A Study on Symbiotic Systems of Cicadas Provides New Insights into Distribution of Microbial Symbionts and Improves Fluorescence In Situ Hybridization Technique
Abstract
1. Introduction
2. Results
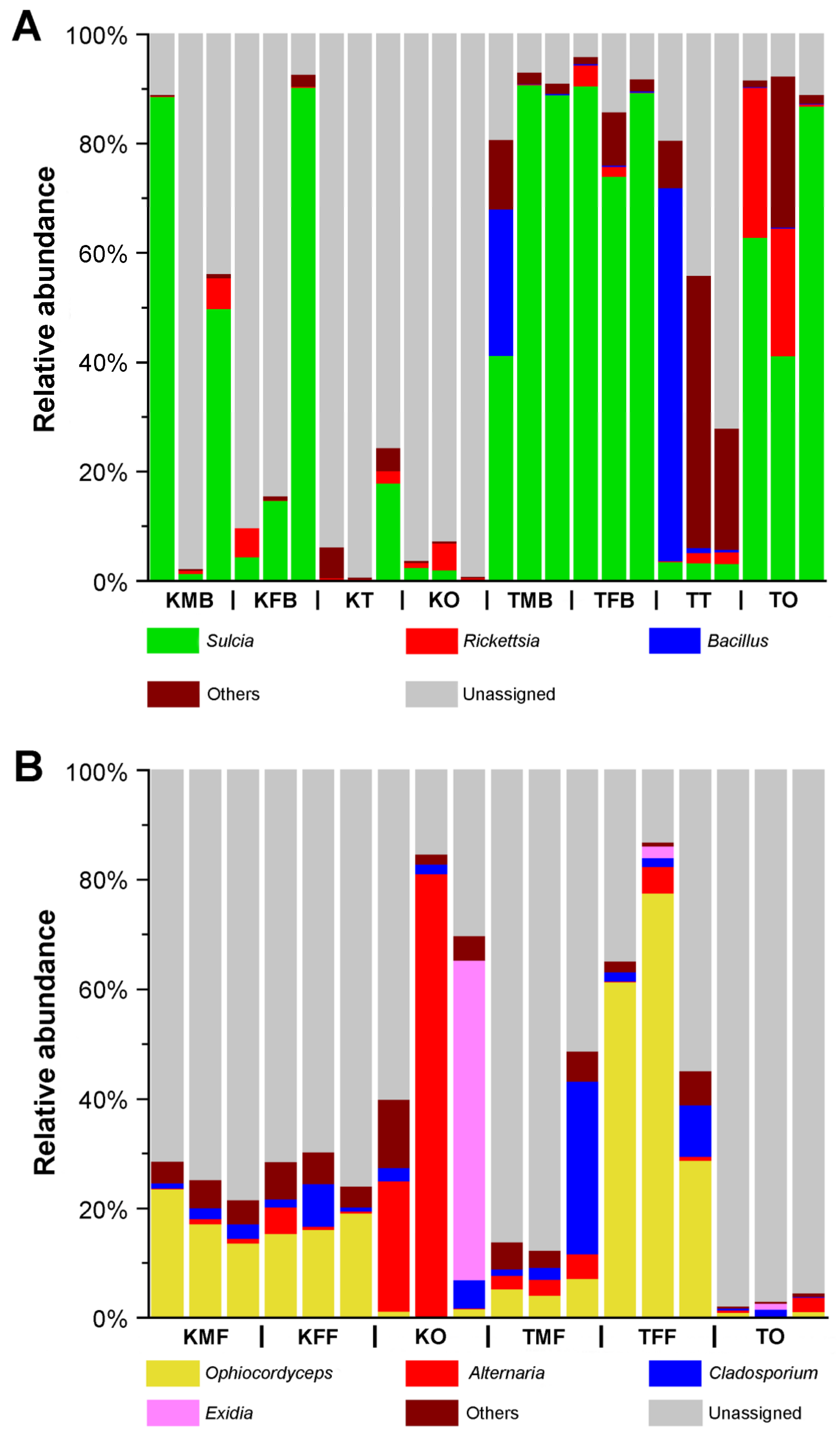
2.1. Microbial Composition in the Reproductive Organs, Fat Bodies, and Bacteriomes of K. caelatata and Ta. sp.
2.2. Diagnostic Pcr of Dominant Symbionts Obtained from K. caelatata and Ta. Sp.
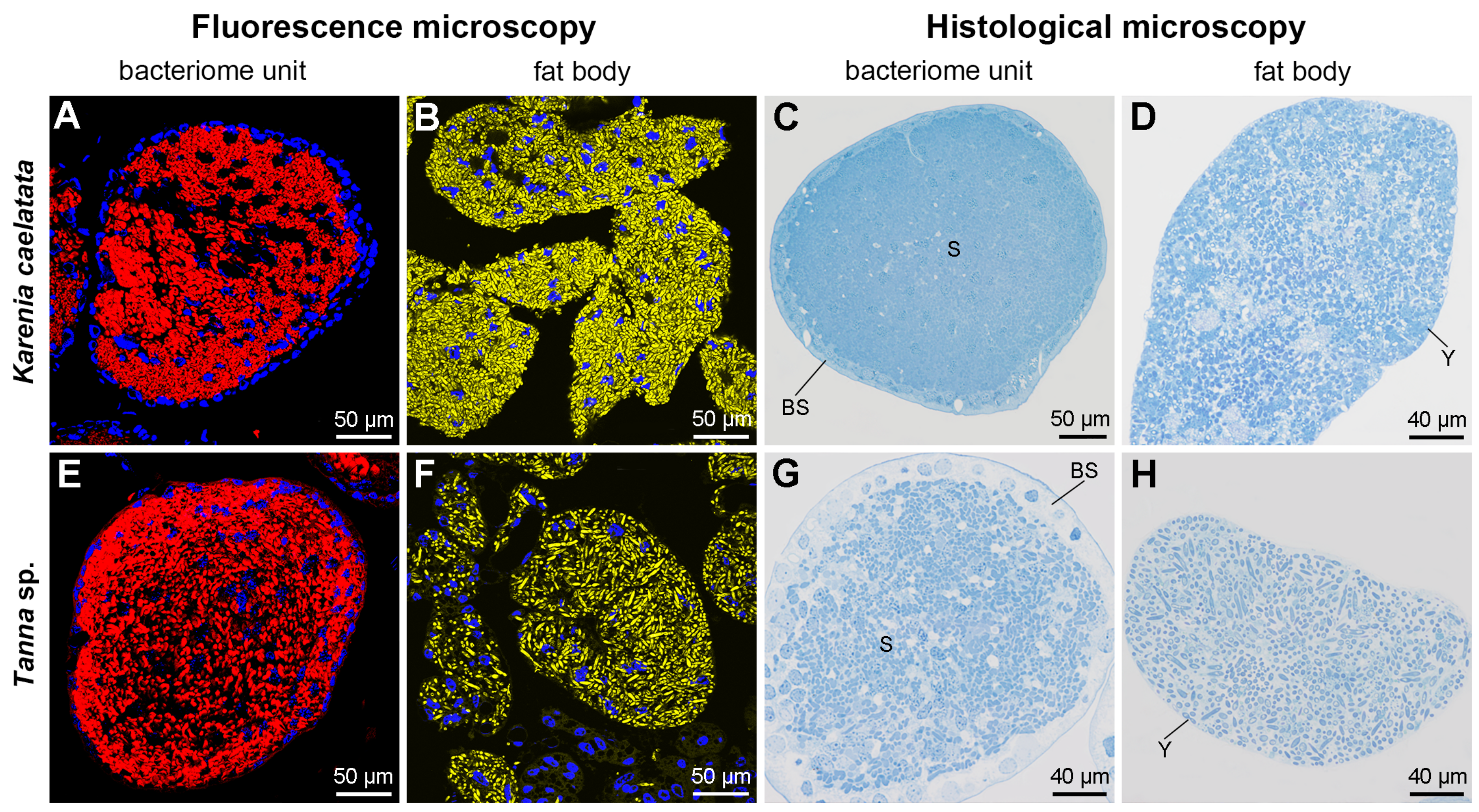
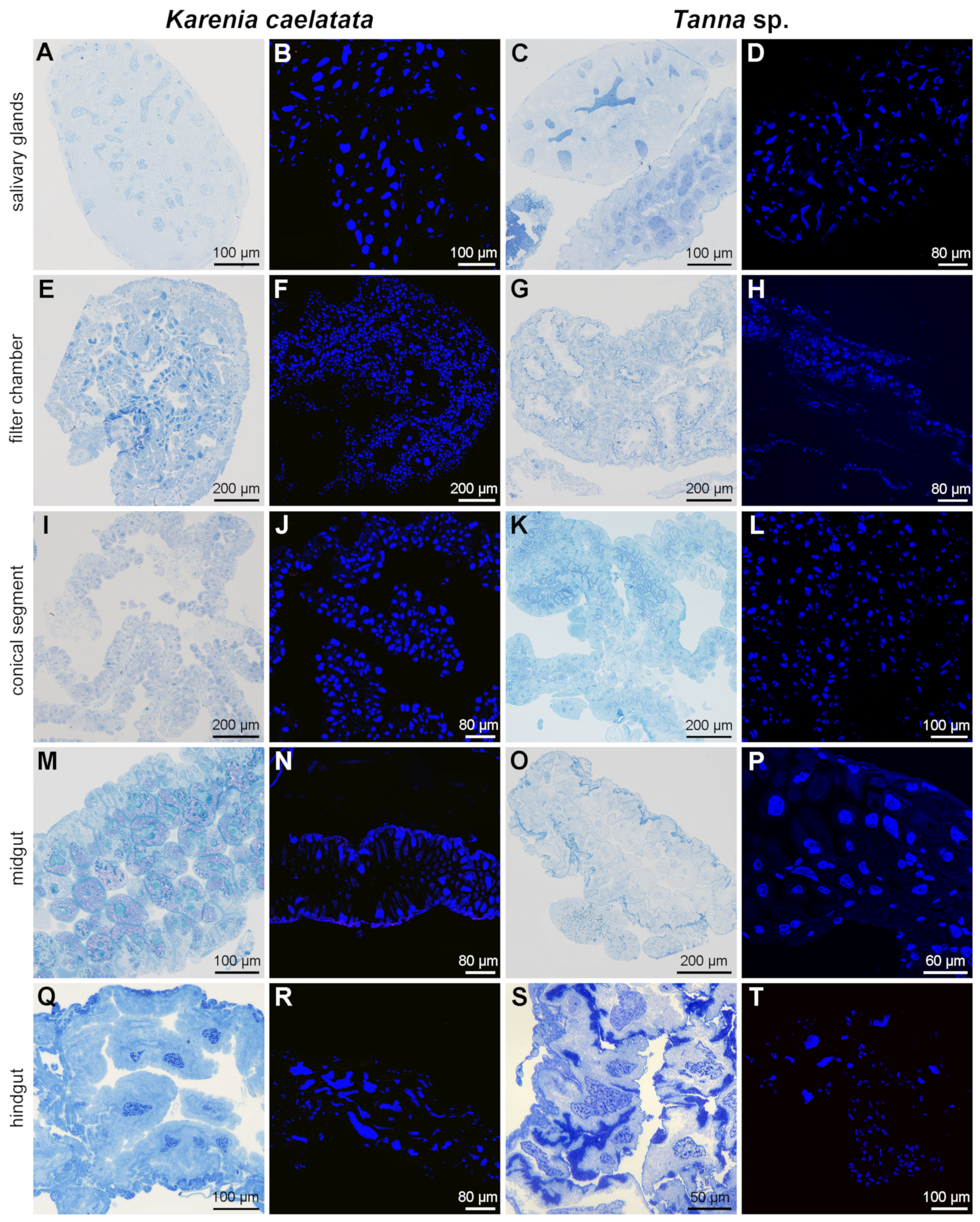
2.3. Distribution of Sulcia and YLS in Salivary Glands, Gut Tissues, Reproductive Organs, Bacteriomes, and Fat Bodies
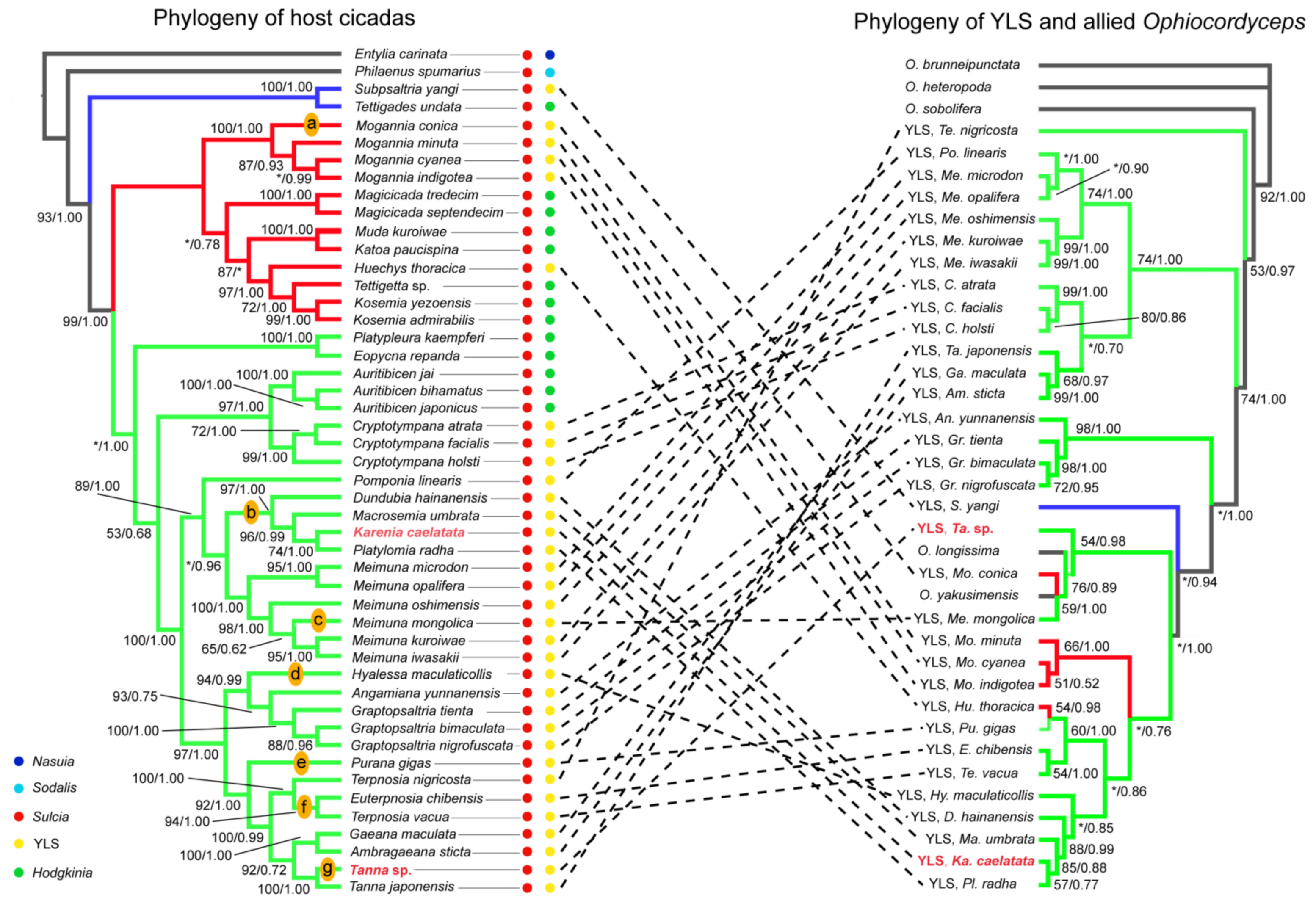
2.4. Phylogenetic Relationships of Sulcia and YLS in Cicadas
3. Discussion
3.1. Distribution of Symbionts in Different Tissues of Cicadas
3.2. Cophylogeny of Sulcia and Host Cicadas Reflecting Host–Symbiont Codiversification
3.3. The Phylogeny of YLS and Cicadas Revealing Complex Evolutionary Trajectories of YLS
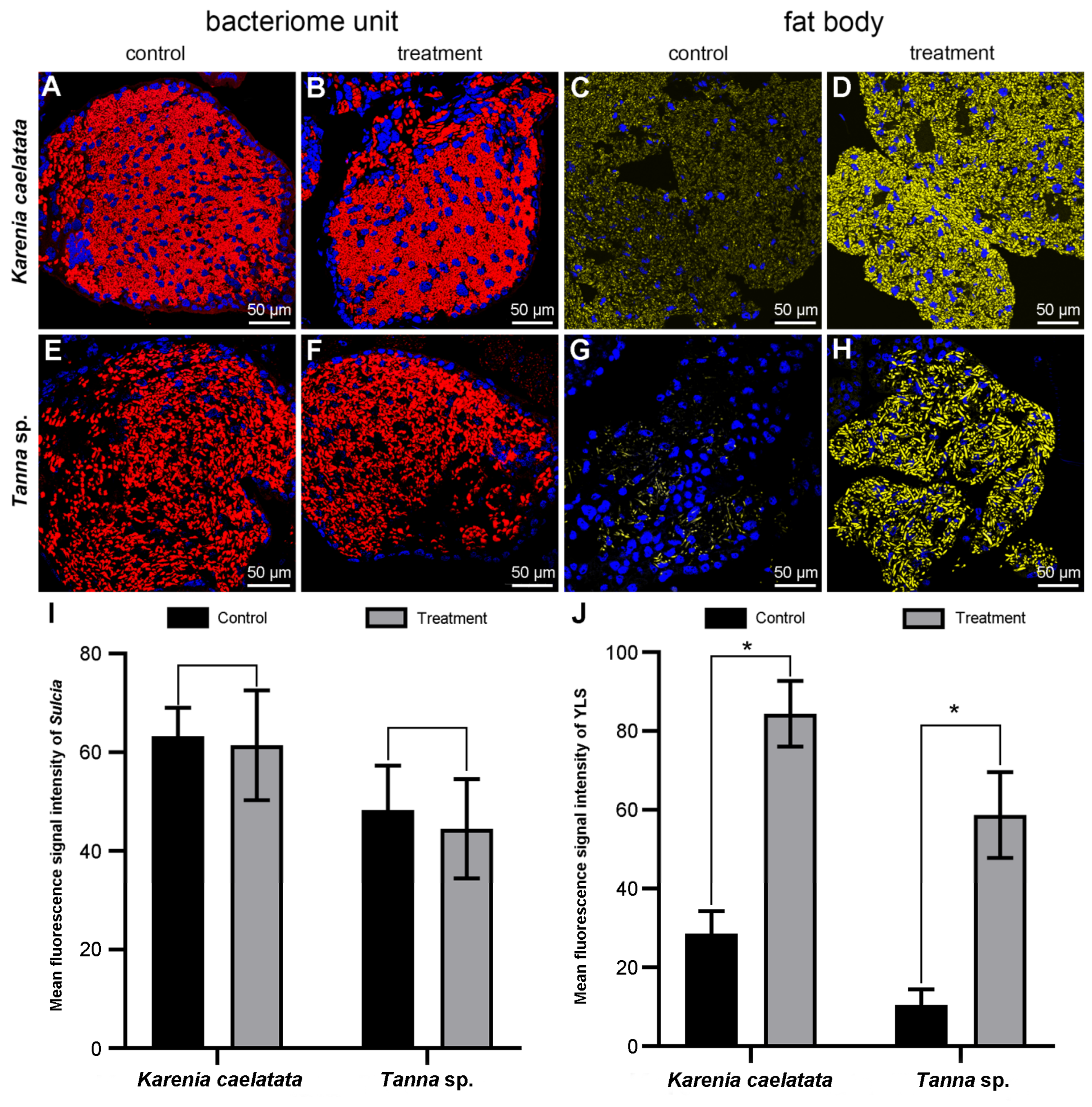
3.4. The Improvement of FISH Signal Intensity for Related Symbiont(s)
4. Materials and Methods
4.1. Samples Collection and Tissues Dissection
4.2. DNA Extraction and IIIumina High-Throughput Sequencing of Bacterial 16S rRNA and Fungal ITS Gene
4.3. Diagnostic PCR Analyses of Dominant Symbionts in Different Tissues
4.4. Histological Microscopy Revealing the Distribution of Sulcia and YLS in Different Tissues of Cicadas
4.5. Fluorescence In Situ Hybridization Confirming the Distribution of Sulcia and YLS in Different Tissues of Cicadas
4.6. Phylogenetic Relationships of Sulcia and YLS in Cicadas
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dale, C.; Moran, N.A. Molecular interactions between bacterial symbionts and their hosts. Cell 2006, 126, 53–465. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. The microbial dimension in insect nutritional ecology. Funct. Ecol. 2009, 23, 38–47. [Google Scholar] [CrossRef]
- Oliver, K.M.; Degnan, P.H.; Burke, G.R.; Moran, N.A. Aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 2010, 55, 247–266. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005, 59, 155–189. [Google Scholar] [CrossRef]
- Moran, N.A.; Plague, G.R.; Sandström, J.P.; Wilcox, J.L. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc. Natl. Acad. Sci. USA 2003, 100, 14543–14548. [Google Scholar] [CrossRef]
- Matsuura, Y.; Kikuchi, Y.; Miura, T.; Fukatsu, T. Ultrabithorax is essential for bacteriocyte development. Proc. Natl. Acad. Sci. USA 2015, 112, 9376–9381. [Google Scholar] [CrossRef]
- Moran, N.A.; Tran, P.; Gerardo, N.M. Symbiosis and insect diversification: An ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 2005, 71, 8802–8810. [Google Scholar] [CrossRef]
- Kobiałka, M.; Michalik, A.; Walczak, M.; Szklarzewicz, T. Dual “bacterial-fungal” symbiosis in Deltocephalinae leafhoppers (Insecta, Hemiptera, Cicadomorpha: Cicadellidae). Microb. Ecol. 2018, 75, 771–782. [Google Scholar] [CrossRef]
- Matsuura, Y.; Moriyama, M.; Łukasik, P.; Vanderpool, D.; Tanahashi, M.; Meng, X.Y.; McCutcheon, J.P.; Fukatsu, T. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc. Natl. Acad. Sci. USA 2018, 115, E5970–E5979. [Google Scholar] [CrossRef]
- Wang, D.; Huang, Z.; Billen, J.; Zhang, G.; He, H.; Wei, C. Structural diversity of symbionts and related cellular mechanisms underlying vertical symbiont transmission in cicadas. Environ. Microbiol. 2021, 23, 6603–6621. [Google Scholar] [CrossRef]
- Koga, R.; Bennett, G.M.; Cryan, J.R.; Moran, N.A. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ. Microbiol. 2013, 15, 2073–2081. [Google Scholar] [CrossRef]
- McCutcheon, J.P.; Moran, N.A. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2010, 2, 708–718. [Google Scholar] [CrossRef]
- Kobiałka, M.; Michalik, A.; Świerczewski, D.; Szklarzewicz, T. Complex symbiotic systems of two treehopper species: Centrotus cornutus (Linnaeus, 1758) and Gargara genistae (Fabricius, 1775) (Hemiptera: Cicadomorpha: Membracoidea: Membracidae). Protoplasma 2020, 257, 819–831. [Google Scholar] [CrossRef]
- Kobiałka, M.; Michalik, A.; Walczak, M.; Junkiert, Ł.; Szklarzewicz, T. Sulcia symbiont of the leafhopper Macrosteles laevis (Ribaut, 1927) (Insecta, Hemiptera, Cicadellidae: Deltocephalinae) harbors Arsenophonus bacteria. Protoplasma 2016, 253, 903–912. [Google Scholar] [CrossRef]
- Michalik, A.; Szwedo, J.; Stroiński, A.; Świerczewski, D.; Szklarzewicz, T. Symbiotic cornucopia of the monophagous planthopper Ommatidiotus dissimilis (Fallen, 1806) (Hemiptera: Fulgoromorpha: Caliscelidae). Protoplasma 2018, 255, 1317–1329. [Google Scholar] [CrossRef]
- Michalik, A.; Franco, D.; Kobiałka, M.; Stroinski, A.; Łukasik, P. Alternative transmission patterns in independently acquired nutritional co-symbionts of Dictyopharidae planthoppers. mBio 2021, 12, e0122821. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, D.; Li, J.; Wei, C.; He, H. Transovarial transmission of bacteriome-associated symbionts in the cicada Pycna repanda (Hemiptera: Cicadidae). Appl. Environ. Microbiol. 2020, 86, e02957-19. [Google Scholar] [CrossRef]
- Lloyd, M.; White, J.A. Xylem feeding by periodical cicada nymphs on pine and grass roots, with novel suggestions for pest-control in conifer plantations and orchards. Ohio J. Sci. 1987, 87, 50–54. [Google Scholar]
- Williams, K.S.; Simon, C. The ecology, behavior, and evolution of periodical cicadas. Annu. Rev. Entomol. 1995, 40, 269–295. [Google Scholar] [CrossRef]
- McCutcheon, J.P.; McDonald, B.R.; Moran, N.A. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. USA 2009, 106, 15394–15399. [Google Scholar] [CrossRef]
- Van Leuven, J.T.; Meister, R.C.; Simon, C.; McCutcheon, J.P. Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one. Cell 2014, 158, 1270–1280. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Van Leuven, J.T.; Meister, R.C.; Carey, K.M.; Simon, C.; McCutcheon, J.P. Genome expansion via lineage splitting and genome reduction in the cicada endosymbiont Hodgkinia. Proc. Natl. Acad. Sci. USA 2015, 112, 10192–10199. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Łukasik, P.; Meyer, M.M.; Buckner, M.; Simon, C.; Veloso, C.; Michalik, A.; McCutcheon, J.P. Changes in endosymbiont complexity drive host-level compensatory adaptations in cicadas. mBio 2018, 9, e02104–e02118. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Huang, Z.; Billen, J.; Zhang, G.; He, H.; Wei, C. Complex co-evolutionary relationships between cicadas and their symbionts. Environ. Microbiol 2021, 24, 195–211. [Google Scholar] [CrossRef]
- Cao, Y.; Dietrich, C.H. Phylogenomics of flavobacterial insect nutritional endosymbionts with implications for Auchenorrhyncha phylogeny. Cladistics 2021, 38, 38–58. [Google Scholar] [CrossRef]
- Simon, C.; Gordon, E.R.L.; Moulds, M.S.; Cole, J.A.; Haji, D.; Lemmon, A.; Lemmon, E.M.; Kortyna, M.; Nazario, K.; Wade, E.; et al. Off-target capture data, endosymbiont genes and morphology reveal a relict lineage that is sister to all other singing cicadas. Bio. J. Linn. Soc. 2019, 128, 865–886. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, D.; He, H.; Wei, C. Bacterial diversity of bacteriomes and organs of reproductive, digestive and excretory systems in two cicada species (Hemiptera: Cicadidae). PLoS ONE 2017, 12, e0175903. [Google Scholar] [CrossRef]
- Wang, D.; Wei, C. Bacterial communities in digestive and excretory organs of cicadas. Arch. Microbiol. 2019, 202, 539–553. [Google Scholar] [CrossRef]
- Zheng, H.; Nishida, A.; Kwong, W.K.; Koch, H.; Engel, P.; Steele, M.I.; Moran, N.A. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio 2016, 7, e01326-16. [Google Scholar] [CrossRef]
- Shan, H.; Luan, J.; Liu, Y.; Douglas, A.E.; Liu, S. The inherited bacterial symbiont Hamiltonella influences the sex ratio of an insect host. Proc. R Soc. B Biol. Sci. 2019, 286, 20191677. [Google Scholar] [CrossRef]
- Zhou, X.; Ling, X.; Guo, H.; Zhu-Salzman, K.; Ge, F.; Sun, Y. Serratia symbiotica enhances fatty acid metabolism of pea aphid to promote host development. Int. J. Mol. Sci. 2021, 22, 5951. [Google Scholar] [CrossRef]
- Rothman, J.A.; Russell, K.A.; Leger, L.; McFrederick, Q.S. The direct and indirect effects of environmental toxicants on the heath of bumblebees and their microbiomes. Proc. Biol. Sci. 2020, 287, 20200980. [Google Scholar]
- Palmer-Young, E.C.; Raffel, T.R.; McFrederick, Q.S. PH-mediated inhibition of a bumble bee parasite by an intestinal symbiont. Parasitology 2019, 146, 380–388. [Google Scholar] [CrossRef]
- Thai, P.H.; Yang, J.T. First record of the cicada genus Karenia Distant, 1888 (Hemiptera: Cicadidae) from Vietnam, with description of a new species. Zootaxa 2012, 3153, 32–38. [Google Scholar] [CrossRef]
- Yuan, F.M.; Wei, C. Phylogenetic status of the cicada tribe Sinosenini based on mitochondrial genomes. Acta Entomol. Sinica 2021, 64, 1205–1217. [Google Scholar]
- Hill, K.B.; Marshall, D.C.; Marathe, K.; Moulds, M.S.; Lee, Y.J.; Pham, T.H.; Mohagan, A.B.; Sarkar, V.; Price, B.W.; Duffels, P.J.; et al. The molecular systematics and diversification of a taxonomically unstable group of Asian cicada tribes related to Cicadini Latreille, 1802 (Hemiptera: Cicadidae). Invertebr. Syst. 2021, 35, 570–601. [Google Scholar] [CrossRef]
- Biedermann, P.H.W.; Vega, F.E. Ecology and evolution of insect-fungal mutualisms. Annu. Rev. Entomol. 2020, 65, 431–455. [Google Scholar] [CrossRef]
- Carbonero, F.; Strobel, G. Fungal ecology special issue: Editorial. Microb. Ecol. 2021, 82, 1–4. [Google Scholar] [CrossRef]
- Stefanini, I. Yeast-insect associations: It takes guts. Yeast 2018, 35, 315–330. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef]
- Engel, P.; James, R.R.; Koga, R.; Kwong, W.K.; McFrederick, Q.S.; Moran, N.A. Standard methods for research on Apis mellifera gut symbionts. J. Apicult. Res. 2013, 52, 1–24. [Google Scholar] [CrossRef]
- Moter, A.; Göbel, U.B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 2000, 41, 85–112. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, B.M.; Glöckner, F.O.; Wulf, J.; Amann, R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 2000, 66, 3603–3607. [Google Scholar] [CrossRef] [PubMed]
- Sacchi, L.; Genchi, M.; Clementi, E.; Bigliardi, E.; Avanzati, A.M.; Pajoro, M.; Negri, I.; Marzorati, M.; Gonella, E.; Alma, A.; et al. Multiple symbiosis in the leafhopper Scaphoideus titanus (Hemiptera: Cicadellidae): Details of transovarial transmission of Cardinium sp. and a yeast-like endosymbionts. Tissue Cell 2008, 40, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi-Man, O.; Davenport, E.R.; Gilad, Y. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: Evaluation of effective study designs. PLoS ONE 2013, 8, e53608. [Google Scholar] [CrossRef]
- Scibetta, S.; Schena, L.; Abdelfattah, A.; Pangallo, S.; Cacciola, S.O. Selection and experimental evaluation of universal primers to study the fungal microbiome of higher plants. Phytobiomes J. 2018, 2, 225–236. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproductive, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotech. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Kolasa, M.; Ścibior, R.; Mazur, M.A.; Kubisz, D.; Dudek, K.; Kajtoch, Ł. How hosts taxonomy, trophy, and endosymbionts shape microbiome diversity in beetles. Microb. Ecol. 2019, 78, 995–1013. [Google Scholar] [CrossRef]
- Xu, S.; Jiang, L.; Qiao, G.; Chen, J. Diversity of bacterial symbionts associated with Myzus persicae (Sulzer) (Hemiptera: Aphididae: Aphidinae) revealed by 16S rRNA Illumina sequencing. Microb. Ecol. 2021, 81, 784–794. [Google Scholar] [CrossRef]
- Chaitra, H.S.; Singh, A.; Pandiyan, K.; Kalia, V.K. Sex biased variance in the structural and functional diversity of the midgut bacterial community of last instar larvae of Pectinophora gossypiella (Lepidoptera: Gelechiidae). Microb. Ecol. 2022, 83, 1112–1122. [Google Scholar] [CrossRef]
- McFrederick, Q.S.; Thomas, J.M.; Neff, J.L.; Vuong, H.Q.; Russell, K.A.; Hale, A.R.; Mueller, U.G. Flowers and wild megachilid bees share microbes. Microb. Ecol. 2017, 73, 188–200. [Google Scholar] [CrossRef]
- Ma, M.; Chen, X.; Li, S.; Luo, J.; Han, R.; Xu, L. Composition and diversity of gut bacterial community in different life stages of a leaf beetle Gastrolina depressa. Microb. Ecol. 2022. [Google Scholar] [CrossRef]
- Łukasik, P.; Nazario, K.; Van Leuven, J.T.; Campbell, M.A.; Meyer, M.; Michalik, A.; Pessacq, P.; Simon, C.; Veloso, C.; McCutcheon, J.P. Multiple origins of interdependent endosymbiotic complexes in a genus of cicadas. Proc. Natl. Acad. Sci. USA 2018, 115, E226–E235. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]



| Probe Name | Fluorophore | Primer Sequence (5′–3′) | References |
|---|---|---|---|
| Sulcia | CY3 | CCACACATTCCAGTTACTCC | [53] |
| Sulcia-Lhelper | unlabelled | GTTCTGTGTGATCTCTATGCATTTCACCGCT | |
| Sulcia-Rhelper | unlabelled | CCTCACTCTAGTTTATCAGTATCAATAGCACTT | |
| YLS | CY5 | CCTGCCTGGAGCACTCT | [9] |
| YLS-Lhelper | unlabelled | CTAATGTATTCGAGCAT | This study |
| YLS-Rhelper | unlabelled | TTTTTCAAAGTAAAAGTCCCGT |
| Hybridization Solution | Final Concentration | Volume (100 μL) |
|---|---|---|
| 25% dextran sulfate | 10% | 40 μL |
| 10% bovine serum albumin | 0.25% | 2.5 μL |
| 20 × SSC | 2.5× | 12.5 μL |
| ssDNA (200 ng/mL) | 10 ng/μL | 5 μL |
| Sulcia probe (2 μM) | 200 nM | 10 μL |
| Sulcia-Lhelper (100 μM) | 2 μM | 2 μL |
| Sulcia-Rhelper (100 μM) | 2 μM | 2 μL |
| YLS probe (2 μM) | 200 nM | 10 μL |
| YLS-Lhelper (100 μM) | 2 μM | 2 μL |
| YLS-Rhelper (100 μM) | 2 μM | 2 μL |
| diH2O | / | 12 μL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Zhou, J.; Zhang, Z.; He, H.; Wei, C. A Study on Symbiotic Systems of Cicadas Provides New Insights into Distribution of Microbial Symbionts and Improves Fluorescence In Situ Hybridization Technique. Int. J. Mol. Sci. 2023, 24, 2434. https://doi.org/10.3390/ijms24032434
Huang Z, Zhou J, Zhang Z, He H, Wei C. A Study on Symbiotic Systems of Cicadas Provides New Insights into Distribution of Microbial Symbionts and Improves Fluorescence In Situ Hybridization Technique. International Journal of Molecular Sciences. 2023; 24(3):2434. https://doi.org/10.3390/ijms24032434
Chicago/Turabian StyleHuang, Zhi, Jinrui Zhou, Zhijun Zhang, Hong He, and Cong Wei. 2023. "A Study on Symbiotic Systems of Cicadas Provides New Insights into Distribution of Microbial Symbionts and Improves Fluorescence In Situ Hybridization Technique" International Journal of Molecular Sciences 24, no. 3: 2434. https://doi.org/10.3390/ijms24032434
APA StyleHuang, Z., Zhou, J., Zhang, Z., He, H., & Wei, C. (2023). A Study on Symbiotic Systems of Cicadas Provides New Insights into Distribution of Microbial Symbionts and Improves Fluorescence In Situ Hybridization Technique. International Journal of Molecular Sciences, 24(3), 2434. https://doi.org/10.3390/ijms24032434






