Mutation of OsLPR3 Enhances Tolerance to Phosphate Starvation in Rice
Abstract
1. Introduction
2. Results
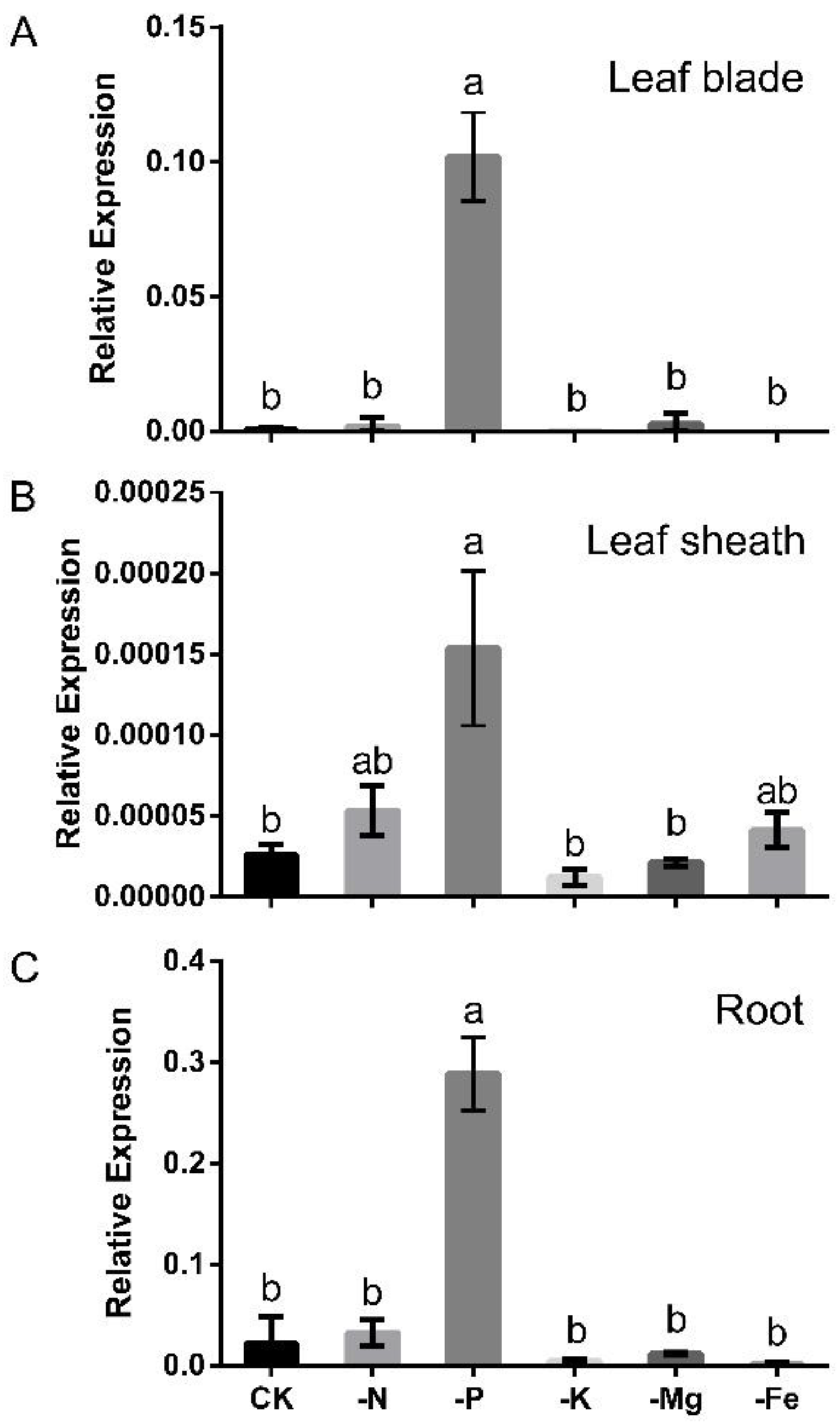
2.1. OsLPR3 Was Responsive to Pi Deprivation
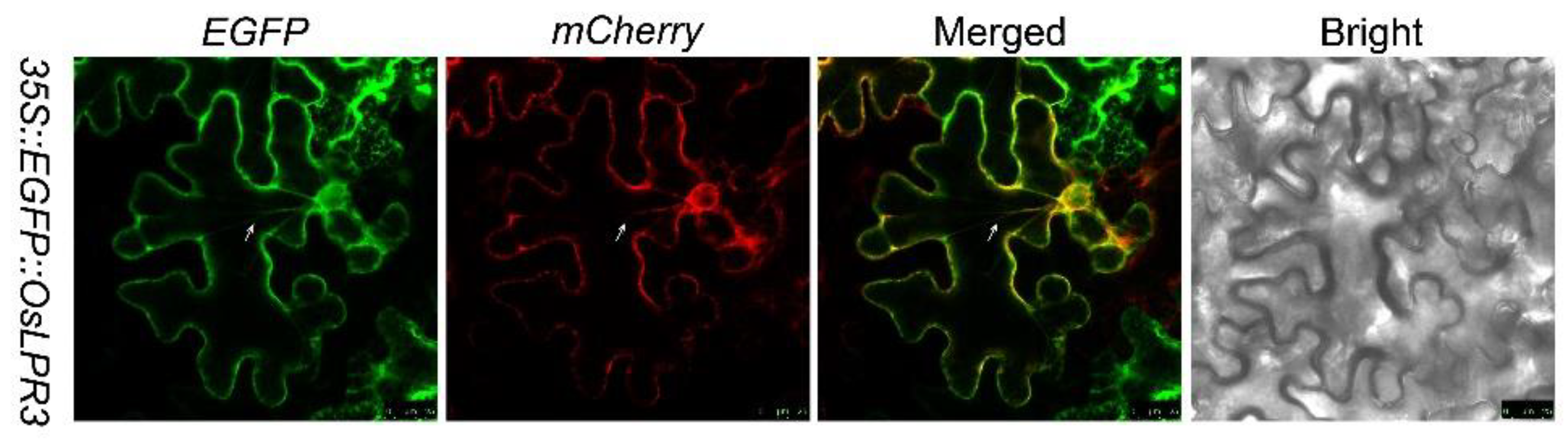
2.2. OsLPR3 Was Localized to the ER
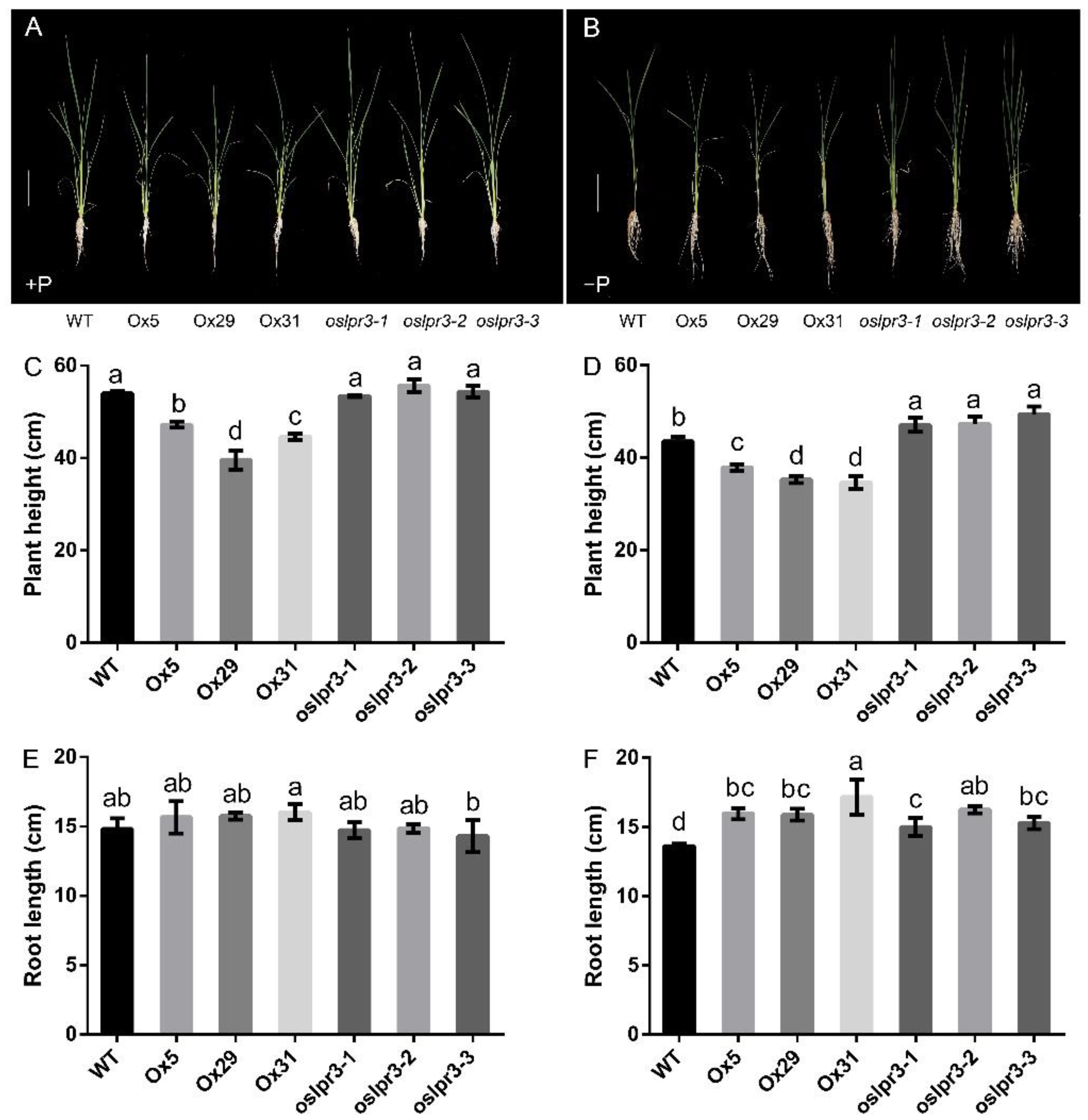
2.3. OsLPR3 Was Involved in Vegetative Growth and RSA of Rice
2.4. Alteration of OsLPR3 Expression Affected the Agronomic Traits of Rice during the Reproductive Stage
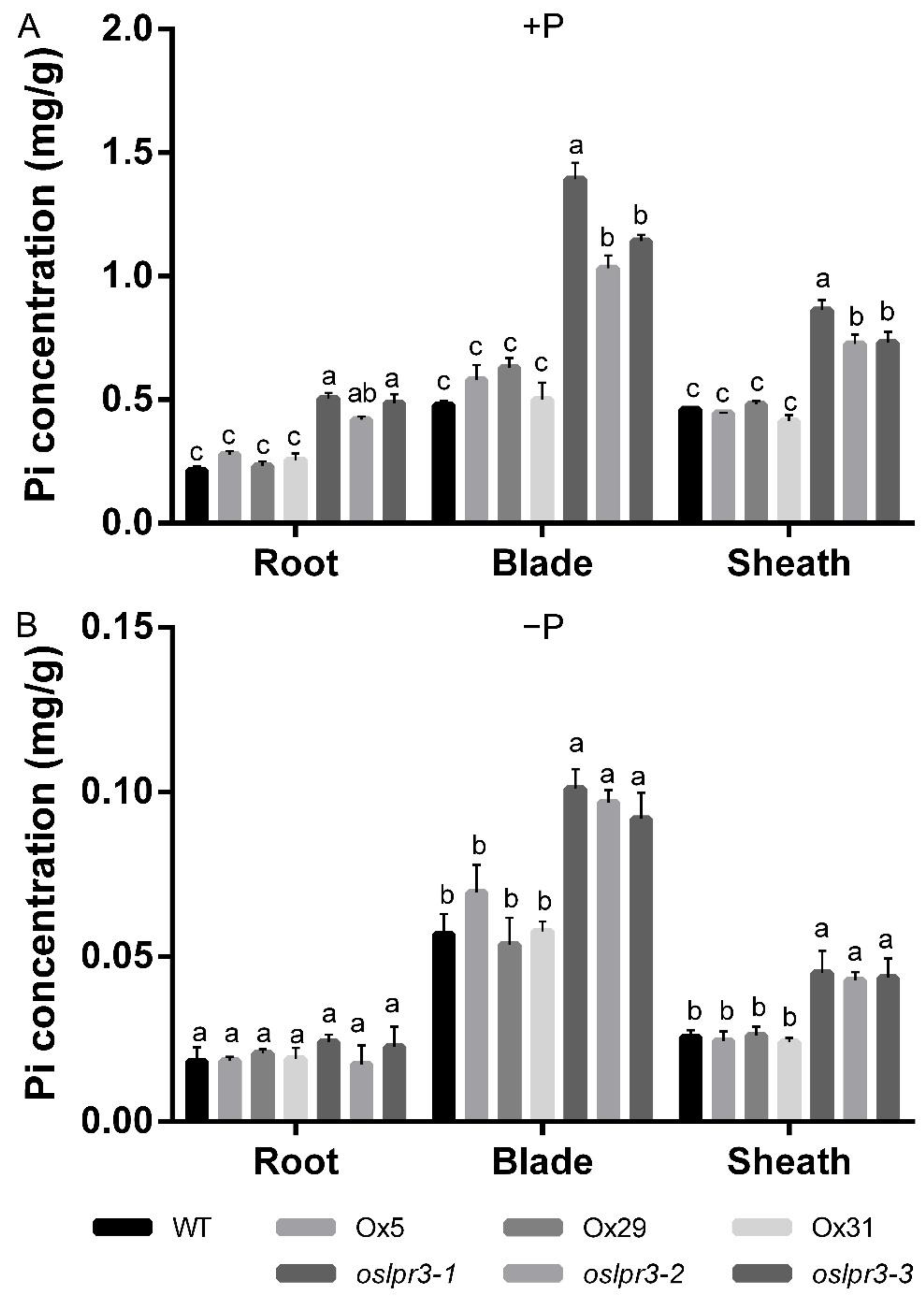
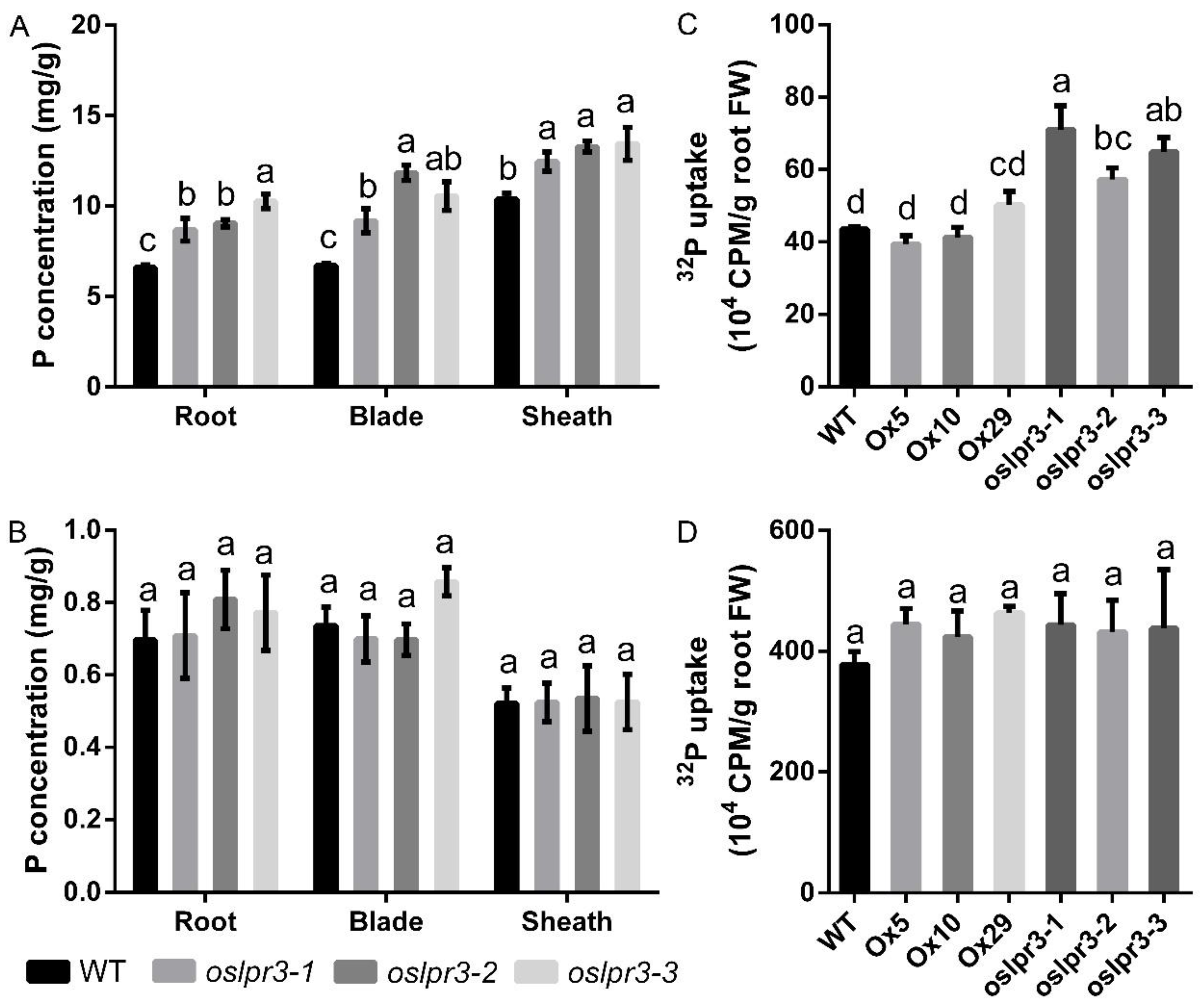
2.5. Alteration of OsLPR3 Expression Affected the Pi Status
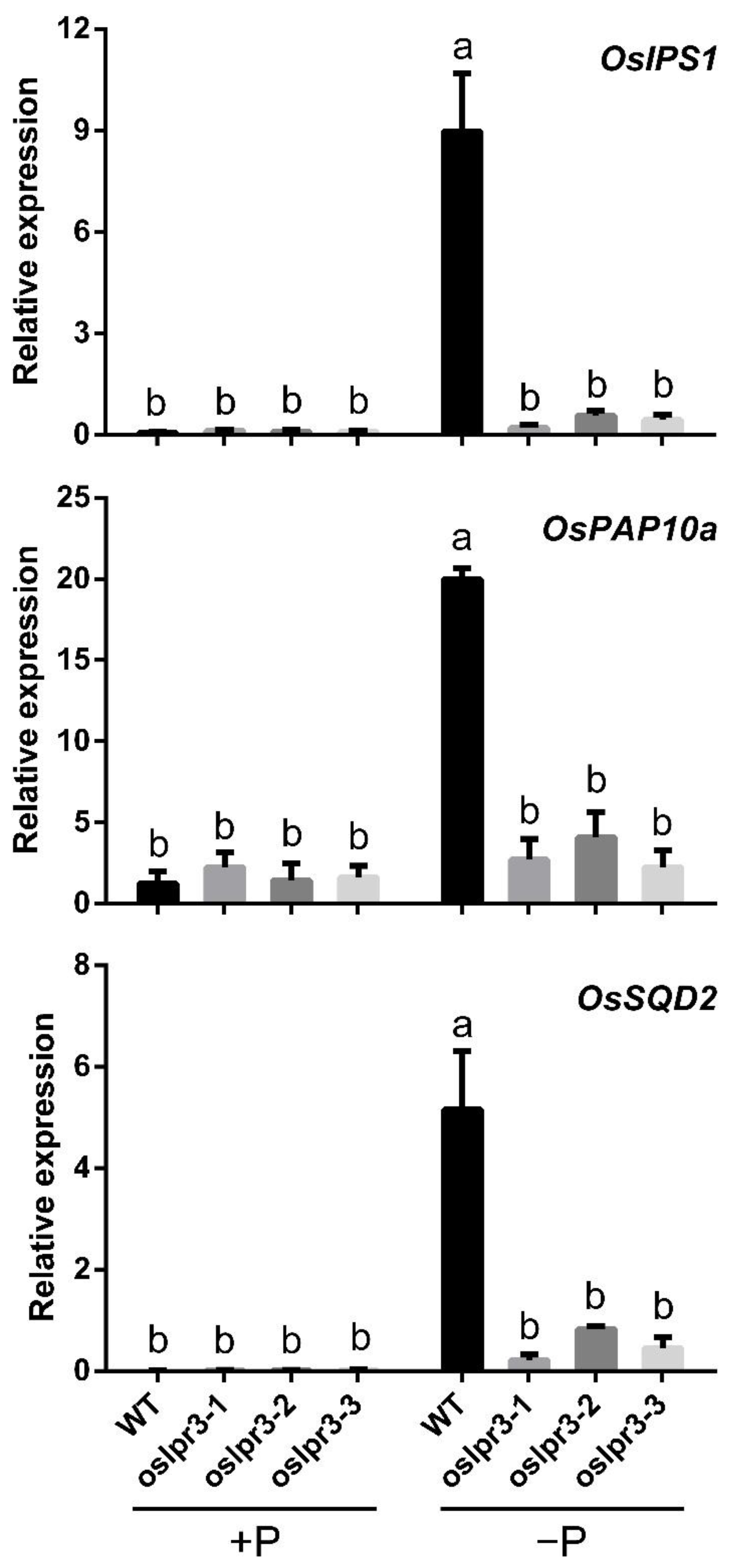
2.6. Alteration of OsLPR3 Expression Affected the Relative Expressions of OsPTs and Pi-Starvation-Induced Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. qRT-PCR
4.3. Transient Expression of OsLPR3 in N. benthamiana Leaves for Subcellular Localization
4.4. Construction of OsLPR3 Overexpression and Mutation Vectors and Generation of Transgenic Plants
4.5. Southern Blot Analyses
4.6. Quantification of Pi and Total P Concentrations
4.7. 32Pi Uptake Assay
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plaxton, W.C.; Carswell, M.C. Metabolic aspects of the phosphate starvation response in plants. In Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization; Lerner, H.R., Ed.; CRC Press: New York, NY, USA, 1999; pp. 349–372. [Google Scholar]
- Poirier, Y.; Bucher, M. Phosphate transport and homeostasis in Arabidopsis. Arab. Book Am. Soc. Plant Biol. 2002, 1, e0024. [Google Scholar] [CrossRef] [PubMed]
- Marschner, H. Mineral Nutrition of Higher Plants; Academic Press: London, UK, 1995. [Google Scholar]
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef]
- Lynch, J.P. Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Thibaud, M.C.; Arrighi, J.F.; Bayle, V.; Chiarenza, S.; Creff, A.; Bustos, R.; Paz-Ares, J.; Poirier, Y.; Nussaume, L. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 2010, 64, 775–789. [Google Scholar] [CrossRef]
- Gutiérrez-Alanís, D.; Ojeda-Rivera, J.O.; Yong-Villalobos, L.; Cárdenas-Torres, L.; Herrera-Estrella, L. Adaptation to phosphate scarcity: Tips from Arabidopsis roots. Trends Plant Sci. 2018, 23, 721–730. [Google Scholar] [CrossRef]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef]
- Péret, B.; Clément, M.; Nussaume, L.; Desnos, T. Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci. 2011, 16, 442–450. [Google Scholar] [CrossRef]
- Chien, P.S.; Chiang, C.P.; Leong, S.J.; Chiou, T.J. Sensing and Signaling of Phosphate Starvation: From Local to Long Distance. Plant Cell Physiol. 2018, 59, 1714–1722. [Google Scholar] [CrossRef]
- Sánchez-Calderón, L.; López-Bucio, J.; Chacón-López, A.; Cruz-Ramírez, A.; Nieto-Jacobo, F.; Dubrovsky, J.G.; Herrera-Estrella, L. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 174–184. [Google Scholar] [CrossRef]
- Svistoonoff, S.; Creff, A.; Reymond, M.; Sigoillot-Claude, C.; Ricaud, L.; Blanchet, A.; Nussaume, L.; Desnos, T. Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 2007, 39, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Brown, K.M. Topsoil foraging-an architectural adaptation of plants to low phosphorus availability. Plant Soil 2001, 237, 225–237. [Google Scholar] [CrossRef]
- Williamson, L.C.; Ribrioux, S.P.; Fitter, A.H.; Leyser, H.M. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 2001, 126, 875–882. [Google Scholar] [CrossRef] [PubMed]
- López-Bucio, J.; Hernández-Abreu, E.; Sánchez-Calderón, L.; Nieto-Jacobo, M.F.; Simpson, J.; Herrera-Estrella, L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002, 129, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.D.; Giehl, R.F.; Friedel, S.; von Wirén, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Mora-Macías, J.; Ojeda-Rivera, J.O.; Gutiérrez-Alanís, D.; Yong-Villalobos, L.; Oropeza-Aburto, A.; Raya-González, J.; Jiménez-Domínguez, G.; Chávez-Calvillo, G.; Rellán-Álvarez, R.; Herrera-Estrella, L. Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc. Natl. Acad. Sci. USA 2017, 114, E3563–E3572. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Toev, T.; Heisters, M.; Teller, J.; Moore, K.L.; Hause, G.; Dinesh, D.C.; Bürstenbinder, K.; Abel, S. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 2015, 33, 216–230. [Google Scholar] [CrossRef] [PubMed]
- Ticconi, C.A.; Delatorre, C.A.; Lahner, B.; Salt, D.E.; Abel, S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 2004, 37, 801–814. [Google Scholar] [CrossRef]
- Reymond, M.; Svistoonoff, S.; Loudet, O.; Nussaume, L.; Desnos, T. Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ. 2006, 29, 115–125. [Google Scholar] [CrossRef]
- Balzergue, C.; Dartevelle, T.; Godon, C.; Laugier, E.; Meisrimler, C.; Teulon, J.M.; Creff, A.; Bissler, M.; Brouchoud, C.; Hagège, A. Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commun. 2017, 8, 15300. [Google Scholar] [CrossRef]
- Shen, N.; Hou, S.; Tu, G.; Lan, W.; Jing, Y. Transcription Factor WRKY33 Mediates the Phosphate Deficiency-Induced Remodeling of Root Architecture by Modulating Iron Homeostasis in Arabidopsis Roots. Int. J. Mol. Sci. 2021, 22, 9275. [Google Scholar] [CrossRef] [PubMed]
- Ticconi, C.A.; Lucero, R.D.; Sakhonwasee, S.; Adamson, A.W.; Creff, A.; Nussaume, L.; Desnos, T.; Abel, S. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. USA 2009, 106, 14174–14179. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wang, Z.; Wang, X.; Liu, D. Blue Light-Triggered Chemical Reactions Underlie Phosphate Deficiency-Induced Inhibition of Root Elongation of Arabidopsis Seedlings Grown in Petri Dishes. Mol. Plant 2019, 12, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Bu, L.; Han, M.; Wang, Y.; Li, Z.; Liu, H.; Chao, D. Long-distance blue light signalling regulates phosphate deficiency-induced primary root growth inhibition. Long-distance blue light signalling regulates phosphate deficiency-induced primary root growth inhibition. Mol. Plant 2021, 14, 1539–1553. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Zhu, Q.; Ye, J.; Zhu, Y.; Jing, X.; Du, W.; Zhou, M.; Lin, X.; Zheng, S.; et al. Phloem iron remodels root development in response to ammonium as the major nitrogen source. Nat. Commun. 2022, 13, 561. [Google Scholar] [CrossRef]
- Mai, C.D.; Phung, N.T.P.; To, H.T.M.; Gonin, M.; Hoang, G.T.; Nguyen, K.L.; Do, V.N.; Courtois, B.; Gantet, P. Genes controlling root development in rice. Rice 2014, 7, 30. [Google Scholar] [CrossRef]
- Wu, W.; Cheng, S. Root genetic research, an opportunity and challenge to rice improvement. Field Crop. Res. 2014, 165, 111–124. [Google Scholar] [CrossRef]
- Cao, Y.; Ai, H.; Jain, A.; Wu, X.; Zhang, L.; Pei, W.; Chen, A.; Xu, G.; Sun, S. Identification and expression analysis of OsLPR family revealed the potential roles of OsLPR3 and 5 in maintaining phosphate homeostasis in rice. BMC Plant Biol. 2016, 16, 210. [Google Scholar] [CrossRef]
- Rapoport, T.A.; Jungnickel, B.; Kutay, U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 1996, 65, 271–303. [Google Scholar] [CrossRef]
- Gaut, J.R.; Hendershot, L.M. The immunoglobulin-binding protein in vitro autophosphorylation site maps to a threonine within the ATP binding cleft but is not a detectable site of in vivo phosphorylation. J. Biol. Chem. 1993, 268, 12691–12698. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenfűhr, A. Amulticolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Chen, X.; Bao, Y.; Dong, J.; Zhang, Z.; Tao, X. Nucleocapsid of Tomato spotted wilt tospovirus forms mobile particles that traffic on an actin/endoplasmic reticulum network driven by myosin XI-K. New Phytol. 2013, 200, 1212–1224. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Shou, H.; Xu, G.; Lian, X. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 2013, 16, 205–212. [Google Scholar] [CrossRef]
- Secco, D.; Jabnoune, M.; Walker, H.; Shou, H.; Wu, P.; Poirier, Y.; Whelan, J. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 2013, 25, 4285–4304. [Google Scholar] [CrossRef] [PubMed]
- LaBonte, M.L. Blobel and Sabatini’s “Beautiful Idea”: Visual Representations of the Conception and Refinement of the Signal Hypothesis. J. Hist. Biol. 2017, 50, 797–833. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.; Cao, Y.; Jain, A.; Wang, X.; Hu, Z.; Zhao, G.; Hu, S.; Shen, X.; Yan, Y.; Liu, X.; et al. The ferroxidase LPR5 functions in the maintenance of phosphate homeostasis and is required for normal growth and development of rice. J. Exp. Bot. 2020, 71, 4828–4842. [Google Scholar] [CrossRef]
- Xu, Z.R.; Cai, M.L.; Yang, Y.; You, T.T.; Ma, J.F.; Wang, P.; Zhao, F.J. The ferroxidases LPR1 and LPR2 control iron translocation in the xylem of Arabidopsis plants. Mol. Plant 2022, 15, 1–14. [Google Scholar] [CrossRef]
- Tsuji, H.; Nakamura, H.; Taoka, K.; Shimamoto, K. Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 2013, 54, 385–397. [Google Scholar] [CrossRef]
- Hu, Y.; Li, S.; Fan, X.; Song, S.; Zhou, X.; Weng, X.; Xiao, J.; Li, X.; Xiong, L.; You, A.; et al. OsHOX1 and OsHOX28 Redundantly Shape Rice Tiller Angle by Reducing HSFA2D Expression and Auxin Content. Plant Physiol. 2020, 184, 1424–1437. [Google Scholar] [CrossRef]
- Wang, J.; Bao, J.; Zhou, B.; Li, M.; Li, X.; Jin, J. The osa-miR164 target OsCUC1 functions redundantly with OsCUC3 in controlling rice meristem/organ boundary specification. New Phytol. 2021, 229, 1566–1581. [Google Scholar] [CrossRef]
- Cao, Y.; Jain, A.; Ai, H.; Liu, X.; Wang, X.; Hu, Z.; Sun, Y.; Hu, S.; Shen, X.; Lan, X.; et al. OsPDR2 mediates the regulation on the development response and maintenance of Pi homeostasis in rice. Plant Physiol. Biochem. 2020, 149, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F.; Pata, M.; Nacry, P.; Doumas, P.; Rossignol, M. Effects of phosphate availability on the root system architecture: Large-scale analysis of the natural variation between Arabidopsis accessions. Plant Cell Environ. 2003, 26, 1839–1850. [Google Scholar] [CrossRef]
- Scheible, W.R.; Rojas-Triana, M. Sensing, signalling, and control of phosphate starvation in plants: Molecular players and applications. Ann. Plant Rev. 2015, 48, 23–63. [Google Scholar]
- IRRI. Annual Report for 1995; International Rice Research Institute: Los Banos, The Philippines, 1996. [Google Scholar]
- Dobermann, A.; Fairhurst, T. Rice: Nutrient Disorders & Nutrient Management; Handbook Series; Potash & Phosphate Institute (PPI): Los Baños, Philippine; Potash & Phosphate Institute of Canada (PPIC): Los Baños, Philippine; International Rice Research Institute: Los Baños, Philippine, 2000; p. 191. [Google Scholar]
- Swamy Mahadeva, H.K.; Anila, M.; Kale, R.R.; Bhadana, V.P.; Anantha, M.S.; Brajendra, P.; Hajira, S.K.; Balachiranjeevi, C.H.; Prasanna, B.L.; Pranathi, K.; et al. Phenotypic and molecular characterization of rice germplasm lines and identification of novel source for low soil phosphorus tolerance in rice. Euphytica 2019, 215, 18. [Google Scholar]
- López-Bucio, J.; Cruz-Ramírez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Den Herder, G.; Van Isterdael, G.; Beeckman, T.; De Smet, I. The roots of a new green revolution. Trends Plant Sci. 2010, 15, 600–607. [Google Scholar] [CrossRef]
- Wang, X.; Yan, X.; Liao, H. Genetic improvement for phosphorus efficiency in soybean: A radical approach. Ann. Bot. 2010, 106, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.L.; Wu, P.; Jiao, F.C.; Jia, Q.J.; Chen, H.M.; Yu, J.; Song, X.W.; Yi, K.K. Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ. 2005, 28, 353–364. [Google Scholar] [CrossRef]
- Zhou, J.; Jiao, F.; Wu, Z.; Li, Y.; Wang, X.; He, X.; Zhong, W.; Wu, P. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 2008, 146, 1673–1686. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Xu, C.; Benning, C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 2002, 99, 5732–5737. [Google Scholar] [CrossRef]
- Wang, H.D.; Sun, R.; Cao, Y.; Pei, W.X.; Sun, Y.F.; Zhou, H.M.; Wu, X.; Zhang, F.; Luo, L.; Shen, Q.; et al. OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice. Plant Cell Physiol. 2015, 56, 2381–2395. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Ren, H.; Gu, M.; Zhao, J.; Sun, S.; Zhang, X.; Chen, J.; Wu, P.; Xu, G. The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 2011, 156, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Bürstenbinder, K.; Savchenko, T.; Müller, J.; Adamson, A.W.; Stamm, G.; Kwong, R.; Zipp, B.J.; Dinesh, D.C.; Abel, S. Arabidopsis calmodulin-binding protein IQ67-domain 1 localizes to microtubules and interacts with kinesin light chain-related protein-1. J. Biol. Chem. 2013, 288, 1871–1882. [Google Scholar] [CrossRef]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, N.M.; Surin, B.; Ramm, K.; Gaudron, J.; Schünmann, P.H.D.; Taylor, W.; Waterhouse, P.M.; Wang, M.-B. Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Aust. J. Plant Physiol. 2000, 27, 201–210. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ai, H.; Liu, X.; Hu, Z.; Cao, Y.; Kong, N.; Gao, F.; Hu, S.; Shen, X.; Huang, X.; Xu, G.; et al. Mutation of OsLPR3 Enhances Tolerance to Phosphate Starvation in Rice. Int. J. Mol. Sci. 2023, 24, 2437. https://doi.org/10.3390/ijms24032437
Ai H, Liu X, Hu Z, Cao Y, Kong N, Gao F, Hu S, Shen X, Huang X, Xu G, et al. Mutation of OsLPR3 Enhances Tolerance to Phosphate Starvation in Rice. International Journal of Molecular Sciences. 2023; 24(3):2437. https://doi.org/10.3390/ijms24032437
Chicago/Turabian StyleAi, Hao, Xiuli Liu, Zhi Hu, Yue Cao, Nannan Kong, Feiyan Gao, Siwen Hu, Xing Shen, Xianzhong Huang, Guohua Xu, and et al. 2023. "Mutation of OsLPR3 Enhances Tolerance to Phosphate Starvation in Rice" International Journal of Molecular Sciences 24, no. 3: 2437. https://doi.org/10.3390/ijms24032437
APA StyleAi, H., Liu, X., Hu, Z., Cao, Y., Kong, N., Gao, F., Hu, S., Shen, X., Huang, X., Xu, G., & Sun, S. (2023). Mutation of OsLPR3 Enhances Tolerance to Phosphate Starvation in Rice. International Journal of Molecular Sciences, 24(3), 2437. https://doi.org/10.3390/ijms24032437





