BoD–Books on Demand: Croatia, Republic of Croatia, 2012; pp. 1–12. ISBN 978-953-51-5201-9. [Google Scholar]
- Pršić, J.; Ongena, M. Elicitors of Plant Immunity Triggered by Beneficial Bacteria. Front. Plant Sci. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Ongena, M.; Jourdan, E.; Adam, A.; Paquot, M.; Brans, A.; Joris, B.; Arpigny, J.L.; Thonart, P. Surfactin and Fengycin Lipopeptides of Bacillus Subtilis as Elicitors of Induced Systemic Resistance in Plants. Environ. Microbiol. 2007, 9, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Green, S.K.; Schroth, M.N.; Cho, J.J.; Kominos, S.D.; Vitanza-Jack, V.B. Agricultural Plants and Soil as a Reservoir for Pseudomonas Aeruginosa. Appl. Microbiol. 1974, 28, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Chuberre, C.; Plancot, B.; Driouich, A.; Moore, J.P.; Bardor, M.; Gügi, B.; Vicré, M. Plant Immunity Is Compartmentalized and Specialized in Roots. Front. Plant Sci. 2018, 871, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denoux, C.; Galletti, R.; Mammarella, N.; Gopalan, S.; Werck, D.; De Lorenzo, G.; Ferrari, S.; Ausubel, F.M.; Dewdney, J. Activation of Defense Response Pathways by OGs and Flg22 Elicitors in Arabidopsis Seedlings. Mol Plant. 2008, 1, 423–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platel, R.; Lucau-Danila, A.; Baltenweck, R.; Maia-Grondard, A.; Chaveriat, L.; Magnin-Robert, M.; Randoux, B.; Trapet, P.; Halama, P.; Martin, P.; et al. Bioinspired Rhamnolipid Protects Wheat Against Zymoseptoria Tritici Through Mainly Direct Antifungal Activity and Without Major Impact on Leaf Physiology. Front. Plant Sci. 2022, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wen, K.S.; Ruan, X.; Zhao, Y.X.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [Green Version]
- Kessler, A.; Kalske, A. Plant Secondary Metabolite Diversity and Species Interactions. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy1 [OPEN]. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Zeiss, D.R.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Altered Metabolomic States Elicited by Flg22 and FlgII-28 in Solanum Lycopersicum: Intracellular Perturbations and Metabolite Defenses. BMC Plant Biol. 2021, 21, 1–18. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Bischoff, K. Glucosinolates. Nutraceuticals 2021, 903–909. [Google Scholar]
- Variyar, P.S.; Banerjee, A.; Akkarakaran, J.J.; Suprasanna, P. Role of Glucosinolates in Plant Stress Tolerance; Elsevier Inc.: Amsterdam, The Netherlands, 2014; Volume 1, ISSN 9780128010884. [Google Scholar]
- Kimura, S.; Waszczak, C.; Hunter, K.; Wrzaczek, M. Bound by Fate: The Role of Reactive Oxygen Species in Receptor-like Kinase Signaling. Plant Cell 2017, 29, 638–654. [Google Scholar] [CrossRef]
- Zehra, A.; Raytekar, N.A.; Meena, M.; Swapnil, P. Efficiency of Microbial Bio-Agents as Elicitors in Plant Defense Mechanism under Biotic Stress: A Review. Curr. Res. Microb. Sci. 2021, 2, 100054. [Google Scholar] [CrossRef]
- Lebel, E.; Heifetz, P.; Thorne, L.; Uknes, S.; Ryals, J.; Ward, E. Functional Analysis of Regulatory Sequences Controlling PR-1 Gene Expression in Arabidopsis. Plant J. 1998, 16, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Corina Vlot, A.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a Multifaceted Hormone to Combat Disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leschevin, M.; Marcelo, P.; Ismael, M.; San-Clemente, H.; Jamet, E.; Rayon, C.; Pageau, K. A Tandem Mass Tags (TMTs) Labeling Approach Highlights Differences between the Shoot Proteome of Two Arabidopsis Thaliana Ecotypes, Col-0 and Ws. Proteomics 2021, 21, 2000293. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pierre, E.; Marcelo, P.; Croutte, A.; Dauvé, M.; Bouton, S.; Rippa, S.; Pageau, K. Impact of Rhamnolipids (RLs), Natural Defense Elicitors, on Shoot and Root Proteomes of Brassica napus by a Tandem Mass Tags (TMTs) Labeling Approach. Int. J. Mol. Sci. 2023, 24, 2390. https://doi.org/10.3390/ijms24032390
Pierre E, Marcelo P, Croutte A, Dauvé M, Bouton S, Rippa S, Pageau K. Impact of Rhamnolipids (RLs), Natural Defense Elicitors, on Shoot and Root Proteomes of Brassica napus by a Tandem Mass Tags (TMTs) Labeling Approach. International Journal of Molecular Sciences. 2023; 24(3):2390. https://doi.org/10.3390/ijms24032390
Chicago/Turabian StylePierre, Elise, Paulo Marcelo, Antoine Croutte, Morgane Dauvé, Sophie Bouton, Sonia Rippa, and Karine Pageau. 2023. "Impact of Rhamnolipids (RLs), Natural Defense Elicitors, on Shoot and Root Proteomes of Brassica napus by a Tandem Mass Tags (TMTs) Labeling Approach" International Journal of Molecular Sciences 24, no. 3: 2390. https://doi.org/10.3390/ijms24032390
APA StylePierre, E., Marcelo, P., Croutte, A., Dauvé, M., Bouton, S., Rippa, S., & Pageau, K. (2023). Impact of Rhamnolipids (RLs), Natural Defense Elicitors, on Shoot and Root Proteomes of Brassica napus by a Tandem Mass Tags (TMTs) Labeling Approach. International Journal of Molecular Sciences, 24(3), 2390. https://doi.org/10.3390/ijms24032390






